A Dual Strategy of Breeding for Drought Tolerance and Introducing Drought-Tolerant, Underutilized Crops into Production Systems to Enhance Their Resilience to Water Deficiency
Abstract
1. Introduction
2. General Overview of Physiological Responses of Plants to Drought Stress Conditions
3. Use of Crop Diversity in Plant Breeding for Drought-Tolerance Traits
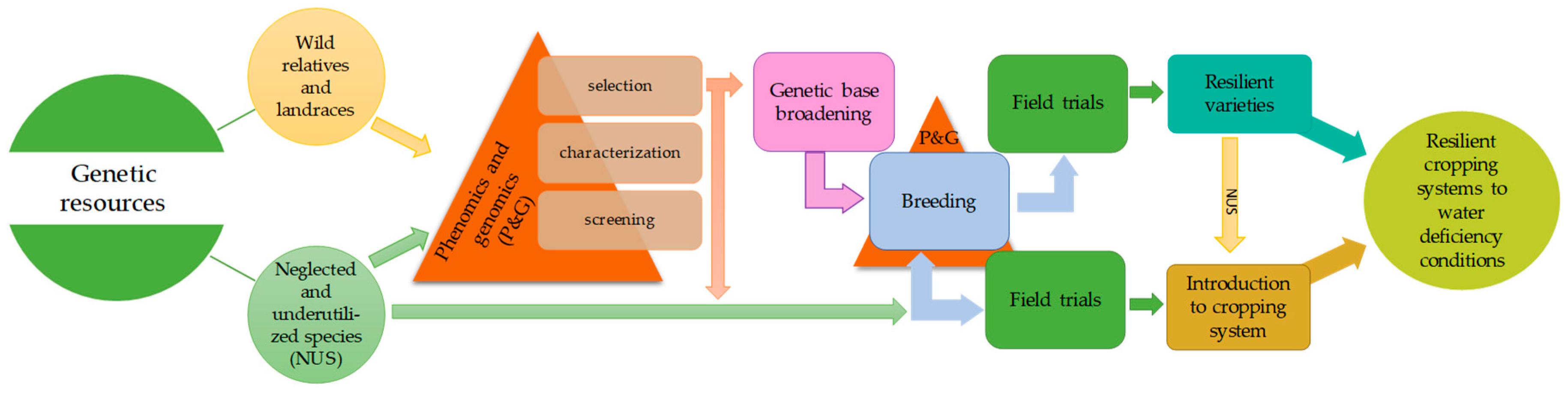
4. Introduction of Neglected and Underutilized Species into Cropping Systems
5. Methods and Approaches to Improve Crop Tolerance to Drought Stress
5.1. Phenotyping Methods for Drought-Tolerance Trait Evaluations
5.2. Potential of Genomic Approaches to Improve Crop Tolerances to Drought Stress
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Simelton, E.S.; Fraser, E.D.G.; Termansen, M.; Benton, T.G.; Gosling, S.N.; South, A.; Arnell, N.W.; Challinor, A.J.; Dougill, A.J.; Forster, P.M.D.F. The socioeconomics of food crop production and climate change vulnerability: A global scale quantitative analysis of how grain crops are sensitive to drought. Food Secur. 2012, 4, 163–179. [Google Scholar] [CrossRef]
- Lipiec, J.; Doussan, C.; Nosalewicz, A.; Kondracka, K. Effect of drought and heat stresses on plant growth and yield: A review. Int. Agrophysics 2013, 27, 463–477. [Google Scholar] [CrossRef]
- Turner, N. Drought resistance and adaptation to water deficits in crop plants. In Stress Physiology in Crop Plants; Mussell, H., Staples, R.C., Eds.; John Wiley & Sons: New York, NY, USA, 1979; pp. 343–372. ISBN 978-0-4710-3809-2. [Google Scholar]
- Passioura, J.B. Drought and drought tolerance. Plant Growth Regul. 1996, 20, 79–83. [Google Scholar] [CrossRef]
- Castañeda-Álvarez, N.P.; Khoury, C.K.; Achicanoy, H.A.; Bernau, V.; Dempewolf, H.; Eastwood, R.J.; Guarino, L.; Harker, R.H.; Jarvis, A.; Maxted, N.; et al. Global conservation priorities for crop wild relatives. Nat. Plants 2016, 2, 16022. [Google Scholar] [CrossRef]
- Vincent, H.; Amri, A.; Castañeda-Álvarez, N.P.; Dempewolf, H.; Dulloo, E.; Guarino, L.; Hole, D.G.; Mba, C.; Toledo, A.; Maxted, N. Modeling of crop wild relative species identifies areas globally for in situ conservation. Commun. Biol. 2019, 2, 136. [Google Scholar] [CrossRef] [PubMed]
- Villa, T.C.C.; Maxted, N.; Scholten, M.; Ford-Lloyd, B. Defining and identifying crop landraces. Plant Genet. Resour. 2005, 3, 373–384. [Google Scholar] [CrossRef]
- Xu, Y.; Skinner, D.J.; Wu, H.; Palacios-Rojas, N.; Araus, J.L.; Yan, J.; Gao, S.; Warburton, M.L.; Crouch, J.H. Advances in maize genomics and their value for enhancing genetic gains from breeding. Int. J. Plant Genom. 2009, 2009, 1–30. [Google Scholar] [CrossRef]
- Chivenge, P.; Mabhaudhi, T.; Modi, A.T.; Mafongoya, P. The potential role of neglected and underutilised crop species as future crops under water scarce conditions in sub-Saharan Africa. Int. J. Environ. Res. Public Health 2015, 12, 5685–5711. [Google Scholar] [CrossRef]
- Mabhaudhi, T.; Chimonyo, V.; Modi, A.T. Status of underutilised crops in South Africa: Opportunities for developing research capacity. Sustainibility 2017, 9, 1569. [Google Scholar] [CrossRef]
- Modi, A.T.; Mabhaudhi, T. Water use and drought tolerance of selected traditional and indigenous crops. In Final Report of Water Research Commission Project K5/1771//4; Water Research Commission: Pretoria, South Africa, 2013; ISBN 978-1-4312-0434-2. [Google Scholar]
- Padulosi, S.; Thompson, J.; Rudebjer, P. Fighting Poverty, Hunger and Malnutrition with Neglected and Underutilized Species (NUS): Needs, Challenges and the Way Forward: Neglected and Underutilized Species; Bioversity International: Rome, Italy, 2013; ISBN 978-92-9043-941-7. [Google Scholar]
- Bazile, D.; Pulvento, C.; Verniau, A.; Al-Nusairi, M.S.; Ba, D.; Breidy, J.; Hassan, L.; Mohammed, M.I.; Mambetov, O.; Otambekova, M.; et al. Worldwide evaluations of quinoa: Preliminary results from post international year of quinoa FAO projects in nine countries. Front. Plant Sci. 2016, 7, 850. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, M.A.; Mayes, S.; Massawe, F. Crop diversification through a wider use of underutilised crops: A strategy to ensure food and nutrition security in the face of climate change. In Sustainable Solutions for Food Security; Springer Science and Business Media LLC.: Berlin, Germany, 2019; pp. 125–149. [Google Scholar]
- Harb, A.; Krishnan, A.; Ambavaram, M.M.; Pereira, A. Molecular and physiological analysis of drought stress in arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 2010, 154, 1254–1271. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Bose, J.; Fuglsang, A.T.; Pottosin, I. On a quest for stress tolerance genes: Membrane transporters in sensing and adapting to hostile soils. J. Exp. Bot. 2015, 67, 1015–1031. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.-S.P. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Zhang, B.; Qin, F. Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 2015, 27, 3228–3244. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Noguchi, K.; Ono, N.; Inoue, S.-I.; Terashima, I.; Kinoshita, T. Overexpression of plasma membrane H+-ATPase in guard cells promotes light-induced stomatal opening and enhances plant growth. Proc. Natl. Acad. Sci. USA 2013, 111, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Daszkowska-Golec, A. The role of abscisic acid in drought stress: How ABA helps plants to cope with drought stress. In Drought Stress Tolerance in Plants; Springer Science and Business Media LLC: Berlin, Germany, 2016; Volume 2, pp. 123–151. [Google Scholar]
- Yang, J.; Zhang, G.; An, J.; Li, Q.; Chen, Y.; Zhao, X.; Wu, J.; Wang, Y.; Hao, Q.; Wang, W.; et al. Expansin gene TaEXPA2 positively regulates drought tolerance in transgenic wheat (Triticum aestivum L.). Plant Sci. 2020, 298, 110596. [Google Scholar] [CrossRef]
- Fang, Q.; Wang, X.; Wang, H.; Tang, X.; Liu, C.; Yin, H.; Ye, S.; Jiang, Y.; Duan, Y.; Luo, K. The poplar R2R3 MYB transcription factor PtrMYB94 coordinates with abscisic acid signaling to improve drought tolerance in plants. Tree Physiol. 2019, 40, 46–59. [Google Scholar] [CrossRef]
- Zhong, M.-S.; Jiang, H.; Cao, Y.; Wang, Y.-X.; You, C.-X.; Li, Y.-Y.; Hao, Y.-J. MdCER2 conferred to wax accumulation and increased drought tolerance in plants. Plant Physiol. Biochem. 2020, 149, 277–285. [Google Scholar] [CrossRef]
- Virlouvet, L.; Fromm, M. Physiological and transcriptional memory in guard cells during repetitive dehydration stress. New Phytol. 2014, 205, 596–607. [Google Scholar] [CrossRef]
- Grondin, A.; Rodrigues, O.; Verdoucq, L.; Merlot, S.; Leonhardt, N.; Maurel, C. Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell 2015, 27, 1945–1954. [Google Scholar] [CrossRef]
- Yang, L.; Fountain, J.C.; Wang, H.; Ni, X.; Ji, P.; Lee, R.D.; Kemerait, R.C.; Scully, B.T.; Guo, B. Stress sensitivity is associated with differential accumulation of reactive oxygen and nitrogen species in maize genotypes with contrasting levels of drought tolerance. Int. J. Mol. Sci. 2015, 16, 24791–24819. [Google Scholar] [CrossRef]
- Laxa, M.; Liebthal, M.; Telman, W.; Chibani, K.; Dietz, K.-J. The role of the plant antioxidant system in drought tolerance. Antioxidants 2019, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Smékalová, V.; Doskočilová, A.; Komis, G.; Šamaj, J. Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnol. Adv. 2014, 32, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones enhanced drought tolerance in plants: A coping strategy. Environ. Sci. Pollut. Res. 2018, 25, 33103–33118. [Google Scholar] [CrossRef]
- Liao, W.-B.; Huang, G.-B.; Yu, J.; Zhang, M. Nitric oxide and hydrogen peroxide alleviate drought stress in marigold explants and promote its adventitious root development. Plant Physiol. Biochem. 2012, 58, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Chai, C.; Qian, Q.; Li, C.; Tang, J.; Sun, L.; Huang, Z.; Guo, X.; Sun, C.; Liu, M.; et al. Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 2008, 54, 177–189. [Google Scholar] [CrossRef]
- Ford-Lloyd, B.V.; Schmidt, M.; Armstrong, S.J.; Barazani, O.; Engels, J.; Hadas, R.; Hammer, K.; Kell, S.P.; Kang, D.; Khoshbakht, K.; et al. Crop Wild Relatives—Undervalued, underutilized and under threat? Bioscience 2011, 61, 559–565. [Google Scholar] [CrossRef]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L. Past and future use of wild relatives in crop breeding. Crop. Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- Mammadov, J.; Buyyarapu, R.; Guttikonda, S.K.; Parliament, K.; Abdurakhmonov, I.Y.; Kumpatla, S.P. Wild relatives of maize, rice, cotton, and soybean: Treasure troves for tolerance to biotic and abiotic stresses. Front. Plant Sci. 2018, 9, 886. [Google Scholar] [CrossRef]
- Ludwig, F.; Rosenthal, D.M.; Johnston, J.A.; Kane, N.C.; Gross, B.L.; Lexer, C.; Dudley, S.A.; Rieseberg, L.H.; Donovan, L.A. Selection on leaf ecophysiological traits in a desert hybrid helianthus species and early-generation hybrids. Evolution 2004, 58, 2682–2692. [Google Scholar] [CrossRef]
- Ndjiondjop, M.N.; Manneh, B.; Cissoko, M.; Dramé, N.; Kakaï, R.G.; Bocco, R.; Baimey, H.; Wopereis, M. Drought resistance in an interspecific backcross population of rice (Oryza spp.) derived from the cross WAB56-104 (O. sativa)×CG14 (O. glaberrima). Plant Sci. 2010, 179, 364–373. [Google Scholar] [CrossRef]
- Budak, H.; Kantar, M.; Kurtoglu, K.Y. Drought tolerance in modern and wild wheat. Sci. World J. 2013, 2013, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Alpha, Y.; Kureh, I.; Menkir, A.; Kartung, P.; Tarfa, B.; Amaza, P. Participatory on-farm evaluation of the performance of drought-tolerant maize varieties in the Guinea savannas of Nigeria. Int. J. Food Agric. Environ. 2006, 4, 192–196. [Google Scholar]
- Xu, J.; Yuan, Y.; Xu, Y.; Zhang, G.; Guo, X.-S.; Wu, F.; Wang, Q.; Rong, T.; Pan, G.; Cao, M.; et al. Identification of candidate genes for drought tolerance by whole-genome resequencing in maize. BMC Plant Biol. 2014, 14, 83. [Google Scholar] [CrossRef] [PubMed]
- Al Sharari, S.; Alsadon, A.; Alharbi, A. Evaluation of drought tolerance of potato cultivars under greenhouse conditions. Acta Hortic. 2007, 747, 67–74. [Google Scholar] [CrossRef]
- International Potato Center (CIP). Annual Report CIP 1993–1994; CIP: Lima, Peru, 1994; p. 192. ISSN 0256-6311. [Google Scholar]
- Monneveux, P.; Ramirez, D.A.; Pino, M.-T. Drought tolerance in potato (S. tuberosum L.). Plant Sci. 2013, 205, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Rolando, J.; Ramirez, D.A.; Yactayo, W.; Monneveux, P.; Quiroz, R. Leaf greenness as a drought tolerance related trait in potato (Solanum tuberosum L.). Environ. Exp. Bot. 2015, 110, 27–35. [Google Scholar] [CrossRef]
- Sharma, N.; Rawal, S.; Kadian, M.; Arya, S.; Bonierbale, M.; Singh, B. Evaluation of advanced potato clones for drought tolerance in arid zone in Rajasthan, India. Potato J. 2014, 41, 189–193. [Google Scholar]
- Reyes, L. Making rice less thirsty. Rice Today 2009, 8, 12–15. [Google Scholar]
- Singh, A.; Singh, A.K.; Singh, V.; Singh, N.; Singh, V.N.; Shamim, M.; Vikram, P.; Singh, S. Genetic variability among traits associated with grain yield of rice (Oryza sativa L.) exposed to drought at flowering stage. Afr. J. Agric. Res. 2014, 9, 1252–1264. [Google Scholar]
- Zheng, X.-G.; Chen, L.; Lou, Q.-J.; Xia, H.; Li, M.-S.; Luo, L. Changes in DNA methylation pattern at two seedling stages in water saving and drought-resistant rice variety after drought stress domestication. Rice Sci. 2014, 21, 262–270. [Google Scholar] [CrossRef]
- Liu, L.; Lafitte, R.; Guan, D. Wild Oryza species as potential sources of drought-adaptive traits. Euphytica 2004, 138, 149–161. [Google Scholar] [CrossRef]
- Thanh, P.T.; Sripichitt, P.; Chanprame, S.; Peyachoknagu, S. Transfer of drought resistant character from wild rice (Oryza meridionalis and Oryza nivara) to cultivated rice (Oryza sativa L.) by backcrossing and immature embryo culture. Kasetsar J. 2006, 40, 582–594. [Google Scholar]
- Cia, M.; Guimaraes, A.; Medici, L.O.; Chabregas, S.; Azevedo, R. Antioxidant responses to water deficit by drought-tolerant and -sensitive sugarcane varieties. Ann. Appl. Biol. 2012, 161, 313–324. [Google Scholar] [CrossRef]
- Da Costa, M.L.M.; Amorim, L.L.B.; Onofre, A.V.C.; De Melo, L.J.O.T.; De Oliveira, M.B.M.; De Carvalho, R.; Benko-Iseppon, A.M. Assessment of genetic diversity in contrasting sugarcane varieties using inter-simple sequence repeat (ISSR) markers. Am. J. Plant Sci. 2011, 2, 425–432. [Google Scholar] [CrossRef]
- Ishaq, M.N.; Olaoye, G.; Akinsanya, T.O. Screening sugar-cane germplasm for drought tolerance in Nigeria. Plant Genetic Resources Newsletter 2008, 154, 48–54. [Google Scholar]
- Srivastava, M.K.; Li, C.N.; Li, Y.R. Development of sequence characterized amplified region (SCAR) marker for identifying drought tolerant sugarcane genotypes. Aust. J. Crop. Sci. 2012, 6, 763–767. [Google Scholar]
- Mohanan, K. Breeding for special purposes. In Essentials of Plant Breeding; Mohanan, K.V., Ed.; Prentice-Hall of India Pvt Limited: New Delhi, India, 2010; p. 107. ISBN 978-812-033-968-2. [Google Scholar]
- Finkel, E. Richard Richards profile: Making every drop count in the buildup to a blue revolution. Science 2009, 323, 1004–1005. [Google Scholar] [CrossRef]
- Sohail, Q.; Inoue, T.; Tanaka, H.; Eltayeb, A.E.; Matsuoka, Y.; Tsujimoto, H. Applicability of Aegilops tauschii drought tolerance traits to breeding of hexaploid wheat. Breed. Sci. 2011, 61, 347–357. [Google Scholar] [CrossRef]
- Turyagyenda, L.F.; Kizito, E.B.; Ferguson, M.; Baguma, Y.; Agaba, M.; Harvey, J.; Osiru, D.S.O. Physiological and molecular characterization of drought responses and identification of candidate tolerance genes in cassava. AoB Plants 2013, 5, plt007. [Google Scholar] [CrossRef]
- Nassar, N.; Abreu, L.; Teodoro, D.; Graciano-Ribeiro, D. Drought tolerant stem anatomy characteristics in Manihot esculenta (Euphorbiaceae) and a wild relative. Genet. Mol. Res. 2010, 9, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Narina, S.; Jasti, M.; Buyyarapu, R.; Bhattacharjee, R. Manihot. In Wild Crop Relatives Genomic and Breeding Resources Industrial Crops; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 133–156. ISBN 978-3-642-21102-7. [Google Scholar]
- Chavez, R.; Reyes, R.; Roca, W. In vitro culture for the conservation of wild Manihot species. In Review of Advances in Plant Biotechnology 1985–1988; Mujeeb-Kazi, A., Sitch, L., Eds.; CYMMYT and IRRI: Ciudad de México, Mexico, 1989; pp. 301–307. ISBN 978-968-612-734-8. [Google Scholar]
- Huang, L.; Zhang, F.; Wang, W.; Zhou, Y.; Fu, B.; Li, Z. Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC Genom. 2014, 15, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Khoury, C.K.; Bjorkman, A.; Dempewolf, H.; Ramirez-Villegas, J.; Guarino, L.; Jarvis, A.; Rieseberg, L.H.; Struik, P.C. Increasing homogeneity in global food supplies and the implications for food security. Proc. Natl. Acad. Sci. USA 2014, 111, 4001–4006. [Google Scholar] [CrossRef] [PubMed]
- Mayes, S.; Massawe, F.; Alderson, P.G.; Roberts, J.A.; Azam-Ali, S.N.; Hermann, M. The potential for underutilized crops to improve security of food production. J. Exp. Bot. 2011, 63, 1075–1079. [Google Scholar] [CrossRef] [PubMed]
- Boote, K.J.; Ibrahim, A.M.H.; Lafitte, R.; McCulley, R.; Messina, C.D.; Murray, S.C.; Specht, J.E.; Taylor, S.; Westgate, M.E.; Glasener, K.; et al. Position statement on crop adaptation to climate change. Crop. Sci. 2011, 51, 2337–2343. [Google Scholar] [CrossRef]
- Bose, J.; Rodrigo-Moreno, A.; Lai, D.; Xie, Y.; Shen, W.; Shabala, S. Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Ann. Bot. 2014, 115, 481–494. [Google Scholar] [CrossRef]
- Yang, A.; Akhtar, S.S.; Amjad, M.; Iqbal, S.; Jacobsen, S.-E. Growth and physiological responses of quinoa to drought and temperature stress. J. Agron. Crop. Sci. 2016, 202, 445–453. [Google Scholar] [CrossRef]
- Shao, H.; Yong, B.; Xu, P.; Zheng, H.; Liao, R.; Wang, X.; Li, X.; Zhang, L.; Shen, J. Phytoene synthase gene (PSY) from sweet potato (Ipomoea batatas Lam) enhances tolerance to abiotic stress. Braz. Arch. Biol. Technol. 2018, 61, e18160558. [Google Scholar] [CrossRef]
- Park, S.; Kim, H.S.; Jung, Y.J.; Kim, S.H.; Ji, C.Y.; Wang, Z.; Jeong, J.C.; Lee, H.-S.; Lee, S.Y.; Kwak, S.-S. Orange protein has a role in phytoene synthase stabilization in sweetpotato. Sci. Rep. 2016, 6, 33563. [Google Scholar] [CrossRef]
- Fang, Z.; Xu, X.; Gao, J.; Wang, P.; Liu, Z.; Feng, B. Characterization of FeDREB1 promoter involved in cold- and drought-inducible expression from common buckwheat (Fagopyrum esculentum). Genet. Mol. Res. 2015, 14, 7990–8000. [Google Scholar] [CrossRef]
- Wu, Q.; Zhao, G.; Bai, X.; Zhao, W.; Xiang, D.; Wan, Y.; Wu, X.; Sun, Y.; Tan, M.; Peng, L.; et al. Characterization of the transcriptional profiles in common buckwheat (Fagopyrum esculentum) under PEG-mediated drought stress. Electron. J. Biotechnol. 2019, 39, 42–51. [Google Scholar] [CrossRef]
- Khoury, C.K.; Achicanoy, H.A.; Bjorkman, A.; Navarro-Racines, C.; Guarino, L.; Flores-Palacios, X.; Engels, J.M.M.; Wiersema, J.H.; Dempewolf, H.; Sotelo, S.; et al. Origins of food crops connect countries worldwide. Proc. R. Soc. B: Biol. Sci. 2016, 283, 20160792. [Google Scholar] [CrossRef]
- Risi, C.; Galwey, N.W. The Chenopodium grains of the Andes: Inca crops for modern agriculture. Adv. Appl. Biol. 1984, 10, 145–216. [Google Scholar]
- Jacobsen, S.-E. The scope for adaptation of quinoa in Northern Latitudes of Europe. J. Agron. Crop. Sci. 2017, 203, 603–613. [Google Scholar] [CrossRef]
- International Union for the Protection of New Varieties of Plants (UPOV). 2020 PLUTO: Plant Variety Database. Available online: https://www.upov.int/pluto/en/ (accessed on 2 September 2020).
- Di Fabio, A.; Parraga, G. Origin, Production and Utilization of Pseudocereals; Wiley: Hoboken, NJ, USA, 2017; pp. 1–27. [Google Scholar]
- Oumar, I.; Mariac, C.; Pham, J.-L.; Vigouroux, Y. Phylogeny and origin of pearl millet (Pennisetum glaucum [L.] R. Br) as revealed by microsatellite loci. Theor. Appl. Genet. 2008, 117, 489–497. [Google Scholar] [CrossRef]
- Zhou, M.; Tang, Y.; Deng, X.; Ruan, C.; Kreft, I.; Tang, Y.; Wu, Y. Overview of buckwheat resources in the world. In Buckwheat Germplasm in the World; Zhou, M., Kreft, I., Suvorova, G., Tang, Y., Wu, Y., Eds.; Academic Press: London, UK, 2018; pp. 1–7. ISBN 978-0-12-811006-5. [Google Scholar]
- Padulosi, S.; Ng, N.Q. Origin, taxonomy, and morphology of Vigna unguiculata (L.) Walp. In Advances in Cowpea Research; Singh, B.B., Mohan Raj, D.R., Dashiell, K.E., Jackai, L.E., Eds.; Sayce Publishing: Devon, UK, 1997; pp. 1–12. ISBN 978-131-110X. [Google Scholar]
- Muñoz-Rodríguez, P.; Carruthers, T.; Wood, J.R.; Williams, B.R.; Weitemier, K.; Kronmiller, B.; Ellis, D.; Anglin, N.L.; Longway, L.; Harris, S.A.; et al. Reconciling conflicting phylogenies in the origin of sweet potato and dispersal to polynesia. Curr. Biol. 2018, 28, 1246–1256.e12. [Google Scholar] [CrossRef] [PubMed]
- Atchison, G.W.; Nevado, B.; Eastwood, R.J.; Contreras-Ortiz, N.; Reynel, C.; Madriñán, S.; Filatov, D.A.; Hughes, C.E. Lost crops of the Incas: Origins of domestication of the Andean pulse crop tarwi, Lupinus mutabilis. Am. J. Bot. 2016, 103, 1592–1606. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Malhotra, N.; Sharma, K. Buckwheat (Fagopyrum sp.) genetic resources: What can they contribute towards nutritional security of changing world? Genet. Resour. Crop. Evol. 2020, 67, 1–20. [Google Scholar] [CrossRef]
- Cuellar-Ortiz, S.M.; Arrieta-Montiel, M.D.L.P.; Acosta-Gallegos, J.; Covarrubias, A.A. Relationship between carbohydrate partitioning and drought resistance in common bean. Plant Cell Environ. 2008, 31, 1399–1409. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef]
- McAusland, L.; Davey, P.A.; Kanwal, N.; Baker, N.R.; Lawson, T. A novel system for spatial and temporal imaging of intrinsic plant water use efficiency. J. Exp. Bot. 2013, 64, 4993–5007. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Monda, K.; Negi, J.; Konishi, F.; Ishikawa, S.; Hashimoto-Sugimoto, M.; Gotô, N.; Iba, K. Natural variation in stomatal responses to environmental changes among arabidopsis thaliana ecotypes. PLoS ONE 2015, 10, e0117449. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.M.; Zhang, H.; Zhou, H.; Du, T.; Wu, Q.; Mockler, T.C.; Berezin, M. Highly sensitive image-derived indices of water-stressed plants using hyperspectral imaging in SWIR and histogram analysis. Sci. Rep. 2015, 5, 15919. [Google Scholar] [CrossRef] [PubMed]
- Parent, B.; Shahinnia, F.; Maphosa, L.; Berger, B.; Rabie, H.; Chalmers, K.; Kovalchuk, A.; Langridge, P.; Fleury, D. Combining field performance with controlled environment plant imaging to identify the genetic control of growth and transpiration underlying yield response to water-deficit stress in wheat. J. Exp. Bot. 2015, 66, 5481–5492. [Google Scholar] [CrossRef] [PubMed]
- Humplík, J.F.; Lazár, D.; Husičková, A.; Spíchal, L. Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses—A review. Plant Methods 2015, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.A.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Prince, S.; Mutava, R.N.; Nguyen, N.; Pathan, S.M.; Shannon, G.J.; Murphy, M.; Zhang, Z.; Kim, Y.H.; Valliyodan, B.; Nguyen, H.T. Evaluation of high yielding soybean germplasm under water limitation. J. Integr. Plant Biol. 2015, 58, 475–491. [Google Scholar] [CrossRef]
- Das, A.; Schneider, H.M.; Burridge, J.D.; Ascanio, A.K.M.; Wojciechowski, T.; Topp, C.N.; Lynch, J.P.; Weitz, J.S.; Bucksch, A. Digital imaging of root traits (DIRT): A high-throughput computing and collaboration platform for field-based root phenomics. Plant Methods 2015, 11, 51. [Google Scholar] [CrossRef]
- Cai, J.; Zeng, Z.; Connor, J.N.; Huang, C.Y.; Melino, V.J.; Kumar, P.; Miklavcic, S.J. RootGraph: A graphic optimization tool for automated image analysis of plant roots. J. Exp. Bot. 2015, 66, 6551–6562. [Google Scholar] [CrossRef]
- Kumar, P.; Cai, J.; Miklavcic, S.J. A complete system for 3D reconstruction of roots for phenotypic analysis. Adv. Exp. Med. Biol. 2014, 823, 249–270. [Google Scholar] [CrossRef]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.A.; Baenziger, P.S.; Börner, A. Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, R. Breeding for increased drought tolerance in wheat: A review. Crop. Pasture Sci. 2018, 69, 223–241. [Google Scholar] [CrossRef]
- Khan, A.; Sovero, V.; Gemenet, D.C. Genome-assisted breeding for drought resistance. Curr. Genom. 2016, 17, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, J.; Joshi, R.; Singh, B.; Bohra, A.; Vijayan, R.; Bhatt, M.; Bisht, S.P.S.; Wani, S.H. Application of Bioinformatics in Understanding of Plant Stress Tolerance; Springer Science and Business Media LLC: Berlin, Germany, 2017; pp. 347–374. [Google Scholar]
- Sharma, V.; Sarkar, I.N. Bioinformatics opportunities for identification and study of medicinal plants. Briefings Bioinform. 2012, 14, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Blum, A. Genomics for drought resistance—Getting down to earth. Funct. Plant Biol. 2014, 41, 1191–1198. [Google Scholar] [CrossRef]
- Nuccio, M.L.; Wu, J.; Mowers, R.; Zhou, H.-P.; Meghji, M.; Primavesi, L.F.; Paul, M.J.; Chen, X.; Gao, Y.; Haque, E.; et al. Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotechnol. 2015, 33, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Daszkowska-Golec, A.; Skubacz, A.; Sitko, K.; Słota, M.; Kurowska, M.; Szarejko, I. Mutation in barley ERA1 (Enhanced Response to ABA1) gene confers better photosynthesis efficiency in response to drought as revealed by transcriptomic and physiological analysis. Environ. Exp. Bot. 2018, 148, 12–26. [Google Scholar] [CrossRef]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.; Pareek, A.; Singla-Pareek, S.L. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J. Exp. Bot. 2012, 63, 5873–5885. [Google Scholar] [CrossRef]
- Jangale, B.L.; Chaudhari, R.S.; Azeez, A.; Sane, P.V.; Sane, A.P.; Krishna, B. Independent and combined abiotic stresses affect the physiology and expression patterns of DREB genes differently in stress-susceptible and resistant genotypes of banana. Physiol. Plant. 2018, 165, 303–318. [Google Scholar] [CrossRef]
- Bundó, M.; Coca, M.A. Calcium-dependent protein kinase OsCPK10 mediates both drought tolerance and blast disease resistance in rice plants. J. Exp. Bot. 2017, 68, 2963–2975. [Google Scholar] [CrossRef] [PubMed]
- Mitula, F.; Tajdel, M.; Cieśla, A.; Kasprowicz-Maluśki, A.; Kulik, A.; Babula-Skowronska, D.; Michalak, M.; Dobrowolska, G.; Sadowski, J.; Ludwików, A. Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol. 2015, 56, 2351–2367. [Google Scholar] [CrossRef] [PubMed]
- Samajova, O.; Plihal, O.; Al-Yousif, M.; Hirt, H.; Šamaj, J. Improvement of stress tolerance in plants by genetic manipulation of mitogen-activated protein kinases. Biotechnol. Adv. 2013, 31, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, Z.; Ran, Q.; Li, P.; Peng, Z.; Zhang, J. ZmNF-YB16 overexpression improves drought resistance and yield by enhancing photosynthesis and the antioxidant capacity of maize plants. Front. Plant Sci. 2018, 9, 709. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhao, Y.; Shen, R.; Wang, B.; Xie, Y.; Ma, X.; Zheng, Z.; Wang, H. Characterization of maize phytochrome-interacting factors in light signaling and photomorphogenesis. Plant Physiol. 2019, 181, 789–803. [Google Scholar] [CrossRef]
- Gao, Y.; Jiang, W.; Dai, Y.; Xiao, N.; Zhang, C.; Li, H.; Lu, Y.; Wu, M.; Tao, X.; Deng, D.; et al. A maize phytochrome-interacting factor 3 improves drought and salt stress tolerance in rice. Plant Mol. Biol. 2015, 87, 413–428. [Google Scholar] [CrossRef]
- Khan, S.; Anwar, S.; Yu, S.; Sun, M.; Yang, Z.; Gao, Z.-Q. Development of drought-tolerant transgenic wheat: Achievements and limitations. Int. J. Mol. Sci. 2019, 20, 3350. [Google Scholar] [CrossRef]
- Yang, S.; Vanderbeld, B.; Wan, J.; Huang, Y. Narrowing down the targets: Towards successful genetic engineering of drought-tolerant crops. Mol. Plant 2010, 3, 469–490. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, H.S.; Gahlaut, V. QTL analysis for drought tolerance in wheat: Present status and future possibilities. Agronomy 2017, 7, 5. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Yin, F.; Gao, J.; Liu, M.; Qin, C.; Zhang, W.; Yang, A.; Xia, M.; Zhang, Z.; Shen, Y.; Lin, H.; et al. Genome-wide analysis of Water-stress-responsive microRNA expression profile in tobacco roots. Funct. Integr. Genom. 2014, 14, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Saad, A.S.I.; Li, X.; Li, H.-P.; Huang, T.; Gao, C.-S.; Guo, M.-W.; Cheng, W.; Zhao, G.-Y.; Liao, Y.-C. A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci. 2013, 203, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.-J.; Jing-Jing, L.; Wang, L.-F.; Cao, Y.-Y.; Ma, J.; Wang, H.; Zhang, D.-F.; Li, H.-Y. Evaluation of drought tolerance in ZmVPP1-overexpressing transgenic inbred maize lines and their hybrids. J. Integr. Agric. 2020, 19, 2177–2187. [Google Scholar] [CrossRef]
- Szalonek, M.; Sierpien, B.; Rymaszewski, W.; Gieczewska, K.; Garstka, M.; Lichocka, M.; Sass, L.; Paul, K.; Vass, I.; Vanková, R.; et al. Potato annexin STANN1 promotes drought tolerance and mitigates light stress in transgenic solanum tuberosum L. plants. PLoS ONE 2015, 10, e0132683. [Google Scholar] [CrossRef]
- You, J.; Hu, H.; Xiong, L. An ornithine δ-aminotransferase gene OsOAT confers drought and oxidative stress tolerance in rice. Plant Sci. 2012, 197, 59–69. [Google Scholar] [CrossRef] [PubMed]
- De Souza, W.R.; Oliveira, N.G.; Vinecky, F.; Ribeiro, A.P.; Basso, M.F.; Casari, R.A.D.C.N.; Cunha, B.A.D.B.; Duarte, K.E.; Santiago, T.R.; Martins, P.K.; et al. Field evaluation of At DREB 2A CA overexpressing sugarcane for drought tolerance. J. Agron. Crop. Sci. 2019, 205, 545–553. [Google Scholar] [CrossRef]
- Ayadi, M.; Brini, F.; Masmoudi, K. Overexpression of a wheat aquaporin gene, TdPIP2;1, enhances salt and drought tolerance in transgenic durum wheat cv. maali. Int. J. Mol. Sci. 2019, 20, 2389. [Google Scholar] [CrossRef]
- Wei, Y.; Liu, W.; Hu, W.; Yan, Y.; Shi, H. The chaperone MeHSP90 recruits MeWRKY20 and MeCatalase1 to regulate drought stress resistance in cassava. New Phytol. 2020, 226, 476–491. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Q.; Nan, H.; Li, X.; Lu, S.; Zhao, X.; Liu, B.; Guo, C.; Kong, F.; Cao, D. Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. PLoS ONE 2017, 12, e0179554. [Google Scholar] [CrossRef]
- Fan, Y.; Shabala, S.; Ma, Y.; Xu, R.; Zhou, M. Using QTL mapping to investigate the relationships between abiotic stress tolerance (drought and salinity) and agronomic and physiological traits. BMC Genom. 2015, 16, 43. [Google Scholar] [CrossRef]
- Wehner, G.; Balko, C.; Enders, M.M.; Humbeck, K.; Ordon, F. Identification of genomic regions involved in tolerance to drought stress and drought stress induced leaf senescence in juvenile barley. BMC Plant Biol. 2015, 15, 125. [Google Scholar] [CrossRef] [PubMed]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Technow, F.; Messina, C.D.; Totir, L.R.; Cooper, M. Integrating crop growth models with whole genome prediction through approximate bayesian computation. PLoS ONE 2015, 10, e0130855. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, R.; Alemzadeh, A.; Razi, H. Microarray analysis of transcriptional responses to salt and drought stress in Arabidopsis thaliana. Heliyon 2019, 5, e02614. [Google Scholar] [CrossRef]
- Hassani-Pak, K.; Singh, A.; Brandizi, M.; Hearnshaw, J.; Amberkar, S.; Phillips, A.L.; Doonan, J.H.; Rawlings, C.J. KnetMiner: A comprehensive approach for supporting evidence-based gene discovery and complex trait analysis across species. bioRxiv 2020, 4, 017004. [Google Scholar]
- Noraziyah, A.A.S.; Swamy, B.P.M.; Wickneswari, R.; Cruz, M.T.S.; Raman, A.; Kumar, A. Marker assisted pyramiding of drought yield QTLs into a popular Malaysian rice cultivar, MR219. BMC Genet. 2016, 17, 30. [Google Scholar] [CrossRef]
- Lopes, M.S.; El-Basyoni, I.S.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef]
- Merchuk-Ovnat, L.; Barak, V.; Fahima, T.; Ordon, F.; Lidzbarsky, G.A.; Krugman, T.; Saranga, Y. Ancestral QTL alleles from wild emmer wheat improve drought resistance and productivity in modern wheat cultivars. Front. Plant Sci. 2016, 7, 452. [Google Scholar] [CrossRef]
- Alqudah, A.M.; Sallam, A.; Baenziger, P.S.; Börner, A. GWAS: Fast-forwarding gene identification and characterization in temperate Cereals: Lessons from Barley—A review. J. Adv. Res. 2020, 22, 119–135. [Google Scholar] [CrossRef]
- Gupta, M.; Chawla, V.; Garg, P.; Yadav, N.; Munjal, R.; Sharma, B. Genetic analysis of yield and heat stress related traits in wheat (Triticum aestivum L. em. Thell) using microsatellite markers. J. Appl. Nat. Sci. 2015, 7, 739–744. [Google Scholar] [CrossRef]
- Qaseem, M.F.; Qureshi, R.; Muqaddasi, Q.H.; Shaheen, H.; Kousar, R.; Röder, M.S. Genome-wide association mapping in bread wheat subjected to independent and combined high temperature and drought stress. PLoS ONE 2018, 13, e0199121. [Google Scholar] [CrossRef]
- Hou, S.; Zhu, G.; Li, Y.; Li, W.; Fu, J.; Niu, E.; Li, L.; Zhang, D.; Guo, W. Genome-wide association studies reveal genetic variation and candidate genes of drought stress related traits in cotton (Gossypium hirsutum L.). Front. Plant Sci. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Kadam, N.N.; Struik, P.C.; Rebolledo, M.C.; Yin, X.; Jagadish, S.V.K. Genome-wide association reveals novel genomic loci controlling rice grain yield and its component traits under water-deficit stress during the reproductive stage. J. Exp. Bot. 2018, 69, 4017–4032. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Cairns, J.E.; Babu, R.; Gowda, M.; Makumbi, D.; Magorokosho, C.; Zhang, A.; Liu, Y.; Wang, N.; Hao, Z.; et al. Genome-wide association mapping and genomic prediction analyses reveal the genetic architecture of grain yield and flowering time under drought and heat stress conditions in maize. Front. Plant Sci. 2019, 9, 9. [Google Scholar] [CrossRef]
- Gurgul, A.; Miksza-Cybulska, A.; Szmatoła, T.; Jasielczuk, I.; Piestrzynska-Kajtoch, A.; Fornal, A.; Semik-Gurgul, E.; Bugno-Poniewierska, M. Genotyping-by-sequencing performance in selected livestock species. Genomics 2019, 111, 186–195. [Google Scholar] [CrossRef]
- Ray, S.; Satya, P. Next generation sequencing technologies for next generation plant breeding. Front. Plant Sci. 2014, 5, 367. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Y.; Hu, Z.; Yang, Z. Genomic selection methods for crop improvement: Current status and prospects. Crop. J. 2018, 6, 330–340. [Google Scholar] [CrossRef]
- Zhao, Y.; Gowda, M.; Liu, W.; Würschum, T.; Maurer, H.P.; Longin, F.H.; Ranc, N.; Reif, J.C. Accuracy of genomic selection in European maize elite breeding populations. Theor. Appl. Genet. 2011, 124, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Vuong, T.; Meinhardt, C.; Tiffin, P.; Denny, R.; Chen, S.; Nguyen, H.T.; Orf, J.H.; Young, N.D. Potential of association mapping and genomic selection to explore PI 88788 derived soybean cyst nematode resistance. Plant Genome 2014, 7, 1–13. [Google Scholar] [CrossRef]
- Lorenzana, R.E.; Bernardo, R. Accuracy of genotypic value predictions for marker-based selection in biparental plant populations. Theor. Appl. Genet. 2009, 120, 151–161. [Google Scholar] [CrossRef]
- Spindel, J.; Begum, H.; Akdemir, D.; Virk, P.; Collard, B.C.; Redoña, E.; Atlin, G.; Jannink, J.-L.; McCouch, S.R. Genomic selection and association mapping in rice (Oryza sativa): Effect of trait genetic architecture, training population composition, marker number and statistical model on accuracy of rice genomic selection in elite, tropical rice breeding lines. PLoS Genet. 2015, 11, e1004982. [Google Scholar] [CrossRef]
- Sallam, A.H.; Endelman, J.B.; Jannink, J.-L.; Smith, K.P. Assessing genomic selection prediction accuracy in a dynamic barley breeding population. Plant Genome 2015, 8, 1–15. [Google Scholar] [CrossRef]
- Cerrudo, D.; Cao, S.; Yuan, Y.; Martínez, C.; Suarez, E.A.; Babu, R.; Zhang, X.; Trachsel, S. Genomic selection outperforms marker assisted selection for grain yield and physiological traits in a maize doubled haploid population across water treatments. Front. Plant Sci. 2018, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Shikha, M.; Kanika, A.; Rao, A.R.; Mallikarjuna, M.G.; Gupta, H.S.; Nepolean, T. Genomic selection for drought tolerance using genome-wide SNPs in maize. Front. Plant Sci. 2017, 8, 550. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ruperao, P.; Batley, J.; Edwards, D.; Khan, T.; Colmer, T.D.; Pang, J.; Siddique, K.H.M.; Sutton, T. Investigating drought tolerance in chickpea using genome-wide association mapping and genomic selection based on whole-genome resequencing data. Front. Plant Sci. 2018, 9, 190. [Google Scholar] [CrossRef]
- Zhang, A.; Wang, H.; Beyene, Y.; Semagn, K.; Liu, Y.; Cao, S.; Cui, Z.; Ruan, Y.; Burgueño, J.; Vicente, F.S.; et al. Effect of trait heritability, training population size and marker density on genomic prediction accuracy estimation in 22 bi-parental tropical maize populations. Front. Plant Sci. 2017, 8, 8. [Google Scholar] [CrossRef]
- Zhang, X.; Pérez-Rodríguez, P.; Burgueño, J.; Olsen, M.; Buckler, E.; Atlin, G.; Prasanna, B.M.; Vargas, M.; Vicente, F.M.S.; Crossa, J. Rapid cycling genomic selection in a multiparental tropical maize population. G3 Genes Genomes Genet. 2017, 7, 2315–2326. [Google Scholar] [CrossRef]
- Čermák, T.; Baltes, N.J.; Cegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef]
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Ito, Y.; Nishizawa, A.; Endo, M.; Mikami, M.; Toki, S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 2015, 467, 76–82. [Google Scholar] [CrossRef]
- Feng, Z.; Mao, Y.; Xu, N.; Zhang, B.; Wei, P.; Yang, N.-L.; Wang, Z.; Zhang, Z.; Zheng, R.; Yang, L.; et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632–4637. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zeng, X.; Zhao, M.; Cui, X.; Wang, Q.; Yang, H.; Cheng, H.; Yu, D. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 2016, 217, 90–97. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, X.; Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant 2016, 9, 628–631. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2016, 15, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef]
- Li, R.; Liu, C.; Zhao, R.; Wang, L.; Chen, L.; Yu, W.; Zhang, S.; Sheng, J.; Shen, L. CRISPR/Cas9-mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 2019, 19, 38. [Google Scholar] [CrossRef]
- Osakabe, Y.; Watanabe, T.; Sugano, S.S.; Ueta, R.; Ishihara, R.; Shinozaki, K.; Osakabe, K. Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 2016, 6, 26685. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]

| Crop | No. DT Varieties or Donors | Method | Wild Species as Possible Sources for DT Introgression |
|---|---|---|---|
| Maize | 41 | Thirty-five varieties obtained by conventional breeding ([38,39] results published in the official internet web pages of CYMMYT, KARI, and WEMA projects) | Information not available |
| Potato | 22 | Conventional breeding [40,41,42,43,44] | Solanum juzepczukii B., S. cardiophyllum, and S. gandarillasii, S. tarijense [42] |
| Rice | 16 | Conventional breeding [45,46,47] | Oryza longistaminata, O. rufipogon [48], O. meridionalis, and O. nivara [49], O. glaberrima [36] |
| Sugarcane | 24 | Conventional breeding [50,51,52,53] | Saccharum spontaneum [54], and S. robustum [53] |
| Wheat | 2 | Conventional breeding [55] | Aegilops kotsehyi, A. variabilis, A. speltoides, A. umbellulata, A. squarrosa [54] and A. tauschii [56] |
| Cassava | 2 | Conventional breeding [57] | Manihot glaziovii [58], M. pseudoglaziovii, M. stipularis, M. caerulenscens [59], M. carthaginensis, and M. dichotoma [60] |
| Crop | Crop Origin | Status | Countries with Registered Varieties |
|---|---|---|---|
| Quinoa (Chenopodium quinoa Willd.) | Andean region [72] | Introduced | The Netherlands (5), Denmark (4), France (1), Canada(4) USA (3), Australia (2), Germany (1), Ukraine (2) [73,74] |
| Amaranth (Amaranthus hypochondriacus, Amaranthus cruentus L., and Amaranthus caudatus L.) | High tropical and subtropical lands of America [75] | Introduced | Russian (7 Ah, 8 Ac, 6 Acr), Germany (1 Ah, 1 Acr), Slovakia (1 Ac, 2 Acr), The Netherlands (1 Ac), Romania (1 Ac, 1 Acr), Ukraine (1 Ac), Poland (2 Acr), New Zealand (1 Acr), Czech Republic (1 Acr) [74] |
| Millet (Pennisetum glaucum L.) | Africa [76] | Introduced | Brazil (13), Russia (5), USA (3), Ukraine (1), Mexico (3), Australia (1) [74] |
| Buckwheat (Fagopyrum sp.) | China [77] | Introduced | Ukraine (19), Denmark (10), USA (4,) Moldova (3), Canada (2), Australia (1) [74] |
| Cowpea (Vigna unguiculata) | Southern Africa [78] | Introduced | Brazil (13), Australia (8), China (7), Turkey (7), Moldova (6), Korea (4), Romania (3), Bulgaria (2), Poland (1), Portugal (1) [74] |
| Sweetpotato (Ipomoea batatas) | Central America and north of South America [79] | Introduced | Switzerland (3), Israel (11,) Romania (2), Slovenia (8), Ukraine (4), USA (89), South Africa (29), China (42) [74] |
| Andean Lupin (Lupinus mutabilis) | Andean region [80] | Introduced | The Netherlands (1), Czech Republic (1), Germany (1) [74] |
| Crop | Genotypes or Varieties Names | Method | DT Source | Reference |
|---|---|---|---|---|
| Maize | PH4CV-T, PH6WC-T, Chang7-2-T, and Zheng58-T | Overexpression | VPP gene | [117] |
| Potato | Cultivar Sante | Overexpression | STANN1 mRNA | [118] |
| Rice | U7, U14 | Overexpression | OsOAT gene | [119] |
| Sugarcane | ZmRab17:AtDREB2A CA | Overexpression | AtDREB2A CA transcription factor | [120] |
| Wheat | Transgenic Durum Wheat cv. Maali | Overexpression | TdPIP2 gene | [121] |
| Cassava | South China 124 (SC124) cassava variety | Silencing | HSP90 protein | [122] |
| Soybean | Transgenic soybean plants | Overexpression | GmFDL19 transcription factor | [123] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosero, A.; Granda, L.; Berdugo-Cely, J.A.; Šamajová, O.; Šamaj, J.; Cerkal, R. A Dual Strategy of Breeding for Drought Tolerance and Introducing Drought-Tolerant, Underutilized Crops into Production Systems to Enhance Their Resilience to Water Deficiency. Plants 2020, 9, 1263. https://doi.org/10.3390/plants9101263
Rosero A, Granda L, Berdugo-Cely JA, Šamajová O, Šamaj J, Cerkal R. A Dual Strategy of Breeding for Drought Tolerance and Introducing Drought-Tolerant, Underutilized Crops into Production Systems to Enhance Their Resilience to Water Deficiency. Plants. 2020; 9(10):1263. https://doi.org/10.3390/plants9101263
Chicago/Turabian StyleRosero, Amparo, Leiter Granda, Jhon A. Berdugo-Cely, Olga Šamajová, Jozef Šamaj, and Radim Cerkal. 2020. "A Dual Strategy of Breeding for Drought Tolerance and Introducing Drought-Tolerant, Underutilized Crops into Production Systems to Enhance Their Resilience to Water Deficiency" Plants 9, no. 10: 1263. https://doi.org/10.3390/plants9101263
APA StyleRosero, A., Granda, L., Berdugo-Cely, J. A., Šamajová, O., Šamaj, J., & Cerkal, R. (2020). A Dual Strategy of Breeding for Drought Tolerance and Introducing Drought-Tolerant, Underutilized Crops into Production Systems to Enhance Their Resilience to Water Deficiency. Plants, 9(10), 1263. https://doi.org/10.3390/plants9101263