Multidimensional Evaluation for Detecting Salt Tolerance of Bread Wheat Genotypes Under Actual Saline Field Growing Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site and Water Source Description
2.2. Plant Materials, Experimental Design, and Agronomic Practices
2.3. Measurements
2.3.1. Physio-Biochemical Parameters
2.3.2. Agronomical Parameters
2.4. Statistical Analysis
3. Results
3.1. Grouping Genotypes Based in Their Salt Tolerance Level
3.2. Physio-Biochemical Attributes
3.3. Agronomical Attributes
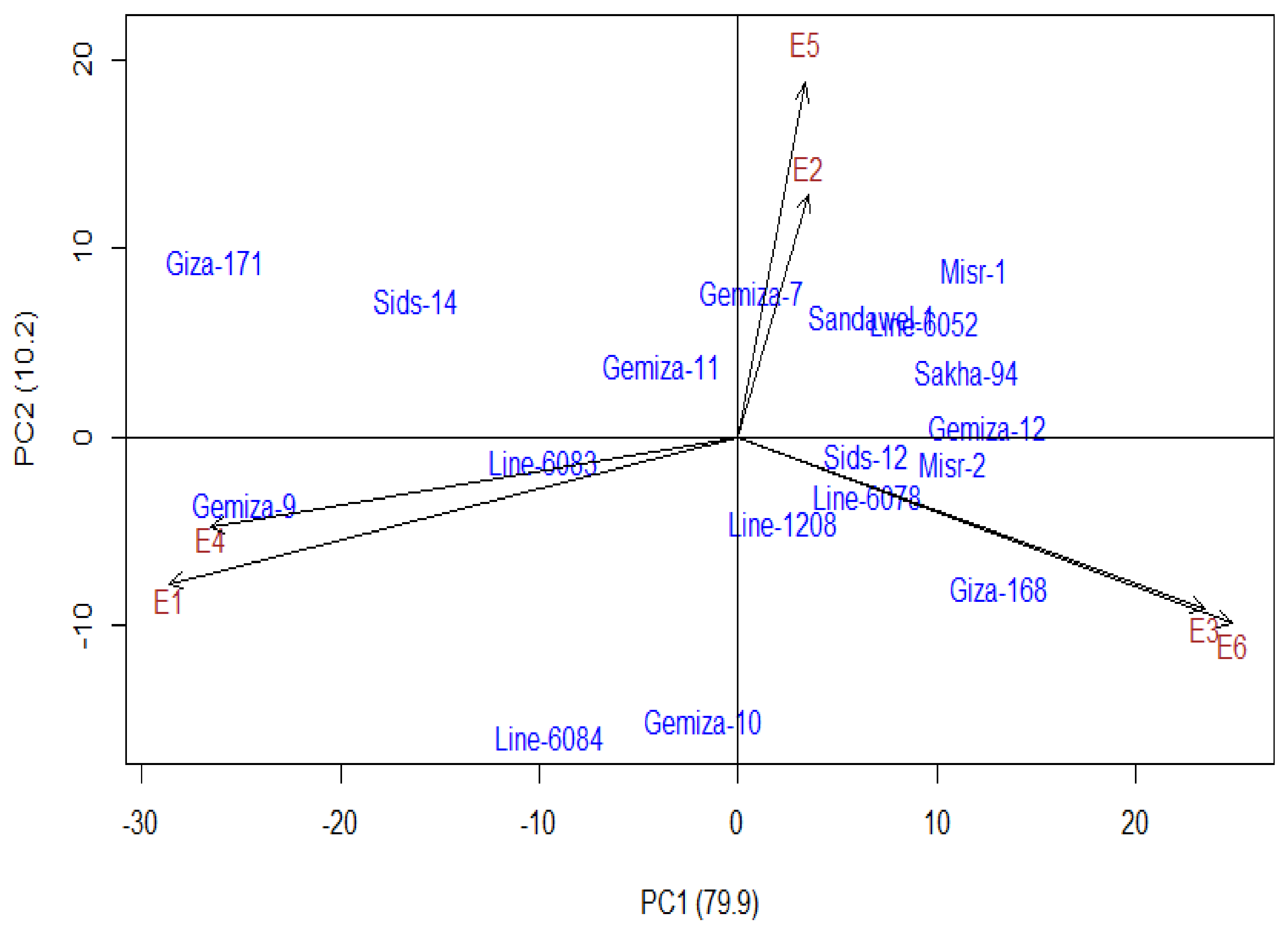
3.4. Additive Main Effect and Multiplicative Interaction Model (AMMI)
3.5. Inter-Relationship Between All Measured Parameters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kumar, P.; Yadava, R.; Gollen, B.; Kumar, S.; Verma, R.K.; Yadav, S. Nutritional contents and medicinal properties of wheat: A review. Life Sci. Med. Res. 2011, 22, 1–10. [Google Scholar]
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Sec. 2015, 4, 178–202. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. Food and Agriculture Organization of the United Nations Statistics Database. 2020. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 21 June 2020).
- Newell, N. Effects of soil salinity on plant growth. Plant Physiol. 2013, 1, 1–4. [Google Scholar]
- Farooq, M.; Hussain, M.; Wakeel, A.; Siddique, K.H. Salt stress in maize: Effects, resistance mechanisms, and management. A review. Agron. Sustain. Dev. 2015, 35, 461–481. [Google Scholar] [CrossRef]
- Allel, D.; Ben-Amar, A.; Abdelly, C. Leaf photosynthesis, chlorophyll fluorescence and ion content of barley (Hordeum vulgare) in response to salinity. J. Plant Nutr. 2018, 41, 497–508. [Google Scholar] [CrossRef]
- Machado, R.M.A.; Serralheiro, R.P. Soil salinity: Effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Kotb, T.H.; Watanabe, T.; Ogino, Y.; Tanji, K.K. Soil salinization in the Nile Delta and related policy issues in Egypt. Agric. Water Manag. 2000, 43, 239–261. [Google Scholar] [CrossRef]
- Chen, J.; Mueller, V. Coastal climate change, soil salinity and human migration in Bangladesh. Nat. Clim. Chang. 2018, 8, 981–985. [Google Scholar] [CrossRef]
- Baker, N.R.; Rosenqvist, E. Applications of chlorophyll fluorescence can improve crop production strategies: An examination of future possibilities. J. Exp. Bot. 2004, 55, 1607–1621. [Google Scholar] [CrossRef]
- Wu, D.; Qiu, L.; Xu, L.; Ye, L.; Chen, M.; Sun, D.; Chen, Z.; Zhang, H.; Jin, X.; Dai, F. Genetic variation of HvCBF genes and their association with salinity tolerance in Tibetan annual wild barley. PLoS ONE 2011, 6, e22938. [Google Scholar] [CrossRef][Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Chen, T.H.; Murata, N. Glycinebetaine protects plants against abiotic stress: Mechanisms and biotechnological applications. Plant Cell Environ. 2011, 34, 1–20. [Google Scholar] [CrossRef] [PubMed]
- El-Hendawy, S.E.; Hassan, W.M.; Al-Suhaibani, N.A.; Refay, Y.; Abdella, K.A. Comparative performance of multivariable agro-physiological parameters for detecting salt tolerance of wheat cultivars under simulated saline field growing conditions. Front. Plant Sci. 2017, 8, 435. [Google Scholar] [CrossRef] [PubMed]
- Desoky, E.-S.M.; Elrys, A.S.; Rady, M.M. Integrative moringa and licorice extracts application improves Capsicum annuum fruit yield and declines its contaminant contents on a heavy metals-contaminated saline soil. Ecotoxicol. Environ. Saf. 2019, 169, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Rady, M.; Desoky, E.-S.; Elrys, A.; Boghdady, M. Can licorice root extract be used as an effective natural biostimulant for salt-stressed common bean plants? S. Afr. J. Bot. 2019, 121, 294–305. [Google Scholar] [CrossRef]
- De Leon, T.B.; Linscombe, S.; Gregorio, G.; Subudhi, P.K. Genetic variation in Southern USA rice genotypes for seedling salinity tolerance. Front. Plant Sci. 2015, 6, 374. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Oxygen processing in photosynthesis: Regulation and signalling. New Phytol. 2000, 146, 359–388. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Hoque, M.A.; Banu, M.N.A.; Okuma, E.; Amako, K.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. Exogenous proline and glycinebetaine increase NaCl-induced ascorbate–glutathione cycle enzyme activities, and proline improves salt tolerance more than glycinebetaine in tobacco Bright Yellow-2 suspension-cultured cells. J. Plant Physiol. 2007, 164, 1457–1468. [Google Scholar] [CrossRef]
- Sudhir, P.; Murthy, S. Effects of salt stress on basic processes of photosynthesis. Photosynthetica 2004, 42, 481–486. [Google Scholar] [CrossRef]
- Kahrizi, S.; Sedighi, M.; Sofalian, O. Effect of salt stress on proline and activity of antioxidant enzymes in ten durum wheat cultivars. Ann. Biol. Res. 2012, 3, 3870–3874. [Google Scholar]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 1–26. [Google Scholar] [CrossRef]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Rady, M.M. Effect of 24-epibrassinolide on growth, yield, antioxidant system and cadmium content of bean (Phaseolus vulgaris L.) plants under salinity and cadmium stress. Sci. Hortic. 2011, 129, 232–237. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, H.; Chen, T.; Pen, J.; Yu, S.; Zhao, X. Morphological and physiological responses of cotton (Gossypium hirsutum L.) plants to salinity. PLoS ONE 2014, 9, e112807. [Google Scholar] [CrossRef]
- Rios, J.J.; Martínez-Ballesta, M.C.; Ruiz, J.M.; Blasco, B.; Carvajal, M. Silicon-mediated improvement in plant salinity tolerance: The role of aquaporins. Front. Plant Sci. 2017, 8, 948. [Google Scholar] [CrossRef]
- Demidchik, V.; Straltsova, D.; Medvedev, S.S.; Pozhvanov, G.A.; Sokolik, A.; Yurin, V. Stress-induced electrolyte leakage: The role of K+ permeable channels and involvement in programmed cell death and metabolic adjustment. J. Exp. Bot. 2014, 65, 1259–1270. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M. Free Radicals in Biology and Medicine; Oxford University Press: New York, NY, USA, 2015; pp. 77–153. [Google Scholar]
- Schutzendubel, A.; Polle, A. Plant responses to abiotic stresses: Heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 2002, 53, 1351–1365. [Google Scholar] [CrossRef]
- Desoky, E.-S.M.; El Sayed, A.I.; Merwad, A.-R.M.; Rady, M.M. Stimulating antioxidant defenses, antioxidant gene expression, and salt tolerance in Pisum sativum seedling by pretreatment using licorice root extract (LRE) as an organic biostimulant. Plant Physiol. Biochem. 2019, 142, 292–302. [Google Scholar] [CrossRef]
- Silveira, J.A.G.; de Almeida Viégas, R.; da Rocha, I.M.A.; Moreira, A.C.d.O.M.; de Azevedo Moreira, R.; Oliveira, J.T.A. Proline accumulation and glutamine synthetase activity are increased by salt-induced proteolysis in cashew leaves. J. Plant Physiol. 2003, 160, 115–123. [Google Scholar] [CrossRef]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotech. 2014, 26, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef] [PubMed]
- Dadshani, S.; Sharma, R.C.; Baum, M.; Ogbonnaya, F.C.; Léon, J.; Ballvora, A. Multi-dimensional evaluation of response to salt stress in wheat. PLoS ONE 2019, 14, e0222659. [Google Scholar] [CrossRef]
- El Hendawy, S.; Ruan, Y.; Hu, Y.; Schmidhalter, U. A comparison of screening criteria for salt tolerance in wheat under field and controlled environmental conditions. J. Agron. Crop Sci. 2009, 195, 356–367. [Google Scholar] [CrossRef]
- Hasan, A.; Hafiz, H.R.; Siddiqui, N.; Khatun, M.; Islam, R.; Mamun, A.-A. Evaluation of wheat genotypes for salt tolerance based on some physiological traits. J. Crop Sci. Biotechnol. 2015, 18, 333–340. [Google Scholar] [CrossRef]
- Oyiga, B.C.; Sharma, R.; Shen, J.; Baum, M.; Ogbonnaya, F.; Léon, J.; Ballvora, A. Identification and characterization of salt tolerance of wheat germplasm using a multivariable screening approach. J. Agron. Crop Sci. 2016, 202, 472–485. [Google Scholar] [CrossRef]
- Ellis, R.; Forster, B.; Gordon, D.; Handley, L.; Keith, R.; Lawrence, P.; Meyer, R.; Powell, W.; Robinson, D.; Scrimgeour, C. Phenotype/genotype associations for yield and salt tolerance in a barley mapping population segregating for two dwarfing genes. J. Exp. Bot. 2002, 53, 1163–1176. [Google Scholar] [CrossRef]
- Bağci, S.A.; Ekiz, H.; Yilmaz, A. Salt tolerance of sixteen wheat genotypes during seedling growth. Turk. J. Agric. For. 2007, 31, 363–372. [Google Scholar]
- Khan, M.; Yasmin, S.; Ansari, R.; Shirazi, M.; Ashraf, M. Screening for salt tolerance in wheat genotypes at an early seedling stage. Pak. J. Bot. 2007, 39, 2501–2509. [Google Scholar]
- El-Hendawy, S.; Hu, Y.; Sakagami, J.-I.; Schmidhalter, U. Screening Egyptian wheat genotypes for salt tolerance at early growth stages. Int. J. Plant Prod. 2011, 5, 283–298. [Google Scholar]
- Muhammad, Z.; Hussain, F. Effect of NaCl salinity on the germination and seedling growth of seven wheat genotypes. Pak. J. Bot. 2012, 44, 1845–1850. [Google Scholar]
- Chahine, K.; Sourour, A.; Youssef, T.; Hajer, S.-A. Salinity effect on plant growth at the seedling stage of durum wheat (Triticum durum Desf.). J. Plant Breed. Crop Sci. 2013, 5, 20–25. [Google Scholar] [CrossRef]
- Hussain, B.; Khan, A.S.; Ali, Z. Genetic variation in wheat germplasm for salinity tolerance atseedling stage: Improved statistical inference. Turk. J. Agric. For. 2015, 39, 182–192. [Google Scholar] [CrossRef]
- Jovović, M.; Tunguz, V.; Mirosavljević, M.; Pržulj, N. Effect of salinity and drought stress on germination and early seedlings growth of bread wheat (Triticum aestivum L.). Genetika 2018, 50, 285–298. [Google Scholar] [CrossRef]
- Guellim, A.; Catterou, M.; Chabrerie, O.; Tetu, T.; Hirel, B.; Dubois, F.; Ben Ahmed, H.; Kichey, T. Identification of phenotypic and physiological markers of salt stress tolerance in durum wheat (Triticum durum Desf.) through integrated server analyses. Agronomy 2019, 9, 844. [Google Scholar] [CrossRef]
- Al-Ashkar, I.; Alderfasi, A.; Ben Romdhane, W.; Seleiman, M.F.; El-Said, R.A.; Al-Doss, A. Morphological and genetic diversity within salt tolerance detection in eighteen wheat genotypes. Plants 2020, 9, 287. [Google Scholar] [CrossRef]
- Igartua, E.; Gracia, M.; Lasa, J. Field responses of grain sorghum to a salinity gradient. Field Crops Res. 1995, 42, 15–25. [Google Scholar] [CrossRef]
- Allel, D.; BenAmar, A.; Badri, M.; Abdelly, C. Evaluation of salinity tolerance indices in North African barley accessions at reproductive stage. Czech J. Genet. Plant Breed. 2019, 55, 61–69. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A. Screening methods for salinity tolerance: A case study with tetraploid wheat. Plant Soil 2003, 253, 201–218. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Blumwald, E. Developing salt-tolerant crop plants: Challenges and opportunities. Trends Plant Sci. 2005, 10, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Mansour, E.; Moustafa, E.S.; Qabil, N.; Abdelsalam, A.; Wafa, H.A.; El Kenawy, A.; Casas, A.M.; Igartua, E. Assessing different barley growth habits under Egyptian conditions for enhancing resilience to climate change. Field Crops Res. 2018, 224, 67–75. [Google Scholar] [CrossRef]
- Mansour, E.; Abdul-Hamid, M.I.; Yasin, M.T.; Qabil, N.; Attia, A. Identifying drought-tolerant genotypes of barley and their responses to various irrigation levels in a Mediterranean environment. Agric. Water Manag. 2017, 194, 58–67. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Weatherley, P. Studies in the water relations of the cotton plant. I. The field measurement of water deficits in leaves. New Phytol. 1950, 87, 81–97. [Google Scholar] [CrossRef]
- Premachandra, G.; Saneoka, H.; Ogata, S. Cell membrane stability, an indicator of drought tolerance as affected by applied nitrogen in soybean. J. Agric. Sci. 1990, 115, 63–66. [Google Scholar] [CrossRef]
- Sullivan, C.Y. Selection for Drought and Heat Tolerance in Grain Sorghum; John Wiley & Sons: New York, NY, USA, 1979; pp. 263–281. [Google Scholar]
- Zhang, Z.L.; Qu, W. Experimental Guidance of Plant Physiology; High Education: Beijing, China, 2004. [Google Scholar]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Irigoyen, J.; Einerich, D.; Sánchez-Díaz, M. Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicago sativa) plants. Physiol. Plant. 1992, 84, 55–60. [Google Scholar] [CrossRef]
- Mukherjee, S.; Choudhuri, M. Implications of water stress-induced changes in the levels of endogenous ascorbic acid and hydrogen peroxide in Vigna seedlings. Physiol. Plant. 1983, 58, 166–170. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Aebi, H. Catalase In Vitro; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Maechlay, A.; Chance, B. The Assay of Catalase and Peroxidase; John Wiley & Sons, Inc.: New York, NY, USA, 1954; pp. 357–424. [Google Scholar]
- Klapheck, S.; Zimmer, I.; Cosse, H. Scavenging of hydrogen peroxide in the endosperm of Ricinus communis by ascorbate peroxidase. Plant Cell Physiol. 1990, 31, 1005–1013. [Google Scholar]
- Wolf, B. A comprehensive system of leaf analyses and its use for diagnosing crop nutrient status. Commun. Soil Sci. Plant Anal. 1982, 13, 1035–1059. [Google Scholar] [CrossRef]
- Gavuzzi, P.; Rizza, F.; Palumbo, M.; Campanile, R.; Ricciardi, G.; Borghi, B. Evaluation of field and laboratory predictors of drought and heat tolerance in winter cereals. Can. J. Plant Sci. 1997, 77, 523–531. [Google Scholar] [CrossRef]
- Ward, J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Gauch, H.G., Jr. Statistical analysis of yield trials by AMMI and GGE. Crop Sci. 2006, 46, 1488–1500. [Google Scholar] [CrossRef]
- Aragüés, R.; Urdanoz, V.; Çetin, M.; Kirda, C.; Daghari, H.; Ltifi, W.; Lahlou, M.; Douaik, A. Soil salinity related to physical soil characteristics and irrigation management in four Mediterranean irrigation districts. Agric. Water Manag. 2011, 98, 959–966. [Google Scholar] [CrossRef]
- El-Hendawy, S.E.; Hu, Y.; Yakout, G.M.; Awad, A.M.; Hafiz, S.E.; Schmidhalter, U. Evaluating salt tolerance of wheat genotypes using multiple parameters. Eur. J. Agron. 2005, 22, 243–253. [Google Scholar] [CrossRef]
- Genc, Y.; Tester, M.; McDonald, G. Calcium requirement of wheat in saline and non-saline conditions. Plant Soil 2010, 327, 331–345. [Google Scholar] [CrossRef]
- Tavakkoli, E.; Rengasamy, P.; McDonald, G.K. The response of barley to salinity stress differs between hydroponic and soil systems. Funct. Plant Biol. 2010, 37, 621–633. [Google Scholar] [CrossRef]
- Houshmand, S.; Arzani, A.; Maibody, S.A.M.; Feizi, M. Evaluation of salt-tolerant genotypes of durum wheat derived from in vitro and field experiments. Field Crops Res. 2005, 91, 345–354. [Google Scholar] [CrossRef]
- Shafi, M.; Bakhat, J.; Khan, M.J.; Khan, M.A.; Anwar, S. Effect of salinity on yield and ion accumulation of wheat genotypes. Pak. J. Bot. 2010, 42, 4113–4121. [Google Scholar]
- Sheng, M.; Tang, M.; Chen, H.; Yang, B.; Zhang, F.; Huang, Y. Influence of arbuscular mycorrhizae on photosynthesis and water status of maize plants under salt stress. Mycorrhiza 2008, 18, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Srinieng, K.; Saisavoey, T.; Karnchanatat, A. Effect of salinity stress on antioxidative enzyme activities in tomato cultured in vitro. Pak. J. Bot. 2015, 47, 1–10. [Google Scholar]
- Rezende, R.A.L.S.; Rodrigues, F.A.; Soares, J.D.R.; Silveira, H.R.d.O.; Pasqual, M.; Dias, G.d.M.G. Salt stress and exogenous silicon influence physiological and anatomical features of in vitro-grown cape gooseberry. Cienc. Rural 2018, 48, e20170176. [Google Scholar] [CrossRef]
- Lin, C.C.; Kao, C.H. Cell wall peroxidase activity, hydrogen peroxide level and NaCl-inhibited root growth of rice seedlings. Plant Soil 2001, 230, 135–143. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Hong, C.Y.; Liu, L.F.; Kao, C.H. Relative importance of Na+ and Cl– in NaCl-induced antioxidant systems in roots of rice seedlings. Physiol. Plant. 2004, 122, 86–94. [Google Scholar] [CrossRef]
- Zhao, H.; Liang, H.; Chu, Y.; Sun, C.; Wei, N.; Yang, M.; Zheng, C. Effects of salt stress on chlorophyll fluorescence and the antioxidant system in Ginkgo biloba L. seedlings. HortScience 2019, 54, 2125–2133. [Google Scholar] [CrossRef]
- Stępień, P.; Kłbus, G. Water relations and photosynthesis in Cucumis sativus L. leaves under salt stress. Biol. Plant. 2006, 50, 610–616. [Google Scholar] [CrossRef]
- Trapp, S.; Feificova, D.; Rasmussen, N.F.; Bauer-Gottwein, P. Plant uptake of NaCl in relation to enzyme kinetics and toxic effects. Environ. Exp. Bot. 2008, 64, 1–7. [Google Scholar] [CrossRef]
- Saqib, M.; Akhtar, J.; Abbas, G.; Nasim, M. Salinity and drought interaction in wheat (Triticum aestivum L.) is affected by the genotype and plant growth stage. Acta Physiol. Plant. 2013, 35, 2761–2768. [Google Scholar] [CrossRef]
- Valero, E.; Macià, H.; Ildefonso, M.; Hernández, J.-A.; González-Sánchez, M.-I.; García-Carmona, F. Modeling the ascorbate-glutathione cycle in chloroplasts under light/dark conditions. BMC Syst. Biol. 2015, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Sairam, R.K.; Rao, K.V.; Srivastava, G. Differential response of wheat genotypes to long term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Sci. 2002, 163, 1037–1046. [Google Scholar] [CrossRef]
- Rady, M.M.; Kuşvuran, A.; Alharby, H.F.; Alzahrani, Y.; Kuşvuran, S. Pretreatment with proline or an organic bio-stimulant induces salt tolerance in wheat plants by improving antioxidant redox state and enzymatic activities and reducing the oxidative stress. J. Plant Growth Regul. 2019, 38, 449–462. [Google Scholar] [CrossRef]
- Zhu, J.-K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Alzahrani, Y.; Rady, M.M. Compared to antioxidants and polyamines, the role of maize grain-derived organic biostimulants in improving cadmium tolerance in wheat plants. Ecotoxicol. Environ. Saf. 2019, 182, 109378. [Google Scholar] [CrossRef]
- Farhangi-Abriz, S.; Torabian, S. Antioxidant enzyme and osmotic adjustment changes in bean seedlings as affected by biochar under salt stress. Ecotoxicol. Environ. Saf. 2017, 137, 64–70. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Howladar, S.M. A novel Moringa oleifera leaf extract can mitigate the stress effects of salinity and cadmium in bean (Phaseolus vulgaris L.) plants. Ecotoxicol. Environ. Saf. 2014, 100, 69–75. [Google Scholar] [CrossRef]
- Semida, W.M.; Rady, M.M. Presoaking application of propolis and maize grain extracts alleviates salinity stress in common bean (Phaseolus vulgaris L.). Sci. Hortic. 2014, 168, 210–217. [Google Scholar] [CrossRef]
- Elrys, A.S.; Abdo, A.I.; Abdel-Hamed, E.M.; Desoky, E.-S.M. Integrative application of licorice root extract or lipoic acid with fulvic acid improves wheat production and defenses under salt stress conditions. Ecotoxicol. Environ. Saf. 2020, 190, 110144. [Google Scholar] [CrossRef]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, J.; Chow, W.; Sun, L.; Chen, J.; Chen, Y.; Peng, C. The influence of low temperature on photosynthesis and antioxidant enzymes in sensitive banana and tolerant plantain (Musa sp.) cultivars. Photosynthetica 2011, 49, 201–208. [Google Scholar] [CrossRef]
- Xu, R.; Yamada, M.; Fujiyama, H. Lipid peroxidation and antioxidative enzymes of two turfgrass species under salinity stress. Pedosphere 2013, 23, 213–222. [Google Scholar] [CrossRef]
- Abogadallah, G.M. Insights into the significance of antioxidative defense under salt stress. Plant Signal. Behav. 2010, 5, 369–374. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Exogenous silicon attenuates cadmium-induced oxidative stress in Brassica napus L. by modulating AsA-GSH pathway and glyoxalase system. Front. Plant Sci. 2017, 8, 1061. [Google Scholar] [CrossRef]
- Zhou, Y.; Wen, Z.; Zhang, J.; Chen, X.; Cui, J.; Xu, W.; Liu, H.-Y. Exogenous glutathione alleviates salt-induced oxidative stress in tomato seedlings by regulating glutathione metabolism, redox status, and the antioxidant system. Sci. Hortic. 2017, 220, 90–101. [Google Scholar] [CrossRef]
- Hernández, J.A.; Almansa, M.S. Short-term effects of salt stress on antioxidant systems and leaf water relations of pea leaves. Physiol. Plant. 2002, 115, 251–257. [Google Scholar] [CrossRef]
- Yadavi, A.; Aboueshaghi, R.; Dehnavi, M.; Balouchi, H. Effect of micronutrients foliar application on grain qualitative characteristics and some physiological traits of bean (Phaseolus vulgaris L.) under drought stress. Indian J. Fundam. Appl. Life Sci. 2014, 4, 124–131. [Google Scholar]
- Allen, D.; McKee, I.; Farage, P.; Baker, N. Analysis of limitations to CO2 assimilation on exposure of leaves of two Brassica napus cultivars to UV-B. Plant Cell Environ. 1997, 20, 633–640. [Google Scholar] [CrossRef]
- Sitohy, M.Z.; Desoky, E.-S.M.; Osman, A.; Rady, M.M. Pumpkin seed protein hydrolysate treatment alleviates salt stress effects on Phaseolus vulgaris by elevating antioxidant capacity and recovering ion homeostasis. Sci. Hortic. 2020, 271, 109495. [Google Scholar] [CrossRef]
- Borsani, O.; Valpuesta, V.; Botella, M. Developing salt tolerant plants in a new century: A molecular biology approach. Plant Cell Tiss. Org. 2003, 73, 101–115. [Google Scholar] [CrossRef]
- Wahome, P.; Jesch, H.; Grittner, I. Mechanisms of salt stress tolerance in two rose rootstocks: Rosa chinensis ‘Major’and R. rubiginosa. Sci. Hortic. 2001, 87, 207–216. [Google Scholar] [CrossRef]
- Wang, M.; Zheng, Q.; Shen, Q.; Guo, S. The critical role of potassium in plant stress response. Int. J. Mol. Sci. 2013, 14, 7370–7390. [Google Scholar] [CrossRef]
- Meloni, D.A.; Martínez, C.A. Glycinebetaine improves salt tolerance in vinal (Prosopis ruscifolia Griesbach) seedlings. Braz. J. Plant Physiol. 2009, 21, 233–241. [Google Scholar] [CrossRef]
- Vysotskaya, L.; Hedley, P.E.; Sharipova, G.; Veselov, D.; Kudoyarova, G.; Morris, J.; Jones, H.G. Effect of salinity on water relations of wild barley plants differing in salt tolerance. AoB Plant. 2010, 2010, 1–8. [Google Scholar] [CrossRef]
- Mansour, E.; Merwad, A.; Yasin, M.; Abdul-Hamid, M.; El-Sobky, E.; Oraby, H. Nitrogen use efficiency in spring wheat: Genotypic variation and grain yield response under sandy soil conditions. J. Agric. Sci. 2017, 155, 1407–1423. [Google Scholar] [CrossRef]
- Zeng, L.; Shannon, M.; Grieve, C. Evaluation of salt tolerance in rice genotypes by multiple agronomic parameters. Euphytica 2002, 127, 235–245. [Google Scholar] [CrossRef]
- Hammami, Z.; Sbei, H.; Kadri, K.; Jmel, Z.; Sahli, A.; Belhaj Fraj, M.; Naser, H.; Teixeira da Silva, J.; Trifa, Y. Evaluation of performance of different barley genotypes irrigated with saline water in South Tunisian Saharan conditions. Environ. Exp. Bot. 2016, 14, 15–21. [Google Scholar] [CrossRef]
- Mansour, E.; Moustafa, E.S.; El-Naggar, N.Z.; Abdelsalam, A.; Igartua, E. Grain yield stability of high-yielding barley genotypes under Egyptian conditions for enhancing resilience to climate change. Crop Pasture Sci. 2018, 69, 681–690. [Google Scholar] [CrossRef]

| Genotypes | Grain Yield (kg ha−1) | Yield Index (YI) | Cluster Group | ||||||
|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | ||
| Gemiza-11 | 4960 | 3973 | 2992 | 3975 | 1.30 | 1.40 | 1.47 | 1.39 | A |
| Gemiza-9 | 5392 | 3549 | 2359 | 3767 | 1.42 | 1.25 | 1.16 | 1.27 | B |
| Gemiza-10 | 4263 | 2849 | 2460 | 3191 | 1.12 | 1.00 | 1.20 | 1.11 | B |
| Giza-171 | 5209 | 3680 | 2050 | 3646 | 1.37 | 1.29 | 1.00 | 1.22 | B |
| Sids-14 | 4602 | 3339 | 1976 | 3306 | 1.21 | 1.17 | 0.97 | 1.12 | B |
| Line-6084 | 4718 | 3080 | 2525 | 3441 | 1.24 | 1.08 | 1.24 | 1.19 | B |
| Line-6083 | 4210 | 2892 | 1937 | 3013 | 1.11 | 1.02 | 0.95 | 1.02 | B |
| Giza-168 | 3182 | 2442 | 2130 | 2585 | 0.84 | 0.86 | 1.04 | 0.91 | C |
| Gemiza-7 | 3560 | 2768 | 1809 | 2712 | 0.94 | 0.97 | 0.89 | 0.93 | C |
| Sakha-94 | 3185 | 2681 | 2007 | 2624 | 0.84 | 0.94 | 0.98 | 0.92 | C |
| Sids-12 | 3613 | 2854 | 2193 | 2887 | 0.95 | 1.00 | 1.07 | 1.01 | C |
| Misr-1 | 3254 | 2840 | 2075 | 2723 | 0.86 | 1.00 | 1.02 | 0.96 | C |
| Line-6052 | 3185 | 2677 | 1884 | 2582 | 0.84 | 0.94 | 0.92 | 0.90 | C |
| Line-6078 | 3360 | 2551 | 1947 | 2619 | 0.88 | 0.90 | 0.95 | 0.91 | C |
| Line-1208 | 3669 | 2690 | 2041 | 2800 | 0.96 | 0.95 | 1.00 | 0.97 | C |
| Gemiza-12 | 2786 | 2225 | 1668 | 2226 | 0.73 | 0.78 | 0.82 | 0.78 | D |
| Shandawel-1 | 2680 | 2097 | 1246 | 2008 | 0.70 | 0.74 | 0.61 | 0.68 | D |
| Misr-2 | 2659 | 1983 | 1462 | 2034 | 0.70 | 0.70 | 0.72 | 0.70 | D |
| Genotypes | Total Chlorophyll (mg g−1 Fresh Weight) | Pn (µmol CO2 m−2 s−1) | E (mmol H2O m−2 s−1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 2.96 | 2.22 | 1.66 | 2.28 a† | 12.72 | 10.14 | 6.89 | 9.92 a | 7.05 | 4.73 | 3.55 | 5.11 a |
| Group B n = 6 | 2.92 | 2.09 | 1.39 | 2.13 b | 12.50 | 9.91 | 6.42 | 9.61 b | 7.01 | 4.52 | 3.22 | 4.92 b |
| Group C n = 8 | 2.67 | 1.90 | 1.30 | 1.95 c | 11.24 | 9.18 | 6.07 | 8.83 c | 6.16 | 4.17 | 3.05 | 4.46 c |
| Group D n = 3 | 2.50 | 1.81 | 1.15 | 1.82 d | 10.60 | 8.39 | 5.24 | 8.07 d | 5.37 | 3.72 | 2.80 | 3.96 d |
| Mean | 2.76 A | 2.00 B | 1.37 C | 11.77 A | 9.40 B | 6.16 C | 6.40 A | 4.29 B | 3.15 C | |||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | |||||
| Salinity (S) | 2 | < 0.001 | 0.011 | < 0.001 | 0.07 | < 0.001 | 0.15 | |||||
| Group (G) | 3 | < 0.001 | 0.029 | < 0.001 | 0.11 | < 0.001 | 0.16 | |||||
| S × G | 6 | < 0.001 | 0.047 | < 0.001 | 0.20 | 0.003 | 0.29 | |||||
| Genotypes | Gs (mmol H2O m−2 s−1) | RWC (%) | MSI (%) | |||||||||
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 0.59 | 0.40 | 0.30 | 0.43 a | 65.81 | 51.56 | 42.04 | 53.13 a | 60.78 | 49.01 | 36.22 | 48.67 a |
| Group B n = 6 | 0.56 | 0.39 | 0.27 | 0.41 b | 63.15 | 50.44 | 38.71 | 50.77 b | 59.96 | 48.07 | 33.90 | 47.31 b |
| Group C n = 8 | 0.49 | 0.36 | 0.26 | 0.37 c | 56.99 | 47.89 | 39.48 | 48.12 c | 55.45 | 44.35 | 32.60 | 44.13 c |
| Group D n = 3 | 0.44 | 0.33 | 0.21 | 0.33 d | 53.93 | 46.39 | 36.36 | 45.56 d | 51.18 | 40.71 | 26.90 | 39.60 d |
| Mean | 0.52 A | 0.37 B | 0.26 C | 59.97 A | 49.07 B | 39.14 C | 56.84 A | 45.54 B | 32.40 C | |||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | |||||
| Salinity (S) | 2 | < 0.001 | 0.003 | < 0.001 | 1.71 | < 0.001 | 0.16 | |||||
| Group (G) | 3 | < 0.001 | 0.006 | < 0.001 | 1.42 | < 0.001 | 0.32 | |||||
| S × G | 6 | < 0.001 | 0.010 | 0.002 | 2.77 | < 0.001 | 0.55 | |||||
| Genotypes | MDA (nmol g−1 Fresh Weight) | EL (%) | Soluble Sugars (mg g−1 Dry Weight) | Free Proline (µg g−1 Dry Weight) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 49.33 | 60.18 | 69.00 | 59.50 d† | 2.80 | 6.60 | 10.46 | 6.62 d | 25.95 | 40.53 | 56.80 | 41.09 a | 127.7 | 192.5 | 242.8 | 187.7 a |
| Group B n = 6 | 50.07 | 61.55 | 72.64 | 61.42 c | 3.05 | 6.93 | 12.51 | 7.49 c | 25.89 | 39.57 | 53.26 | 39.57 b | 123.5 | 183.7 | 230.5 | 179.2 b |
| Group C n = 8 | 53.65 | 64.26 | 74.94 | 64.28 b | 4.20 | 7.63 | 13.12 | 8.32 b | 24.11 | 35.23 | 50.03 | 36.46 c | 98.6 | 164.1 | 223.7 | 162.2 c |
| Group D n = 3 | 57.61 | 67.92 | 82.56 | 69.36 a | 5.36 | 8.69 | 15.07 | 9.71 a | 21.79 | 30.62 | 44.66 | 32.36 d | 84.7 | 154.7 | 207.5 | 148.9 d |
| Mean | 52.67 C | 63.48 B | 74.78 A | 3.85 C | 7.46 B | 12.79 A | 24.43 C | 36.48 B | 51.19 A | 108.6 C | 173.8 B | 226.1 A | ||||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | p-value | LSD | |||||||
| Salinity (S) | 2 | < 0.001 | 0.57 | < 0.001 | 0.20 | < 0.001 | 1.44 | < 0.001 | 1.64 | |||||||
| Group (G) | 3 | < 0.001 | 0.62 | < 0.001 | 0.28 | < 0.001 | 1.29 | < 0.001 | 1.82 | |||||||
| S × G | 6 | < 0.001 | 1.04 | < 0.001 | 0.49 | < 0.001 | 2.93 | < 0.001 | 2.31 | |||||||
| Genotypes | AsA (µmol g−1 dry weight) | SOD (U µg−1 protein) | CAT (U mg−1 min−1) | POD (µg g−1 fresh weight min−1) | ||||||||||||
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 1.74 | 2.53 | 3.03 | 2.43 a | 52.41 | 69.93 | 87.46 | 69.93 a | 0.50 | 0.70 | 0.96 | 0.72 a | 90.33 | 131.57 | 177.93 | 133.28 a |
| Group B n = 6 | 1.72 | 2.46 | 2.86 | 2.35 b | 51.92 | 67.42 | 82.62 | 67.32 b | 0.50 | 0.65 | 0.85 | 0.67 b | 88.94 | 126.67 | 167.34 | 127.65 b |
| Group C n = 8 | 1.61 | 2.25 | 2.77 | 2.21 c | 47.43 | 63.49 | 79.78 | 63.57 c | 0.46 | 0.60 | 0.81 | 0.62 c | 82.22 | 113.58 | 161.78 | 119.19 c |
| Group D n = 3 | 1.53 | 2.08 | 2.56 | 2.06 d | 42.27 | 58.10 | 73.71 | 58.03 d | 0.42 | 0.57 | 0.73 | 0.57 d | 77.53 | 102.46 | 143.91 | 107.97 d |
| Mean | 1.65 C | 2.33 B | 2.80 A | 48.51 C | 64.74 B | 80.89 A | 0.47 C | 0.63 B | 0.84 A | 84.76 C | 118.57 B | 162.74 A | ||||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | p-value | LSD | |||||||
| Salinity (S) | 2 | < 0.001 | 0.009 | < 0.001 | 0.27 | < 0.001 | 0.009 | < 0.001 | 0.94 | |||||||
| Group (G) | 3 | < 0.001 | 0.013 | < 0.001 | 0.56 | < 0.001 | 0.012 | < 0.001 | 1.41 | |||||||
| S × G | 6 | < 0.001 | 0.023 | < 0.001 | 0.95 | < 0.001 | 0.015 | < 0.001 | 2.45 | |||||||
| Genotypes | Cl− (%) | Na+ (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 1.55 | 2.05 | 3.30 | 2.30 c† | 4.35 | 8.21 | 11.15 | 7.90 d |
| Group B n = 6 | 1.57 | 2.21 | 3.18 | 2.32 c | 4.56 | 8.77 | 12.18 | 8.50 c |
| Group C n = 8 | 1.75 | 2.61 | 3.51 | 2.62 b | 5.84 | 9.76 | 12.87 | 9.49 b |
| Group D n = 3 | 1.87 | 2.87 | 3.38 | 2.71 a | 6.78 | 10.24 | 15.24 | 10.75 a |
| Mean | 1.69 C | 2.43 B | 3.34 A | 5.38 C | 9.24 B | 12.86 A | ||
| ANOVA | df | p-value | LSD | p-value | LSD | |||
| Salinity (S) | 2 | < 0.001 | 0.02 | < 0.001 | 0.15 | |||
| Group (G) | 3 | < 0.001 | 0.05 | < 0.001 | 0.14 | |||
| S × G | 6 | < 0.001 | 0.10 | < 0.001 | 0.27 | |||
| Genotypes | K+ (%) | K+/Na+ ratio | ||||||
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 19.97 | 16.50 | 11.32 | 15.93 a | 4.59 | 2.01 | 1.01 | 2.54 a |
| Group B n = 6 | 19.77 | 15.09 | 10.00 | 14.95 b | 4.37 | 1.73 | 0.83 | 2.31 b |
| Group C n = 8 | 18.30 | 13.41 | 9.41 | 13.71 c | 3.15 | 1.38 | 0.74 | 1.75 c |
| Group D n = 3 | 17.27 | 12.36 | 8.29 | 12.64 d | 2.55 | 1.21 | 0.54 | 1.43 d |
| Mean | 18.83 A | 14.34 B | 9.75 C | 3.66 A | 1.58 B | 0.78 C | ||
| ANOVA | df | p-value | LSD | p-value | LSD | |||
| Salinity (S) | 2 | < 0.001 | 0.04 | < 0.001 | 0.14 | |||
| Group (G) | 3 | < 0.001 | 0.17 | < 0.001 | 0.07 | |||
| S × G | 6 | < 0.001 | 0.28 | < 0.001 | 0.17 | |||
| Genotypes | Plant Height (cm) | Spike Length (cm) | Number of Spikes m−2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 92.50 | 88.67 | 74.00 | 85.06 a† | 12.31 | 10.85 | 9.89 | 11.02 a | 182.9 | 161.9 | 151.0 | 162.2 a |
| Group B n = 6 | 87.54 | 76.36 | 67.91 | 77.27 b | 11.87 | 10.72 | 9.68 | 10.76 b | 173.6 | 154.3 | 133.3 | 156.9 b |
| Group C n = 8 | 83.40 | 77.02 | 70.23 | 76.88 bc | 11.70 | 10.63 | 9.60 | 10.64 b | 144.9 | 130.0 | 117.8 | 130.9 c |
| Group D n = 3 | 81.89 | 76.72 | 69.61 | 76.07 c | 11.13 | 10.28 | 9.13 | 10.18 c | 126.3 | 110.1 | 98.6 | 111.7 d |
| Mean | 86.33 A | 79.69 B | 70.44 C | 11.75 A | 10.62 B | 9.58 C | 156.9 A | 139.1 B | 125.2 C | |||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | |||||
| Salinity (S) | 2 | < 0.001 | 0.79 | < 0.001 | 0.13 | < 0.001 | 2.25 | |||||
| Group (G) | 3 | < 0.001 | 0.91 | < 0.001 | 0.17 | < 0.001 | 3.64 | |||||
| S × G | 6 | < 0.001 | 2.03 | < 0.001 | 0.29 | < 0.001 | 6.80 | |||||
| Genotypes | Number of grains per spike | 1000-grain weight (g) | Grain yield (kg ha-1) | |||||||||
| S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | S1 | S2 | S3 | Mean | |
| Group A n = 1 | 59.17 | 54.00 | 44.17 | 52.44 a | 55.59 | 49.55 | 41.10 | 48.75 a | 4960.0 | 3973.0 | 2992.0 | 3975.0 a |
| Group B n = 6 | 55.31 | 47.48 | 41.32 | 48.03 b | 53.16 | 47.55 | 40.78 | 47.16 b | 4732.3 | 3231.5 | 2217.8 | 3393.9 b |
| Group C n = 8 | 51.60 | 44.65 | 40.04 | 45.43 c | 50.79 | 46.03 | 40.65 | 45.82 c | 3376.0 | 2687.9 | 2010.8 | 2691.5 c |
| Group D n = 3 | 51.00 | 44.06 | 38.39 | 44.48 d | 48.66 | 44.01 | 39.00 | 43.89 d | 2708.3 | 2101.7 | 1458.7 | 2089.6 d |
| Mean | 54.27 A | 47.54 B | 40.98 C | 52.05 A | 46.78 B | 40.38 C | 3944 A | 2999 B | 2170 C | |||
| ANOVA | df | p-value | LSD | p-value | LSD | p-value | LSD | |||||
| Salinity (S) | 2 | < 0.001 | 0.52 | < 0.001 | 0.40 | < 0.001 | 74.58 | |||||
| Group (G) | 3 | < 0.001 | 0.77 | < 0.001 | 0.62 | < 0.001 | 124.3 | |||||
| S × G | 6 | 0.003 | 1.33 | < 0.001 | 1.03 | < 0.001 | 203.4 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mansour, E.; Moustafa, E.S.A.; Desoky, E.-S.M.; Ali, M.M.A.; Yasin, M.A.T.; Attia, A.; Alsuhaibani, N.; Tahir, M.U.; El-Hendawy, S. Multidimensional Evaluation for Detecting Salt Tolerance of Bread Wheat Genotypes Under Actual Saline Field Growing Conditions. Plants 2020, 9, 1324. https://doi.org/10.3390/plants9101324
Mansour E, Moustafa ESA, Desoky E-SM, Ali MMA, Yasin MAT, Attia A, Alsuhaibani N, Tahir MU, El-Hendawy S. Multidimensional Evaluation for Detecting Salt Tolerance of Bread Wheat Genotypes Under Actual Saline Field Growing Conditions. Plants. 2020; 9(10):1324. https://doi.org/10.3390/plants9101324
Chicago/Turabian StyleMansour, Elsayed, Ehab S. A. Moustafa, El-Sayed M. Desoky, Mohamed M. A. Ali, Mohamed A. T. Yasin, Ahmed Attia, Nasser Alsuhaibani, Muhammad Usman Tahir, and Salah El-Hendawy. 2020. "Multidimensional Evaluation for Detecting Salt Tolerance of Bread Wheat Genotypes Under Actual Saline Field Growing Conditions" Plants 9, no. 10: 1324. https://doi.org/10.3390/plants9101324
APA StyleMansour, E., Moustafa, E. S. A., Desoky, E.-S. M., Ali, M. M. A., Yasin, M. A. T., Attia, A., Alsuhaibani, N., Tahir, M. U., & El-Hendawy, S. (2020). Multidimensional Evaluation for Detecting Salt Tolerance of Bread Wheat Genotypes Under Actual Saline Field Growing Conditions. Plants, 9(10), 1324. https://doi.org/10.3390/plants9101324









