Genome Wide Identification and Comparative Analysis of the Serpin Gene Family in Brachypodium and Barley
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification and Genomic Distribution of Serpin Genes
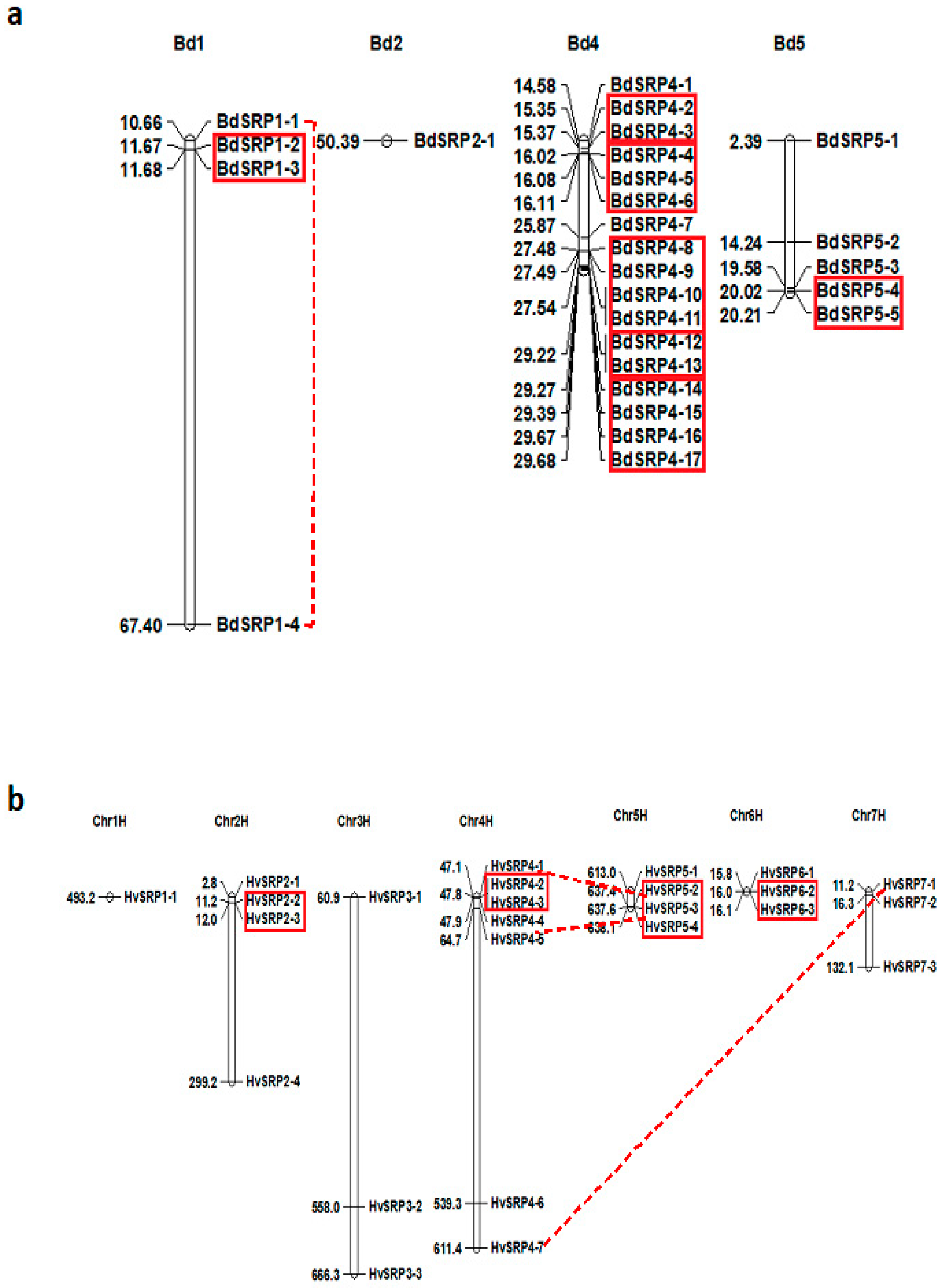
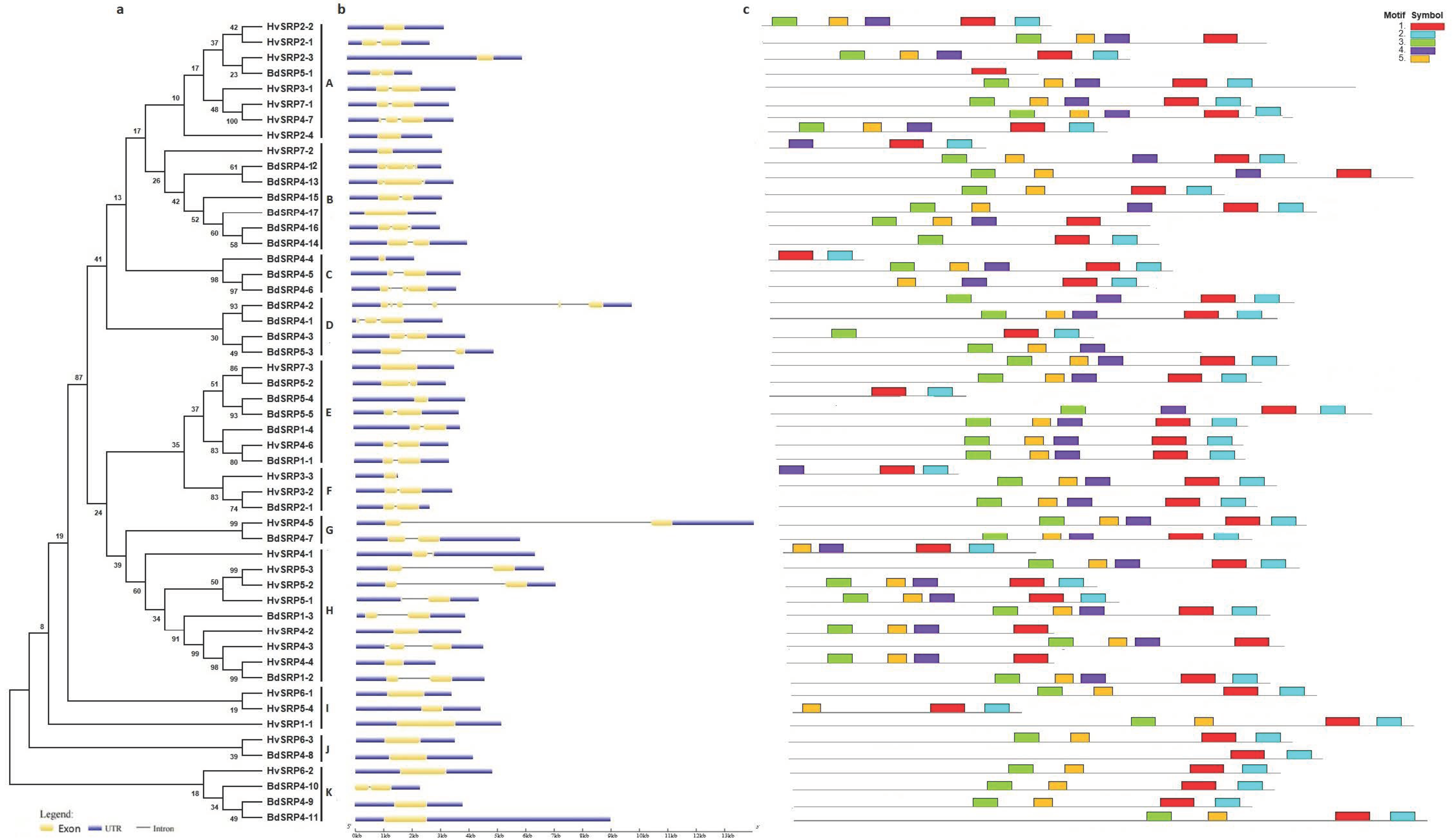
2.2. Chromosomal Distribution, Gene Structure and Conserved Motif Analysis
2.3. Subcellular Localization
2.4. Duplication and Evolutionary Pattern of Serpin Genes
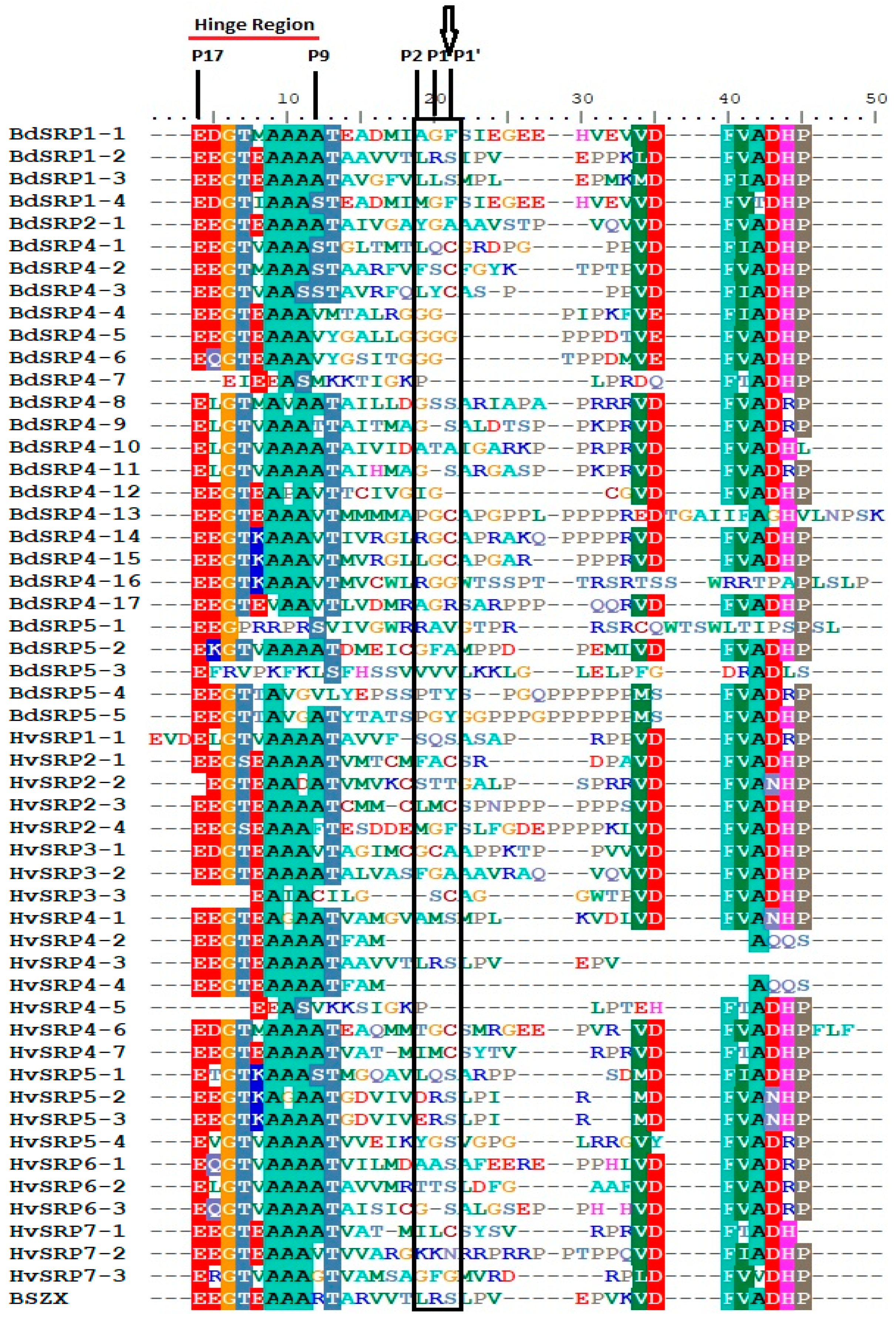
2.5. Domain Analysis
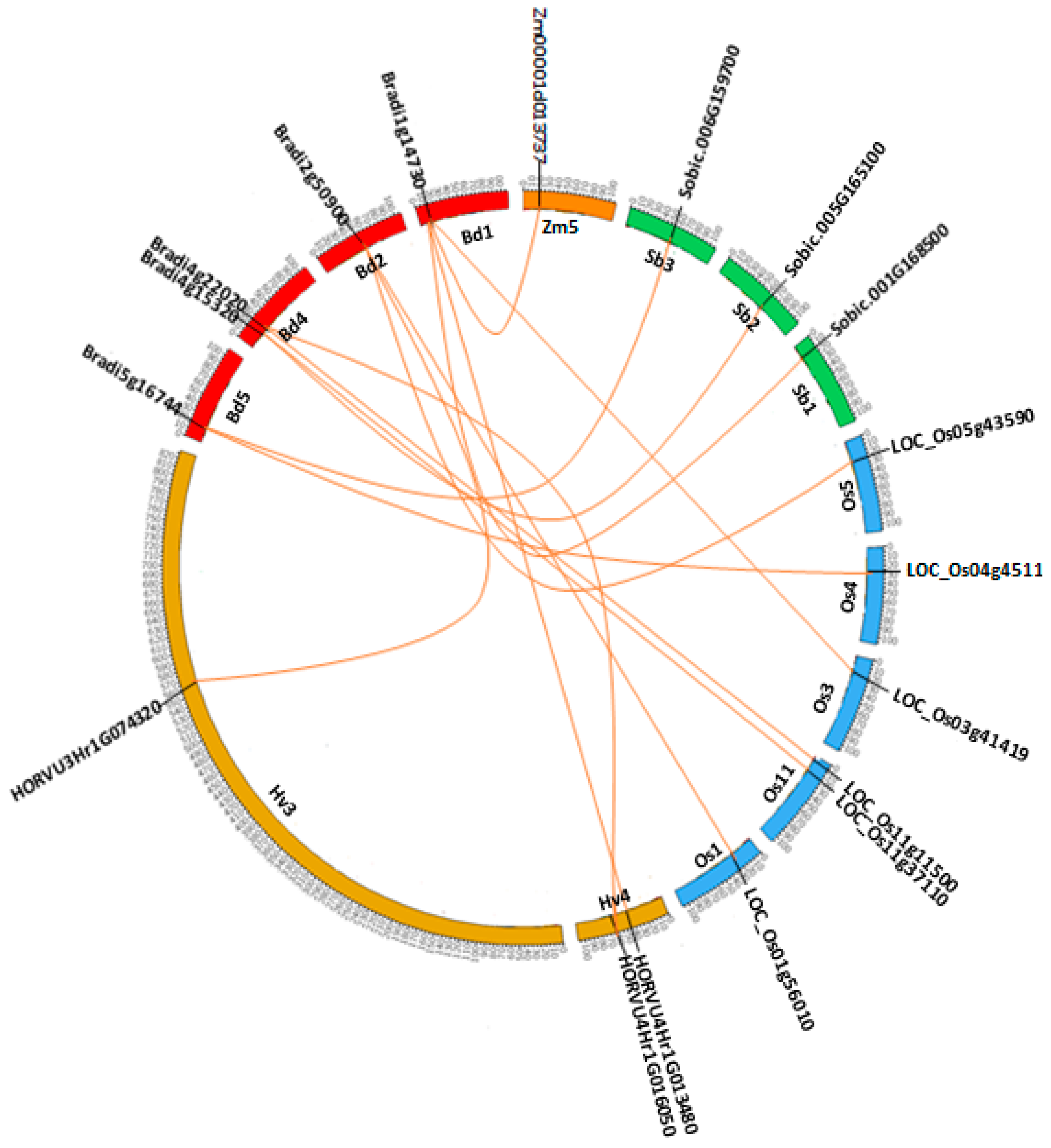
2.6. Synteny and Phylogenetic Analysis
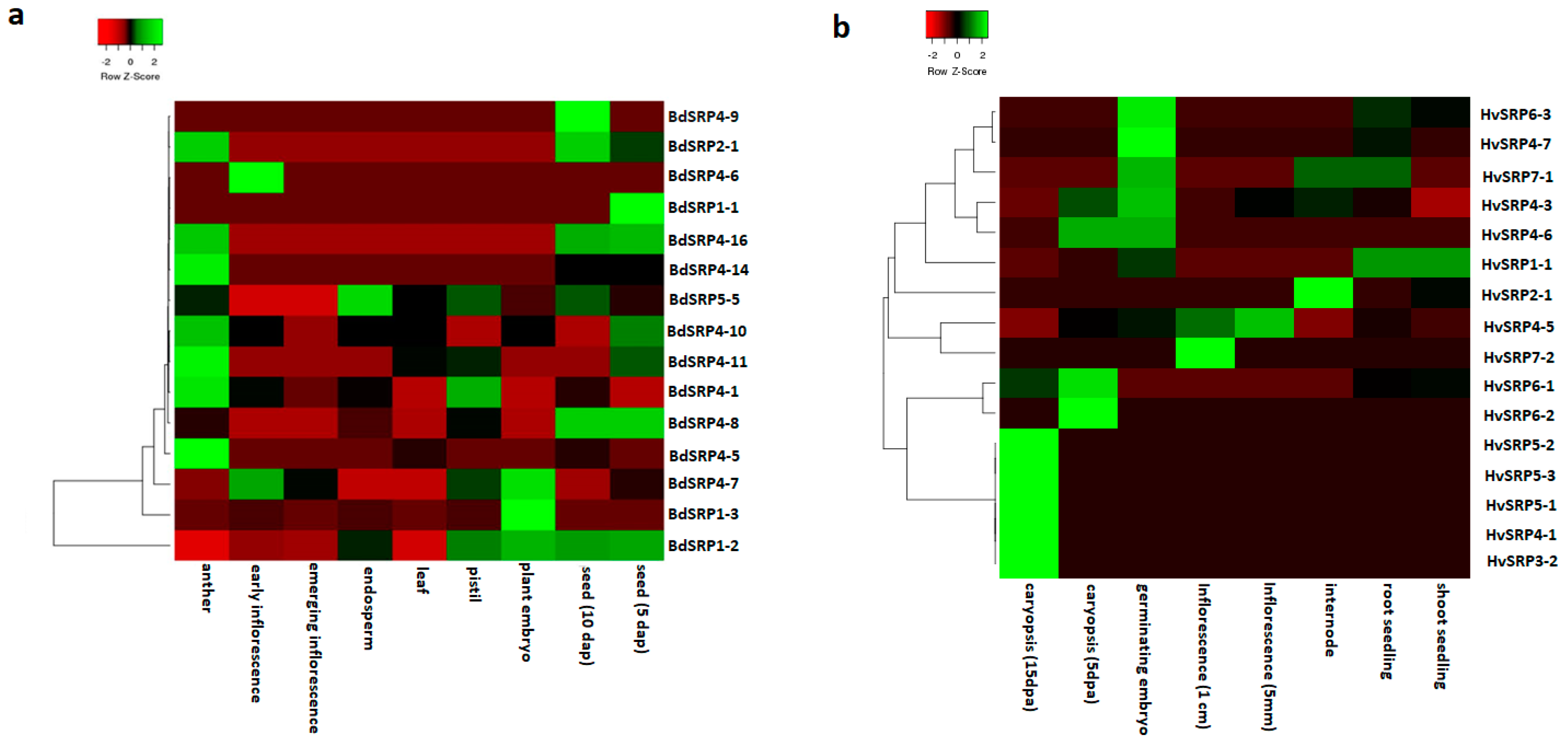
2.7. Development and Tissue Specific Expression Analysis of Serpin Genes
3. Materials and Methods
3.1. Sequence Analysis
3.2. Determination of Chromosomal Location and Synteny Analysis
3.3. Gene Structure and Conserved Motif Identification
3.4. Alignment of Sequences and Phylogenetic Analysis
3.5. Evolutionary Rate Calculations
3.6. Database Search for Expression Data of Serpin Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Roberts, T.H.; Hejgaard, J. Serpins in plants and green algae. Funct. Integr. Genom. 2008, 8, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Fluhr, R.; Lampl, N.; Roberts, T.H. Serpin protease inhibitors in plant biology. Physiol Plant. 2012, 145, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Pan, G.; Poncz, M.; Wei, J.; Ran, M.; Zhou, Z. Serpin functions in host-pathogen interactions. PeerJ 2018, 6, e4557. [Google Scholar] [CrossRef] [PubMed]
- Silverman, G.A.; Bird, P.I.; Carrell, R.W.; Church, F.C.; Coughlin, P.B.; Gettins, P.G.; Irving, J.A.; Lomas, D.A.; Luke, C.J.; Moyer, R.W.; et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001, 276, 33293–33296. [Google Scholar] [CrossRef]
- Gettins, P.G.W. Serpin structure, mechanism, and function. Chem. Rev. 2002, 102, 4751–4804. [Google Scholar] [CrossRef]
- Schick, C.; Brömme, D.; Bartuski, A.; Uemura, Y.; Schechter, N.M.; Silverman, G.A. The reactive site loop of the serpin SCCA1 is essential for cysteine proteinase inhibition. Proc. Natl. Acad. Sci. USA 1998, 95, 13465–13470. [Google Scholar] [CrossRef]
- Rustgi, S.; Boex-Fontvieille, E.; Reinbothe, C.; von Wettstein, D.; Reinbothe, S. The complex world of plant protease inhibitors: Insights into a Kunitz-type cysteine protease inhibitor of Arabidopsis thaliana. Commun. Integ. Biol. 2018, 11, e1368599. [Google Scholar] [CrossRef]
- Huntington, J.A. Serpin structure, function and dysfunction. J. Thromb. Haemost. 2011, 9, 26–34. [Google Scholar] [CrossRef]
- Francis, S.E.; Ersoy, R.A.; Ahn, J.W.; Atwell, B.J.; Roberts, T.H. Serpins in rice: Protein sequence analysis, phylogeny and gene expression during development. BMC Genom. 2012, 13, 449. [Google Scholar] [CrossRef]
- Law, R.H.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; et al. An overview of the serpin superfamily. Genome Biol. 2006, 7, 216. [Google Scholar] [CrossRef][Green Version]
- Laluk, K.; Mengiste, T. The Arabidopsis extracellular unusual serine protease inhibitor functions in resistance to necrotrophic fungi and insect herbivory. Plant. J. 2011, 68, 480–494. [Google Scholar] [CrossRef]
- Ostergaard, H.; Rasmussen, S.K.; Roberts, T.H.; Hejgaard, J. Inhibitory serpins from wheat grain with reactive centers resembling glutamine-rich repeats of prolamin storage proteins. Cloning and characterization of five major molecular forms. J. Biol. Chem. 2000, 275, 33272–33279. [Google Scholar] [CrossRef]
- Hejgaard, J. Inhibitory serpins from rye grain with glutamine as P-1 and P-2 residues in the reactive center. FEBS Lett. 2001, 488, 149–153. [Google Scholar] [CrossRef]
- Hejgaard, J.; Laing, W.A.; Marttila, S.; Gleave, A.P.; Roberts, T.H. Serpins in fruit and vegetative tissues of apple (Malus domestica): Expression of four serpins with distinct reactive centers and characterisation of a major inhibitory seed form, MdZ1b. Funct. Plant. Biol. 2005, 32, 517–527. [Google Scholar] [CrossRef]
- Pekkarinen, A.I.; Longstaff, C.; Jones, B.L. Kinetics of the Inhibition of Fusarium Serine Proteinases by Barley (Hordeum vulgare L.) Inhibitors. J. Agric. Food Chem. 2007, 55, 2736–2742. [Google Scholar] [CrossRef]
- Quilis, J.; Lopez-Garcia, B.; Meynard, D.; Guiderdoni, E.; Segundo, B.S. Inducible expression of a fusion gene encoding two proteinase inhibitors leads to insect and pathogen resistance in transgenic rice. Plant. Biotechnol. J. 2014, 12, 367–377. [Google Scholar] [CrossRef]
- Xia, C.J.; Wang, M.N.; Cornejo, O.E.; Jiwan, D.A.; See, D.R.; Chen, X. Secretome Characterization and Correlation Analysis Reveal Putative Pathogenicity Mechanisms and Identify Candidate Avirulence Genes in the Wheat Stripe Rust Fungus Puccinia striiformis f. sp tritici. Front. Microbiol. 2017, 8, 2394. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.C.; Aoki, K.; Xiang, Y.; Campbell, L.R.; Hull, R.J.; Xoconostle-Cazares, B.; Monzer, J.; Lee, J.Y.; Ullman, D.E.; Lucas, W.J. Characterization of Cucurbita maxima phloem serpin-1 (CmPS-1). A developmentally regulated elastase inhibitor. J. Biol. Chem. 2000, 275, 35122–35128. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Alfageme, F.; Maharramov, J.; Carrillo, L.; Vandenabeele, S.; Vercammen, D.; van Breusegem, F.; Smagghe, G. Potential use of a serpin from Arabidopsis for pest control. PLoS ONE 2011, 6, e20278. [Google Scholar] [CrossRef]
- Johnson, E.T.; Skory, C.D.; Naumann, T.A.; Jairajpuri, M.A.; Dowd, P.F. Three sorghum serpin recombinant proteins inhibit midgut trypsin activity and growth of corn earworm. Agri Gene 2016, 2, 11–16. [Google Scholar] [CrossRef]
- Hejgaard, J.; Rasmussen, S.K.; Brandt, A.; Svendsen, I. Sequence homology between barley endosperm protein Z and protease inhibitors of the α1-antitrypsin family. FEBS Lett. 1985, 180, 89–94. [Google Scholar] [CrossRef]
- Cohen, M.; Davydov, O.; Fluhr, R. Plant serpin protease inhibitors: Specificity and duality of Function. J. Exp. Bot. 2019, 70, 2077–2085. [Google Scholar] [CrossRef]
- Vercammen, D.; Belenghi, B.; van de Cotte, B.; Beunens, T.; Gavigan, J.A.; de Rycke, R.; Brackenier, A.; Inze, D.; Harris, J.L.; van Breusegem, F. Serpin1 of Arabidopsis thaliana is a suicide inhibitor for Metacaspase 9. J. Mol. Biol. 2006, 364, 625–636. [Google Scholar] [CrossRef]
- Lampl, N.; Budai-Hadrian, O.; Davydov, O.; Joss, T.V.; Harrop, S.J.; Curmi, P.M.; Roberts, T.H.; Fluhr, R. Arabidopsis AtSerpin1, crystal structure and in vivo interaction with its target protease responsive to desiccation-21 (RD21). J. Biol. Chem. 2010, 285, 13550–13560. [Google Scholar] [CrossRef]
- Koh, E.; Carmieli, R.; Mor, A.; Fluhr, R. Singlet Oxygen-Induced Membrane Disruption and Serpin- Protease Balance in Vacuolar-Driven Cell Death. Plant. Physiol. 2016, 171, 1616–1625. [Google Scholar] [CrossRef]
- Bhattacharjee, L.; Singh, D.; Gautam, J.K.; Nandi, A.K. Arabidopsis thaliana serpins AtSRP4 and AtSRP5 negatively regulate stress-induced cell death and effector-triggered immunity induced by bacterial effector AvrRpt2. Physiol. Plant. 2017, 159, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.W.; Atwell, B.J.; Roberts, T.H. Serpin genes AtSRP2 and AtSRP3 are required for normal growth sensitivity to a DNA alkylating agent in Arabidopsis. BMC Plant. Biol. 2009, 9, 52. [Google Scholar] [CrossRef]
- Bhattacharjee, L.; Singh, P.K.; Singh, S.; Nandi, A.K. Down-regulation of rice serpin gene OsSRP-LRS exaggerates stress-induced cell death. J. Plant. Biol. 2015, 58, 327–332. [Google Scholar] [CrossRef]
- Dhanushkodi, R.; Matthew, C.; McManus, M.T.; Dijkwel, P.P. Drought induced senescence of Medicago truncatula nodules involves serpin and ferritin to control proteolytic activity and iron levels. New Phytol. 2018, 220, 196–208. [Google Scholar] [CrossRef]
- Pemberton, P.A.; Stein, P.E.; Pepys, M.B.; Potter, J.M.; Carrell, R.W. Hormone binding globulins undergo serpin conformational change in inflammation. Nature 1988, 336, 257–258. [Google Scholar] [CrossRef]
- Nagata, K. Hsp47: A collagen-specific molecular chaperone. Trends Biochem. Sci. 1996, 21, 22–26. [Google Scholar] [CrossRef]
- Zou, Z.; Anisowicz, A.; Hendrix, M.J.; Thor, A.; Neveu, M.; Sheng, S.; Rafidi, K.; Seftor, E.; Sager, R. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science 1994, 263, 526–529. [Google Scholar] [CrossRef]
- Cohen, M.; Fluhr, R. Noncanonical interactions between serpin and β-amylase in barley grain improve β-amylase activity in vitro. Plant. Direct. 2018, 2, 1–12. [Google Scholar] [CrossRef]
- Finnie, C.; Maeda, K.; Østergaard, O.; Bak-Jensen, K.S.; Larsen, J.; Svensson, B. Aspects of the barley seed proteome during development and germination. Biochem. Soc. Trans. 2004, 32, 517–519. [Google Scholar] [CrossRef]
- Luo, H.; Zhang, Q.; Tucek, M.; Zhang, X.Q.; Li, C. Phenotypic and allelic variation for wort protein Z in Australian and Canadian barleys. J. Cereal Sci. 2020, 93, 102935. [Google Scholar] [CrossRef]
- Benbow, H.R.; Jermiin, L.S.; Doohan, F.M. Serpins: Genome-wide characterisation and expression analysis of the serine protease inhibitor family in Triticum aestivum. G3 Genes Genomes Genet. 2019, 9, 2709–2722. [Google Scholar] [CrossRef]
- Hejgaard, J. Free and protein-bound β-amylases of barley grain: Characterization by two-dimensional immunoelectrophoresis. Physiol. Plant. 1976, 38, 293–299. [Google Scholar] [CrossRef]
- Roberts, T.H.; Marttila, S.; Rasmussen, S.K.; Hejgaard, J. Differential gene expression for suicide-substrate serine proteinase inhibitors (serpins) in vegetative and grain tissues of barley. J. Exp. Bot. 2003, 54, 2251–2263. [Google Scholar] [CrossRef]
- Grosse-Holz, F.M.; van der Hoorn, R.A. Juggling jobs: Roles and mechanisms of multifunctional protease inhibitors in plants. New Phytol. 2016, 210, 794–807. [Google Scholar] [CrossRef]
- Pilar, C.; Boulos, C.; Vincent, C.; Garvin, D.F.; Hasterok, R.; Manzaneda, A.J.; Mur, L.A.J.; Pecchioni, N.; Rasmussen, S.K.; Vogel, J.P.; et al. Update on the genomics and basic biology of Brachypodium. Trends Plant. Sci. 2014, 19, 414–415. [Google Scholar]
- Hejgaard, J. Purification and properties of protein Z—A major albumin of barley endosperm. Physiol. Plant. 1982, 54, 174–182. [Google Scholar] [CrossRef]
- Rasmussen, S.K.; Hopp, H.E.; Brandt, A.; Svendsen, I.; Hejgaard, J. A cDNA clone for protein Z, a major barley endosperm albumin. Carlsberg Res. Commun. 1984, 49, 385–390. [Google Scholar] [CrossRef]
- Rasmussen, S.K. A gene coding for a new plant serpin. Biochim. Biophys. Acta 1993, 1172, 151–154. [Google Scholar] [CrossRef]
- Gordon, S.P.; Contreras-Moreira, B.; Woods, D.P.; Des Marais, D.L.; Burgess, D.; Shu, S.; Stritt, C.; Roulin, A.C.; Schackwitz, W.; Tyler, L.; et al. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Specker, C.; Niessen, L.; Vogel, R.F. In vitro studies on the main beer protein Z4 of Hordeum vulgare concerning heat stability, protease inhibition and gushing. J. Inst. Brew. 2014, 120, 85–92. [Google Scholar] [CrossRef]
- Habib, H.; Fazili, K.M. Plant protease inhibitors: A defense strategy in plants. Biotech. Mol. Biol. Rev. 2007, 2, 68–85. [Google Scholar]
- Brandt, A.; Svendsen, I.; Hejgaard, J. A plant serpin gene. Structure, organization and expression of the gene encoding barley protein Z4. Eur. J. Biochem. 1990, 194, 499–505. [Google Scholar] [CrossRef]
- Lcohamy, A.; Boudet, I.; Aubourg, S.; Kreis, M. Introns in, introns out in plant gene families: A genomic approach of the dynamics of gene structure. J. Struct. Funct. Genom. 2003, 3, 111–116. [Google Scholar]
- Lurin, C.; Andrés, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyère, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organdie biogenesis. Plant. Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef]
- Jaill, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar]
- Huntington, J.A.; Fan, B.; Karlsson, K.E.; Deinum, J.; Lawrence, D.A.; Gettins, P.G. Serpin conformational change in ovalbumin. Enhanced reactive center loop insertion through hinge region mutations. Biochemistry 1997, 36, 5432–5440. [Google Scholar] [CrossRef] [PubMed]
- Silverman, G.A.; Whisstock, J.C.; Askew, D.J.; Pak, S.C.; Luke, C.J.; Cataltepe, S.; Irving, J.A.; Bird, P.I. Human clade B serpins (ov-serpins) belong to a cohort of evolutionarily-dispersed intracellular proteinase inhibitor clades that protect cells from promiscuous proteolysis. Cell Mol. Life Sci. 2004, 61, 301–325. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant. Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, H.; Chen, W.; Qian, Y.; Ma, Q.; Cheng, B.; Zhu, S. Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant. Growth Regul. 2011, 63, 225–234. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Gaut, B.S. Evolutionary dynamics of grass genomes. New Phytol. 2002, 154, 15–28. [Google Scholar] [CrossRef]
- The International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef]
- Hejgaard, J.; Hauge, S. Serpins of oat (Avena sativa) grain with distinct reactive centers and inhibitory specificity. Physiol. Plant. 2002, 116, 155–163. [Google Scholar] [CrossRef]
- Dahl, S.W.; Rasmussen, S.K.; Hejgaard, J. Heterologous expression of three plant serpins with distinct inhibitory specificities. J. Biol. Chem. 1996, 271, 25083–25088. [Google Scholar] [CrossRef]
- Santamaría, M.E.; Diaz-Mendoza, M.; Diaz, I.; Martinez, M. Plant protein peptidase inhibitors: An evolutionary overview based on comparative genomics. BMC Genom. 2014, 15, 812. [Google Scholar] [CrossRef]
- Cohen, M.; Roberts, T.H.; Fluhr, R. Serpins in Plants. In The Serpin Family: Proteins with Multiple Functions in Health and Disease; Geiger, M., Wahlmüller, F., Furtmüller, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 15–28. [Google Scholar]
- Ma, D.Y.; Huang, X.; Hou, J.F.; Ma, Y.; Han, Q.X.; Hou, G.G.; Wang, C.Y.; Guo, T.C. Quantitative analysis of the grain amyloplast proteome reveals differences in metabolism between two wheat cultivars at two stages of grain development. BMC Genom. 2018, 19, 768. [Google Scholar] [CrossRef]
- Koller, A.; Washburn, M.P.; Lange, B.M.; Andon, N.L.; Deciu, C.; Haynes, P.A.; Hays, L.; Schieltz, D.; Ulaszek, R.; Wei, J.; et al. Proteomic survey of metabolic pathways in rice. Proc. Natl. Acad. Sci. USA 2002, 99, 11969–11974. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega 6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4 phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Suyama, M.; Torrents, D.; Bork, P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006, 34, W609–W612. [Google Scholar] [CrossRef]

| Motifs | Sequences |
|---|---|
| 1 | RPLYVSSVFHKAVVEVBEEGTEAAAATAA |
| 2 | FVADHPFLFLIVEEVSGAVLF |
| 3 | DTRLVLGNALYFKGKWTEPFD |
| 4 | FSMYIFLPDARDGLWGLADKJ |
| 5 | IACHDGFKVLKLPYKQ |
| Segment Pair | Ka | Ks | Ka/Ks | Estimated Time (MYA) | Mode of Duplication |
|---|---|---|---|---|---|
| BdSRP1-2/BdSRP1-3 | 0.0842 | 0.3685 | 0.22846 | 30.19 | Tandem |
| BdSRP4-5/BdSRP4-6 | 0.2022 | 0.3732 | 0.54167 | 30.59 | Tandem |
| BdSRP4-5/BdSRP4-4 | 0.175 | 0.7633 | 0.2292 | 62.56 | Tandem |
| BdSRP4-8/BdSRP4-10 | 0.2806 | 0.6012 | 0.46676 | 49.27 | Tandem |
| BdSRP4-9/BdSRP4-11 | 0.0883 | 0.223 | 0.39612 | 18.27 | Tandem |
| BdSRP4-12/BdSRP4-13 | 0.2209 | 0.2929 | 0.75396 | 24 | Tandem |
| BdSRP4-15/BdSRP4-16 | 0.0755 | 0.1509 | 0.50059 | 12.36 | Tandem |
| BdSRP4-14/BdSRP4-17 | 1.0413 | 0.1591 | 0.1528 | 13.04 | Tandem |
| BdSRP5-4/BdSRP5-5 | 0.3304 | 0.2925 | 1.12963 | 23.97 | Tandem |
| Mean | 0.3659 | 0.3582 | 0.4887 | 29.3611 |
| Segment Pair | Ka | Ks | Ka/Ks | Estimated Time (MYA) | Mode of Duplication |
|---|---|---|---|---|---|
| BdSRP1-1/BdSRP1-4 | 0.0986 | 0.2347 | 0.42019 | 19.23 | Segmental |
| Segment Pair | Ka | Ks | Ka/Ks | Estimated Time (MYA) | Mode of Duplication |
|---|---|---|---|---|---|
| HvSRP2-2/HvSRP2-3 | 0.1906 | 0.6556 | 0.2906 | 53.7454 | Tandem |
| HvSRP4-2/HvSRP4-3 | 0.0536 | 0.0633 | 0.8479 | 5.1888 | Tandem |
| HvSRP5-2/HvSRP5-3 | 0.0193 | 0.035 | 0.5513 | 2.8756 | Tandem |
| HvSRP6-1/HvSRP6-2 | 0.2058 | 0.4277 | 0.4812 | 35.0602 | Tandem |
| HvSRP6-3/HvSRP6-2 | 0.2133 | 0.3655 | 0.5836 | 29.9666 | Tandem |
| Mean | 0.1365 | 0.3094 | 0.5509 | 25.3673 |
| Segment pair | Ka | Ks | Ka/Ks | Estimated time (MYA) | Mode of duplication |
|---|---|---|---|---|---|
| HvSRP4-1/HvSRP5-2 | 0.2072 | 0.6857 | 0.3022 | 56.2082 | Segmental |
| HvSRP4-4/HvSRP5-3 | 0.1703 | 0.6761 | 0.2518 | 55.4252 | Segmental |
| HvSRP4-7/HvSRP7-1 | 0.0146 | 0.0495 | 0.2948 | 4.0591 | Segmental |
| Mean | 0.1307 | 0.469 | 0.2829 | 38.5641 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehman, S.; Jørgensen, B.; Aziz, E.; Batool, R.; Naseer, S.; Rasmussen, S.K. Genome Wide Identification and Comparative Analysis of the Serpin Gene Family in Brachypodium and Barley. Plants 2020, 9, 1439. https://doi.org/10.3390/plants9111439
Rehman S, Jørgensen B, Aziz E, Batool R, Naseer S, Rasmussen SK. Genome Wide Identification and Comparative Analysis of the Serpin Gene Family in Brachypodium and Barley. Plants. 2020; 9(11):1439. https://doi.org/10.3390/plants9111439
Chicago/Turabian StyleRehman, Shazia, Bodil Jørgensen, Ejaz Aziz, Riffat Batool, Samar Naseer, and Søren K. Rasmussen. 2020. "Genome Wide Identification and Comparative Analysis of the Serpin Gene Family in Brachypodium and Barley" Plants 9, no. 11: 1439. https://doi.org/10.3390/plants9111439
APA StyleRehman, S., Jørgensen, B., Aziz, E., Batool, R., Naseer, S., & Rasmussen, S. K. (2020). Genome Wide Identification and Comparative Analysis of the Serpin Gene Family in Brachypodium and Barley. Plants, 9(11), 1439. https://doi.org/10.3390/plants9111439






