Potential and Challenges of Improving Photosynthesis in Algae
Abstract
:1. Introduction
1.1. Why Study Photosynthesis in Microalgae?
1.2. Microalgal Species of Interest for Research on the Regulatory Mechanisms of Photosynthesis
2. Photosynthesis
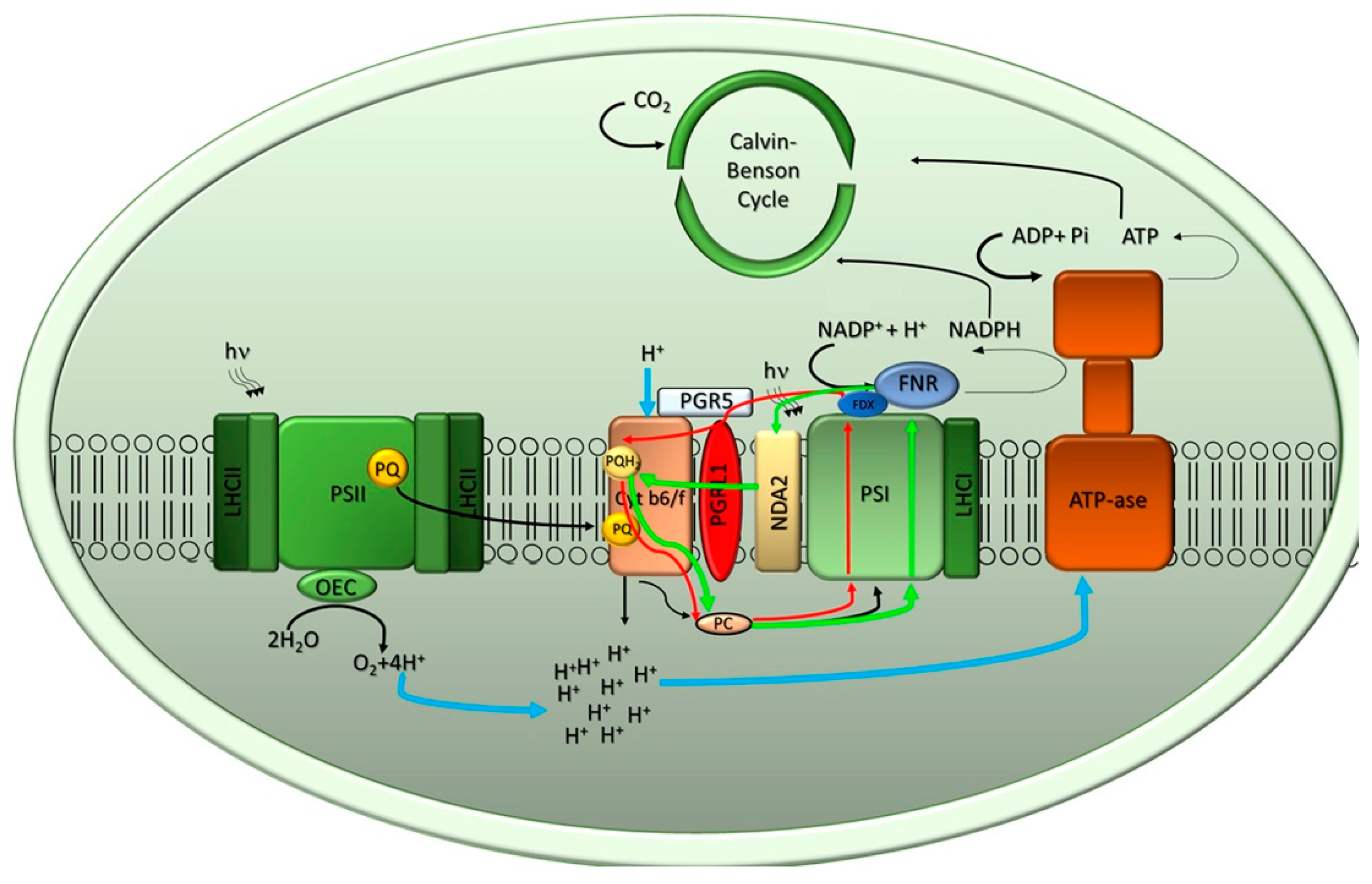
2.1. The Light Phase of Photosynthesis
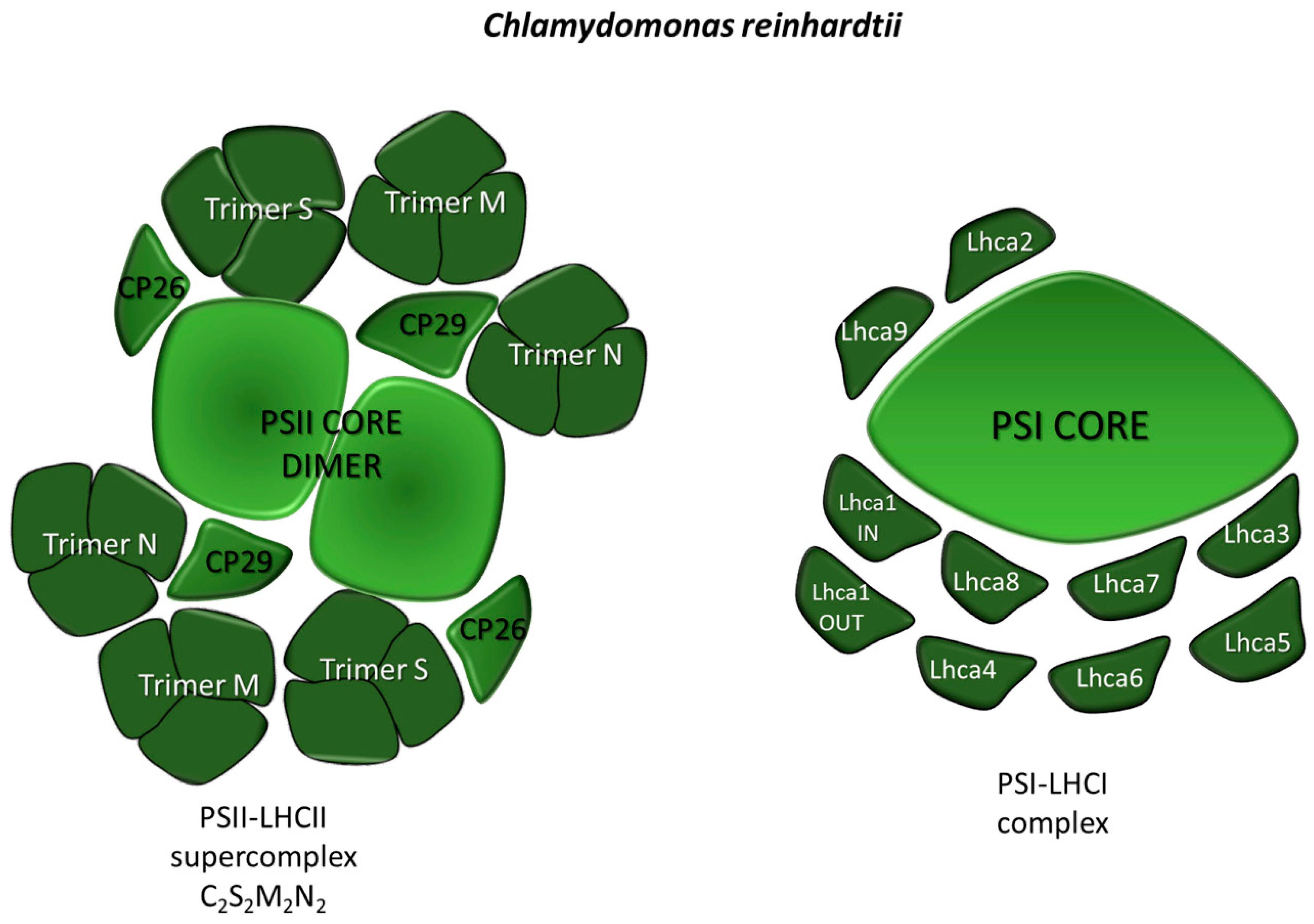
2.1.1. Light-Harvesting Systems: PSI-LHCI and PSII-LHCII Supercomplexes Organization in Microalgae
PSII-LHCII
PSI-LHCI
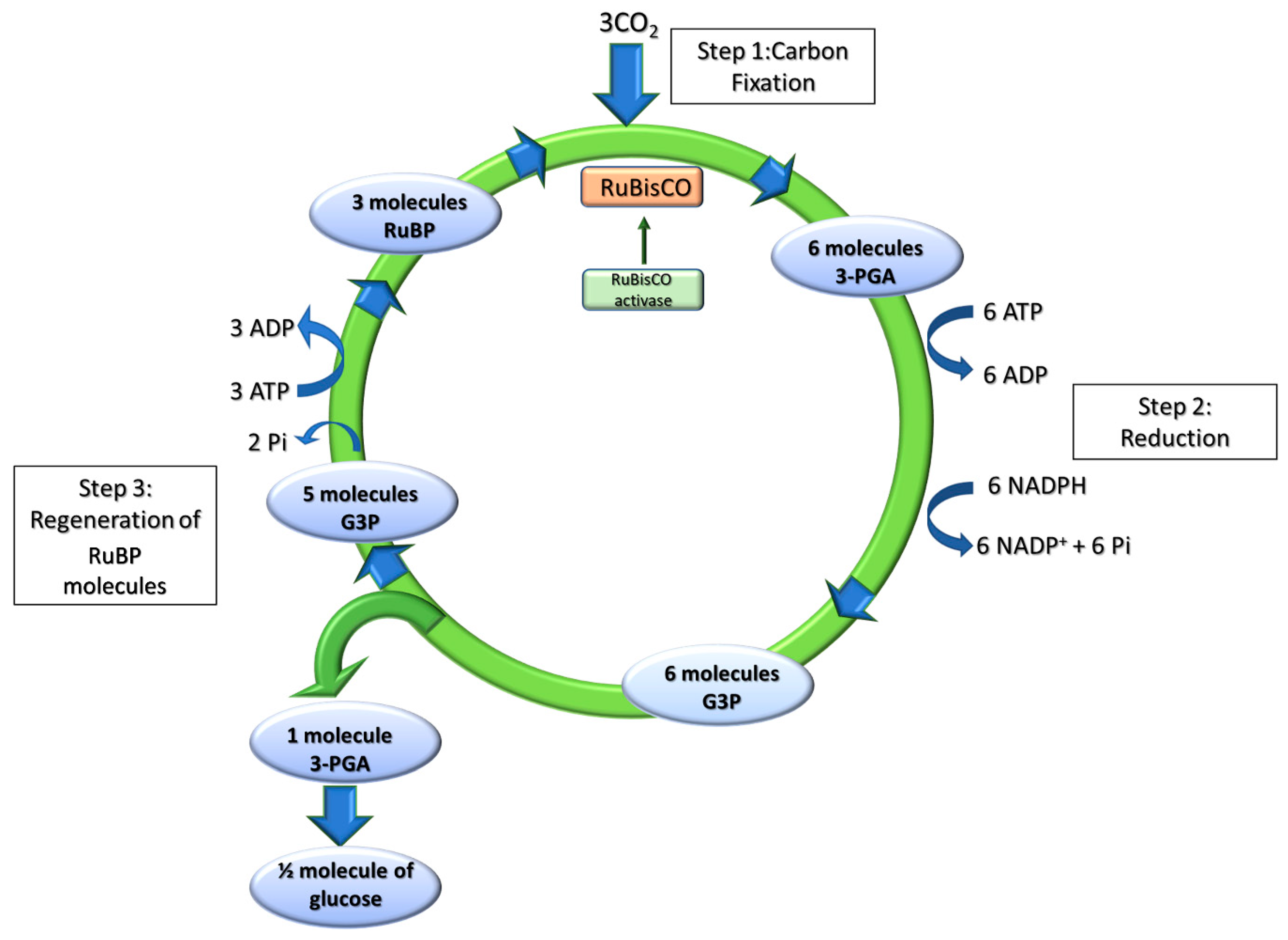
2.2. The Dark Phase of Photosynthesis
2.2.1. Dark Reactions of Photosynthesis: The Calvin-Benson-Bassham Cycle
2.2.2. RuBisCO
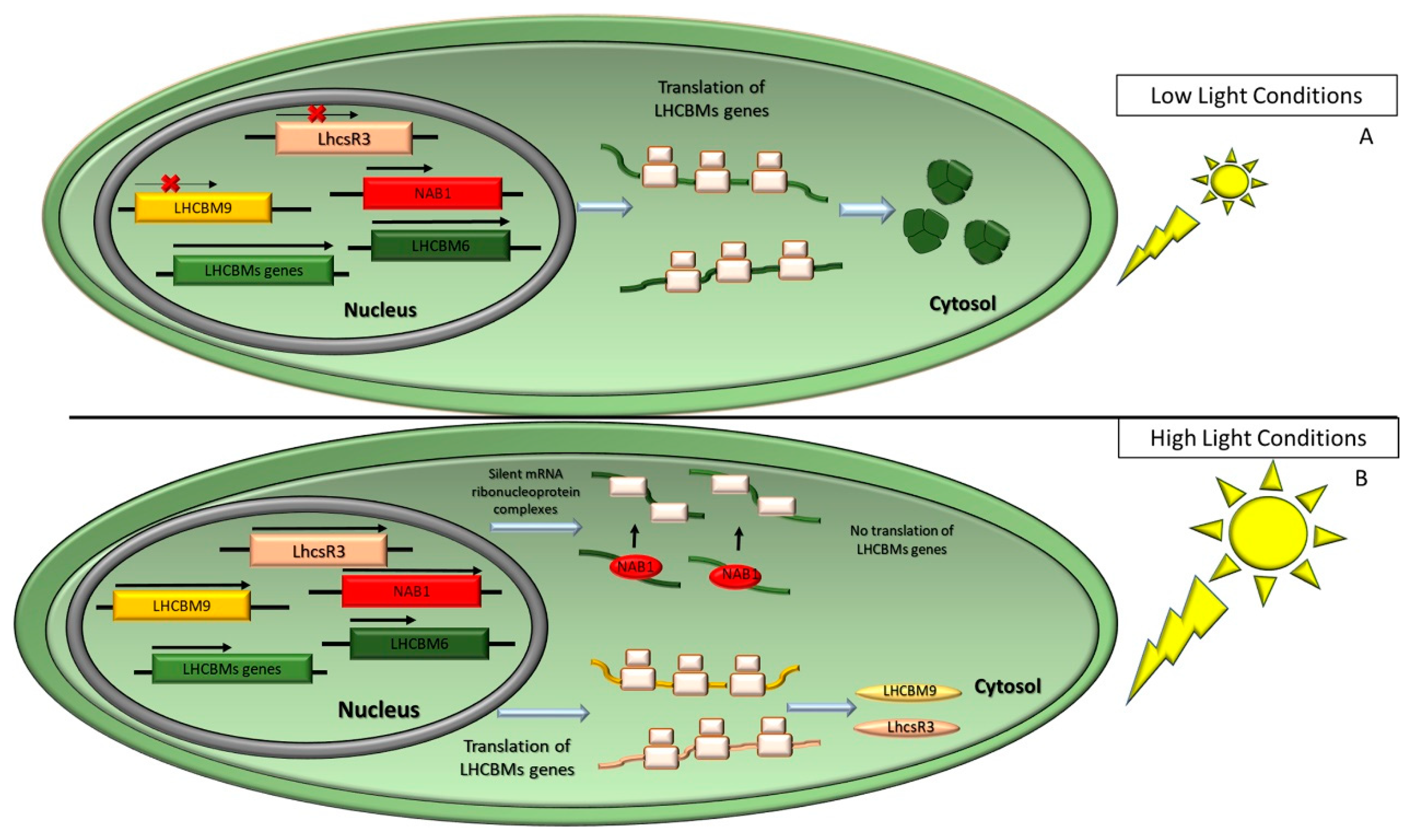
2.3. Dynamics of the Photosynthetic Apparatus in Response to Environmental Conditions: Photoprotective Mechanisms
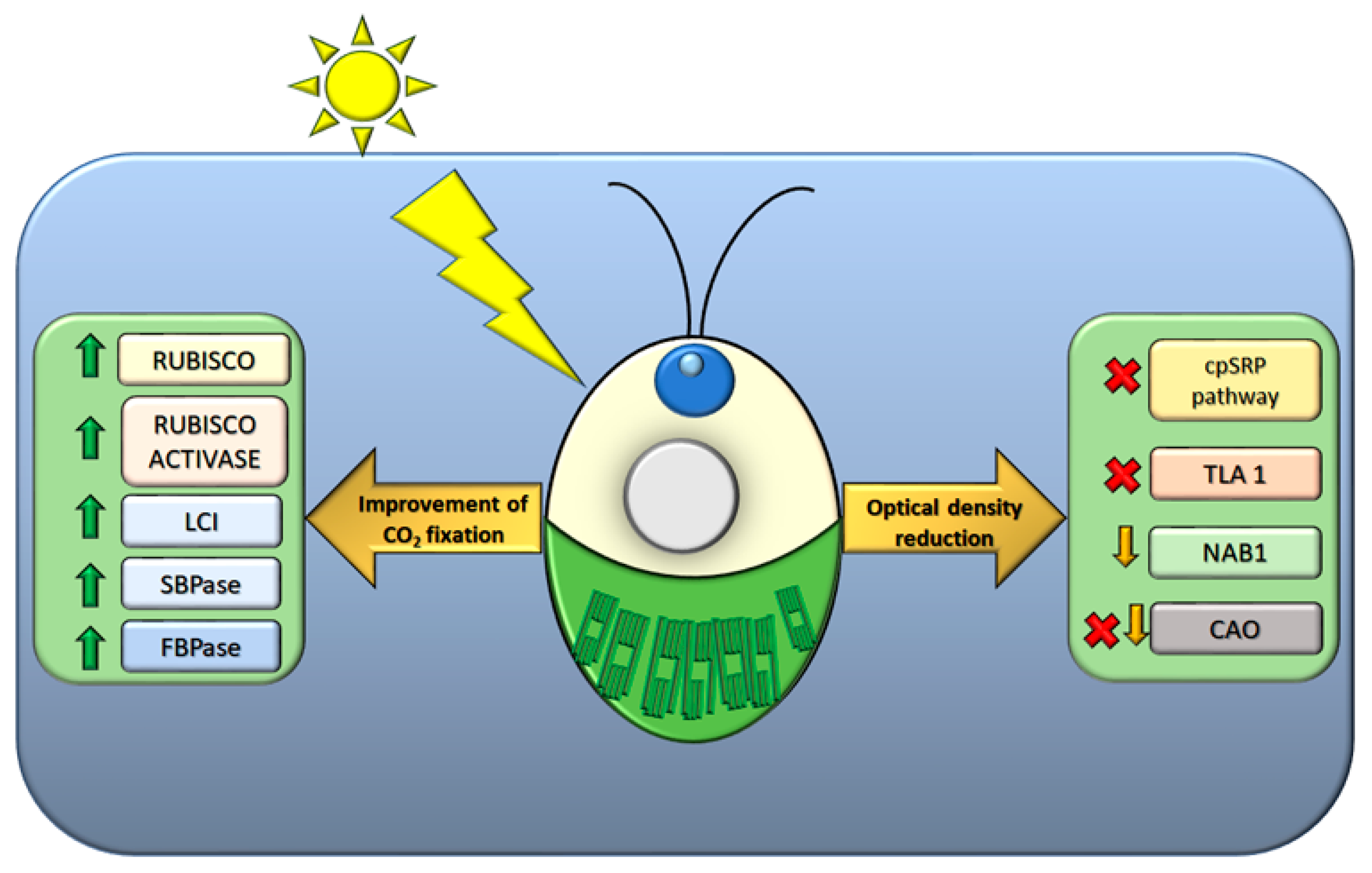
3. Improving Photosynthetic Yield
3.1. Light Harvesting Antenna as Target to Reduce Optical Density in Mass Culture
3.2. Bioengineering Response to Light Fluctuations and Improving Resistance to Photo-Inhibition
3.3. RuBisCO as Target to Improve Carbon Assimilation Efficiency
3.4. Engineering of the Lipid Biosynthesis for Renewable Energies Production
3.5. Endogenous Up-Regulation and Heterologous Expression of Isoprenoid Biosynthetic Pathways in Microalgae
4. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Barber, J. Photosynthetic energy conversion: Natural and artificial. Chem. Soc. Rev. 2009, 38, 185–196. [Google Scholar] [CrossRef]
- Stephenson, P.G.; Moore, C.M.; Terry, M.J.; Zubkov, M.V.; Bibby, T.S. Improving photosynthesis for algal biofuels: Toward a green revolution. Trends Biotechnol. 2011, 29, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, M.; Vecchi, V.; Barera, S.; Dall’Osto, L. Biomass from microalgae: The potential of domestication towards sustainable biofactories. Microb. Cell Fact. 2018, 17, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georgianna, D.R.; Stephen, P. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 2012, 488, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Melis, A. Solar energy conversion efficiencies in photosynthesis: Minimizing the chlorophyll antennae to maximize efficiency. Plant Sci. 2009, 177, 272–280. [Google Scholar] [CrossRef]
- Guiry, M.D. How many species of algae are there? J. Phycol. 2012, 48, 1057–1063. [Google Scholar] [CrossRef]
- Harris, E.H. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 363–406. [Google Scholar] [CrossRef] [Green Version]
- Grossman, A.R. Chlamydomonas reinhardtii and photosynthesis: Genetics to genomics. Curr. Opin. Plant Biol. 2000, 3, 132–137. [Google Scholar] [CrossRef]
- Ip, P.F.; Chen, F. Production of astaxanthin by the green microalga Chlorella zofingiensis in the dark. Process Biochem. 2005, 40, 733–738. [Google Scholar] [CrossRef]
- García, J.L.; de Vicente, M.; Galán, B. Microalgae, old sustainable food and fashion nutraceuticals. Microb. Biotechnol. 2017, 10, 1017–1024. [Google Scholar] [CrossRef] [Green Version]
- Guccione, A.; Biondi, N.; Sampietro, G.; Rodolfi, L.; Bassi, N.; Tredici, M.R. Chlorella for protein and biofuels: From strain selection to outdoor cultivation in a Green Wall Panel photobioreactor. Biotechnol. Biofuels 2014, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cazzaniga, S.; Dall’Osto, L.; Szaub, J.; Scibilia, L.; Ballottari, M.; Purton, S.; Bassi, R. Domestication of the green alga Chlorella sorokiniana: Reduction of antenna size improves light-use efficiency in a photobioreactor. Biotechnol. Biofuels 2014, 7, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Ibrahim, I.; Wosu, C.; Ben-Amotz, A.; Harvey, P. Potential of New Isolates of Dunaliella Salina for Natural β-Carotene Production. Biology 2018, 7, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, R.A.; He, M.; Aftab, R.A.; Zheng, S.; Nagi, M.; Bakri, R.; Wang, C. Bioenergy application of Dunaliella salina SA 134 grown at various salinity levels for lipid production. Sci. Rep. 2017, 7, 8118. [Google Scholar] [CrossRef] [Green Version]
- Shah, M.M.R.; Liang, Y.; Cheng, J.J.; Daroch, M. Astaxanthin-Producing Green Microalga Haematococcus pluvialis: From Single Cell to High Value Commercial Products. Front. Plant Sci. 2016, 7, 531. [Google Scholar] [CrossRef] [Green Version]
- Basso, S.; Simionato, D.; Gerotto, C.; Segalla, A.; Giacometti, G.M.; Morosinotto, T. Characterization of the photosynthetic apparatus of the Eustigmatophycean Nannochloropsis gaditana: Evidence of convergent evolution in the supramolecular organization of photosystem i. Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 306–314. [Google Scholar] [CrossRef]
- Simionato, D.; Block, M.A.; La Rocca, N.; Jouhet, J.; Maréchal, E.; Finazzi, G.; Morosinotto, T. The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus. Eukaryot. Cell 2013, 12, 665–676. [Google Scholar] [CrossRef] [Green Version]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yu, L.-J.; Xu, C.; Tomizaki, T.; Zhao, S.; Umena, Y.; Chen, X.; Qin, X.; Xin, Y.; Suga, M.; et al. Structural basis for blue-green light harvesting and energy dissipation in diatoms. Science 2019, 363, eaav0365. [Google Scholar] [CrossRef] [PubMed]
- Nelson, N.; Yocum, C.F. Structure and function of photosystem Ι and II. Annu. Rev. Plant Biol. 2006, 57, 521–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munekage, Y.; Hashimoto, M.; Miyake, C.; Tomizawa, K.; Endo, T.; Tasaka, M.; Shikanai, T. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 2004, 429, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Jans, F.; Mignolet, E.; Houyoux, P.-A.; Cardol, P.; Ghysels, B.; Cuine, S.; Cournac, L.; Peltier, G.; Remacle, C.; Franck, F. A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc. Natl. Acad. Sci. USA 2008, 105, 20546–20551. [Google Scholar] [CrossRef] [Green Version]
- Jokel, M.; Johnson, X.; Peltier, G.; Aro, E.M.; Allahverdiyeva, Y. Hunting the main player enabling Chlamydomonas reinhardtii growth under fluctuating light. Plant. J. 2018, 94, 822–835. [Google Scholar] [CrossRef] [Green Version]
- Dall’Osto, L.; Bressan, M.; Bassi, R. Biogenesis of light harvesting proteins. Biochim. Biophys. Acta-Bioenerg. 2015, 1847, 861–871. [Google Scholar] [CrossRef] [Green Version]
- Ballottari, M.; Girardon, J.; Dall’Osto, L.; Bassi, R. Evolution and functional properties of Photosystem II light harvesting complexes in eukaryotes. Biochim. Biophys. Acta-Bioenerg. 2012, 1817, 143–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Yan, H.; Wang, K.; Kuang, T.; Zhang, J.; Gui, L.; An, X.; Chang, W. Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 2004, 428, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shem, A.; Frolow, F.; Nelson, N. Crystal structure of plant photosystem I. Nature 2003, 426, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Dall’Osto, L.; Piques, M.; Ronzani, M.; Molesini, B.; Alboresi, A.; Cazzaniga, S.; Bassi, R. The Arabidopsis nox mutant lacking carotene hydroxylase activity reveals a critical role for xanthophylls in photosystem I biogenesis. Plant Cell 2013, 25, 591–608. [Google Scholar] [CrossRef] [Green Version]
- Rochaix, J.-D.; Bassi, R. LHC-like proteins involved in stress responses and biogenesis/repair of the photosynthetic apparatus. Biochem. J. 2019, 476, 581–593. [Google Scholar] [CrossRef]
- Alboresi, A.; Caffarri, S.; Nogue, F.; Bassi, R.; Morosinotto, T. In Silico and Biochemical Analysis of Physcomitrella patens Photosynthetic Antenna: Identification of Subunits which Evolved upon Land Adaptation. PLoS ONE 2008, 3, e2033. [Google Scholar] [CrossRef] [Green Version]
- Peers, G.; Truong, T.B.; Ostendorf, E.; Busch, A.; Elrad, D.; Grossman, A.R.; Hippler, M.; Niyogi, K.K. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 2009, 462, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Miloslavina, Y.; de Bianchi, S.; Dall’Osto, L.; Bassi, R.; Holzwarth, A.R. Quenching in Arabidopsis thaliana Mutants Lacking Monomeric Antenna Proteins of Photosystem II. J. Biol. Chem. 2011, 286, 36830–36840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, X.; Ma, J.; Wei, X.; Cao, P.; Zhu, D.; Chang, W.; Liu, Z.; Zhang, X.; Li, M. Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science (80-) 2017, 357, 815–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drop, B.; Webber-Birungi, M.; Yadav, S.K.N.; Filipowicz-Szymanska, A.; Fusetti, F.; Boekema, E.J.; Croce, R. Light-harvesting complex II (LHCII) and its supramolecular organization in Chlamydomonas reinhardtii. Biochim. Biophys. Acta-Bioenerg. 2014, 1837, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Huang, Z.; Chang, S.; Wang, W.; Wang, J.; Kuang, T.; Han, G.; Shen, J.-R.; Zhang, X. Structure of a C2S2M2N2-type PSII–LHCII supercomplex from the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2019, 2019, 12462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drop, B.; Webber-birungi, M.; Fusetti, F.; Kour, R.; Redding, K.E.; Boekema, E.J.; Croce, R. Photosystem I of Chlamydomonas reinhardtii Contains Nine Light-harvesting Complexes (Lhca) Located on One Side of the core. J. Biol. Chem. 2011, 286, 44878–44887. [Google Scholar] [CrossRef] [Green Version]
- Steinbeck, J.; Ross, I.L.; Rothnagel, R.; Gäbelein, P.; Schulze, S.; Giles, N.; Ali, R.; Drysdale, R.; Sierecki, E.; Gambin, Y.; et al. Structure of a PSI–LHCI–cyt b6f supercomplex in Chlamydomonas reinhardtii promoting cyclic electron flow under anaerobic conditions. Proc. Natl. Acad. Sci. USA 2018, 115, 10517–10522. [Google Scholar] [CrossRef] [Green Version]
- Su, X.; Ma, J.; Pan, X.; Zhao, X.; Chang, W.; Liu, Z.; Zhang, X.; Li, M. Antenna arrangement and energy transfer pathways of a green algal photosystem-I–LHCI supercomplex. Nat. Plants 2019, 5, 273–281. [Google Scholar] [CrossRef]
- Harrison, E.P.; Willingham, N.M.; Lloyd, J.C.; Raines, C.A. Reduced sedoheptulose-1,7-bisphosphatase levels in transgenic tobacco lead to decreased photosynthetic capacity and altered carbohydrate accumulation. Planta 1997, 204, 27–36. [Google Scholar] [CrossRef]
- Curmi, P.M.; Cascio, D.; Sweet, R.M.; Eisenberg, D.; Schreuder, H. Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-A resolution. J. Biol. Chem. 1992, 267, 16980–16989. [Google Scholar]
- Newman, J.; Branden, C.I.; Jones, T.A. Structure determination and refinement of ribulose 1,5-bisphosphate carboxylase/oxygenase from Synechococcus PCC6301. Acta Crystallogr. D Biol. Crystallogr. 1993, 49, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.C.; Backlund, A.; Bjorhall, K.; Spreitzer, R.J.; Andersson, I. First crystal structure of Rubisco from a green alga, Chlamydomonas reinhardtii. J. Biol. Chem. 2001, 276, 48159–48164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uemura, K.; Miyachi, S.; Yokota, A. Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase from Thermophilic Red Algae with a Strong Specificity for CO 2 Fixation. Biochem. Biophys. Res. Commun. 1997, 571, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Kuchitsu, K.; Tsuzuki, M.; Miyachi, S. Polypeptide composition and enzyme activities of the pyrenoid and its regulation by CO2 concentration in unicellular green algae. Can. J. Bot. 1991, 69, 1062–1069. [Google Scholar] [CrossRef]
- Ryan, P.; Forrester, T.J.B.; Wroblewski, C.; Kenney, T.M.G.; Kitova, E.N.; Klassen, J.S.; Kimber, M.S. The small RbcS-like domains of the β-carboxysome structural protein CcmM bind RubisCO at a site distinct from that binding the RbcS subunit. J. Biol. Chem. 2019, 294, 2593–2603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, P.; Ruban, A.V.; Walters, R.G. Regulation of Light Harvesting In Green Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 655–684. [Google Scholar] [CrossRef]
- Allorent, G.; Tokutsu, R.; Roach, T.; Peers, G.; Cardol, P.; Girard-Bascou, J.; Seigneurin-Berny, D.; Petroutsos, D.; Kuntz, M.; Breyton, C.; et al. Dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell 2013, 25, 545–557. [Google Scholar] [CrossRef] [Green Version]
- Dall’Osto, L.; Caffarri, S.; Bassi, R. A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 2005, 17, 1217–1232. [Google Scholar] [CrossRef] [Green Version]
- Quaas, T.; Berteotti, S.; Ballottari, M.; Flieger, K.; Bassi, R.; Wilhelm, C.; Goss, R. Non-photochemical quenching and xanthophyll cycle activities in six green algal species suggest mechanistic differences in the process of excess energy dissipation. J. Plant Physiol. 2015, 172, 92–103. [Google Scholar] [CrossRef]
- Alboresi, A.; Gerotto, C.; Giacometti, G.M.; Bassi, R.; Morosinotto, T. Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc. Natl. Acad. Sci. USA 2010, 107, 11128–11133. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-P.; Björkman, O.; Shih, C.; Grossman, A.R.; Rosenquist, M.; Jansson, S.; Niyogi, K.K. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 2000, 403, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Li, M.; Liu, Z.; Cao, P.; Pan, X.; Zhang, H.; Zhao, X.; Zhang, J.; Chang, W. Crystal structures of the PsbS protein essential for photoprotection in plants. Nat. Struct. AMP Mol. Biol. 2015, 22, 729. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-P.; Gilmore, A.M.; Caffarri, S.; Bassi, R.; Golan, T.; Kramer, D.; Niyogi, K.K. Regulation of Photosynthetic Light Harvesting Involves Intrathylakoid Lumen pH Sensing by the PsbS Protein. J. Biol. Chem. 2004, 279, 22866–22874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballottari, M.; Truong, T.B.; De Re, E.; Erickson, E.; Stella, G.R.; Fleming, G.R.; Bassi, R.; Niyogi, K.K. Identification of pH-sensing Sites in the Light Harvesting Complex Stress-related 3 Protein Essential for Triggering Non-photochemical Quenching in Chlamydomonas reinhardtii. J. Biol. Chem. 2016, 291, 7334–7346. [Google Scholar] [CrossRef] [Green Version]
- Gabilly, S.T.; Baker, C.R.; Wakao, S.; Crisanto, T.; Guan, K.; Bi, K.; Guiet, E.; Guadagno, C.R.; Niyogi, K.K. Regulation of photoprotection gene expression in Chlamydomonas by a putative E3 ubiquitin ligase complex and a homolog of CONSTANS. Proc. Natl. Acad. Sci. USA 2019, 116, 11556–11562. [Google Scholar] [CrossRef] [Green Version]
- Petroutsos, D.; Busch, A.; Janssen, I.; Trompelt, K.; Bergner, S.V.; Weinl, S.; Holtkamp, M.; Karst, U.; Kudla, J.; Hippler, M. The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell 2011, 23, 2950–2963. [Google Scholar] [CrossRef] [Green Version]
- Pinnola, A.; Staleva-Musto, H.; Capaldi, S.; Ballottari, M.; Bassi, R.; Polívka, T. Electron transfer between carotenoid and chlorophyll contributes to quenching in the LHCSR1 protein from Physcomitrella patens. Biochim. Biophys. Acta-Bioenerg. 2016, 1857, 1870–1878. [Google Scholar] [CrossRef]
- Kondo, T.; Pinnola, A.; Chen, W.J.; Dall’Osto, L.; Bassi, R.; Schlau-Cohen, G.S. Single-molecule spectroscopy of LHCSR1 protein dynamics identifies two distinct states responsible for multi-timescale photosynthetic photoprotection. Nat. Chem. 2017, 9, 772. [Google Scholar] [CrossRef]
- Kondo, T.; Gordon, J.B.; Pinnola, A.; Dall’Osto, L.; Bassi, R.; Schlau-Cohen, G.S. Microsecond and millisecond dynamics in the photosynthetic protein LHCSR1 observed by single-molecule correlation spectroscopy. Proc. Natl. Acad. Sci. USA 2019, 2018, 21207. [Google Scholar] [CrossRef] [Green Version]
- Ahn, T.K.; Avenson, T.J.; Ballottari, M.; Cheng, Y.-C.; Niyogi, K.K.; Bassi, R.; Fleming, G.R. Architecture of a Charge-Transfer State Regulating Light Harvesting in a Plant Antenna Protein. Science (80-) 2008, 320, 794–797. [Google Scholar] [CrossRef] [Green Version]
- Holt, N.E.; Zigmantas, D.; Valkunas, L.; Li, X.-P.; Niyogi, K.K.; Fleming, G.R. Carotenoid Cation Formation and the Regulation of Photosynthetic Light Harvesting. Science (80-) 2005, 307, 433–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonente, G.; Ballottari, M.; Truong, T.B.; Morosinotto, T.; Ahn, T.K.; Fleming, G.R.; Niyogi, K.K.; Bassi, R. Analysis of LhcSR3, a Protein Essential for Feedback De-Excitation in the Green Alga Chlamydomonas reinhardtii. PLoS Biol. 2011, 9, e1000577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuciman, M.; Enriquez, M.M.; Polívka, T.; Dall’Osto, L.; Bassi, R.; Frank, H.A. Role of Xanthophylls in Light Harvesting in Green Plants: A Spectroscopic Investigation of Mutant LHCII and Lhcb Pigment–Protein Complexes. J. Phys. Chem. B 2012, 116, 3834–3849. [Google Scholar] [CrossRef] [PubMed]
- Ruban, A.V.; Berera, R.; Ilioaia, C.; van Stokkum, I.H.M.; Kennis, J.T.M.; Pascal, A.A.; van Amerongen, H.; Robert, B.; Horton, P.; van Grondelle, R. Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 2007, 450, 575. [Google Scholar] [CrossRef]
- Li, Z.; Peers, G.; Dent, R.M.; Bai, Y.; Yang, S.Y.; Apel, W.; Leonelli, L.; Niyogi, K.K. Evolution of an atypical de-epoxidase for photoprotection in the green lineage. Nat. Plants 2016, 2, 16140. [Google Scholar] [CrossRef] [Green Version]
- Niyogi, K.K. PHOTOPROTECTION REVISITED: Genetic and Molecular Approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 333–359. [Google Scholar] [CrossRef]
- Shapiguzov, A.; Chai, X.; Fucile, G.; Longoni, P.; Zhang, L.; Rochaix, J.D. Activation of the Stt7/STN7 Kinase through dynamic interactions with the cytochrome b6 f complex. Plant Physiol. 2016, 171, 82–92. [Google Scholar] [CrossRef] [Green Version]
- Gilmore, A.M.; Ball, M.C. Protection and storage of chlorophyll in overwintering evergreens. Proc. Natl. Acad. Sci. USA 2000. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.M.; Chow, W.S.; Park, Y.-I. The grand design of photosynthesis: Acclimation of the photosynthetic apparatus to environmental cues. Photosynth. Res. 1995, 46, 129–139. [Google Scholar] [CrossRef]
- Durnford, D.; Price, J.A.; McKim, S.M.; Sarchfield, M.L. Light-harvesting complex gene expression is controlled by both transcriptional and post-transcriptional mechanisms during photoacclimation in Chlamydomonas reinhardtii. Physiol. Plant. 2003, 118, 193–205. [Google Scholar] [CrossRef]
- Escoubas, J.M.; Lomas, M.; LaRoche, J.; Falkowski, P.G. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. USA 1995, 92, 10237–10241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonente, G.; Pippa, S.; Castellano, S.; Bassi, R.; Ballottari, M. Acclimation of Chlamydomonas reinhardtii to different growth irradiances. J. Biol. Chem. 2012, 287, 5833–5847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mettler, T.; Mühlhaus, T.; Hemme, D.; Schöttler, M.A.; Rupprecht, J.; Idoine, A.; Veyel, D.; Pal, S.K.; Yaneva-Roder, L.; Winck, F.V.; et al. Systems Analysis of the Response of Photosynthesis, Metabolism, and Growth to an Increase in Irradiance in the Photosynthetic Model Organism Chlamydomonas reinhardtii. Plant Cell 2014, 26, 2310–2350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballottari, M.; Dall’Osto, L.; Morosinotto, T.; Bassi, R. Contrasting Behavior of Higher Plant Photosystem I and II Antenna Systems during Acclimation. J. Biol. Chem. 2007, 282, 8947–8958. [Google Scholar] [CrossRef] [Green Version]
- Ferrante, P.; Ballottari, M.; Bonente, G.; Giuliano, G.; Bassi, R. LHCBM1 and LHCBM2/7 polypeptides, components of major LHCII complex, have distinct functional roles in photosynthetic antenna system of Chlamydomonas reinhardtii. J. Biol. Chem. 2012, 287, 16276–16288. [Google Scholar] [CrossRef] [Green Version]
- Elrad, D.; Niyogi, K.K.; Grossman, A.R. A Major Light-Harvesting Polypeptide of Photosystem II Functions in Thermal Dissipation. Plant Cell 2002, 14, 1801–1816. [Google Scholar] [CrossRef] [Green Version]
- Grewe, S.; Ballottari, M.; Alcocer, M.; D’Andrea, C.; Blifernez-Klassen, O.; Hankamer, B.; Mussgnug, J.H.; Bassi, R.; Kruse, O. Light-Harvesting Complex Protein LHCBM9 Is Critical for Photosystem II Activity and Hydrogen Production in Chlamydomonas reinhardtii. Plant Cell 2014, 26, 1598–1611. [Google Scholar] [CrossRef] [Green Version]
- Mussgnug, J.H.; Wobbe, L.; Elles, I.; Claus, C.; Hamilton, M.; Fink, A.; Kahmann, U.; Kapazoglou, A.; Mullineaux, C.W.; Hippler, M.; et al. NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii. Plant Cell 2005, 17, 3409–3421. [Google Scholar] [CrossRef] [Green Version]
- Polle, J.E.W.; Kanakagiri, S.-D.; Melis, A. tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size. Planta 2003, 217, 49–59. [Google Scholar] [CrossRef]
- Mitra, M.; Kirst, H.; Dewez, D.; Melis, A. Modulation of the light-harvesting chlorophyll antenna size in Chlamydomonas reinhardtii by TLA1 gene over-expression and RNA interference. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 3430–3443. [Google Scholar] [CrossRef] [Green Version]
- Kirst, H.; García-Cerdán, J.G.; Zurbriggen, A.; Melis, A. Assembly of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii requires expression of the TLA2-CpFTSY gene. Plant Physiol. 2012, 158, 930–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirst, H.; Melis, A. The chloroplast signal recognition particle (CpSRP) pathway as a tool to minimize chlorophyll antenna size and maximize photosynthetic productivity. Biotechnol. Adv. 2014, 32, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.; Kim, D.H.; Jeong, J.; Sim, S.J.; Melis, A.; Kim, J.S.; Jin, E.; Bae, S. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 2016, 6, 30620. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Baek, K.; Yu, J.; Kirst, H.; Betterle, N.; Shin, W.; Bae, S.; Melis, A.; Jin, E. Deletion of the chloroplast LTD protein impedes LHCI import and PSI-LHCI assembly in Chlamydomonas reinhardtii. J. Exp. Bot. 2018, 69, 1147–1158. [Google Scholar] [CrossRef] [Green Version]
- Perin, G.; Bellan, A.; Segalla, A.; Meneghesso, A.; Alboresi, A.; Morosinotto, T. Generation of random mutants to improve light-use efficiency of Nannochloropsis gaditana cultures for biofuel production. Biotechnol. Biofuels 2015, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Ito, H.; Tanaka, R.; Tanaka, N.K.; Yoshida, K.; Okada, K. Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc. Natl. Acad. Sci. USA 1998, 95, 12719–12723. [Google Scholar] [CrossRef] [Green Version]
- Bujaldon, S.; Kodama, N.; Rappaport, F.; Subramanyam, R.; de Vitry, C.; Takahashi, Y.; Wollman, F.A. Functional Accumulation of Antenna Proteins in Chlorophyll b-Less Mutants of Chlamydomonas reinhardtii. Mol. Plant 2017, 10, 115–130. [Google Scholar] [CrossRef] [Green Version]
- Perrine, Z.; Negi, S.; Sayre, R.T. Optimization of photosynthetic light energy utilization by microalgae. Algal Res. 2012, 1, 134–142. [Google Scholar] [CrossRef] [Green Version]
- Dall’Osto, L.; Cazzaniga, S.; Guardini, Z.; Barera, S.; Benedetti, M.; Mannino, G.; Maffei, M.E.; Bassi, R. Combined resistance to oxidative stress and reduced antenna size enhance light-to-biomass conversion efficiency in Chlorella vulgaris cultures. Biotechnol. Biofuels 2019, 12, 221. [Google Scholar] [CrossRef] [Green Version]
- Kromdijk, J.; Głowacka, K.; Leonelli, L.; Gabilly, S.T.; Iwai, M.; Niyogi, K.K.; Long, S.P. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science (80-) 2016, 354, 857–861. [Google Scholar] [CrossRef] [Green Version]
- Förster, B.; Osmond, C.B.; Pogson, B.J. Improved survival of very high light and oxidative stress is conferred by spontaneous gain-of-function mutations in Chlamydomonas. Biochim. Biophys. Acta-Bioenerg. 2005, 1709, 45–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, B.B.; Ledford, H.K.; Wakao, S.; Huang, S.G.; Casero, D.; Pellegrini, M.; Merchant, S.S.; Koller, A.; Eggen, R.I.L.; Niyogi, K.K. Singlet Oxygen Resistant 1 links reactive electrophile signaling to singlet oxygen acclimation in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2012, 109, E1302–E1311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schierenbeck, L.; Ries, D.; Rogge, K.; Grewe, S.; Weisshaar, B.; Kruse, O. Fast forward genetics to identify mutations causing a high light tolerant phenotype in Chlamydomonas reinhardtii by whole-genome-sequencing. BMC Genom. 2015, 16, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treves, H.; Raanan, H.; Finkel, O.M.; Berkowicz, S.M.; Keren, N.; Shotland, Y.; Kaplan, A. A newly isolated Chlorella sp. from desert sand crusts exhibits a unique resistance to excess light intensity. FEMS Microbiol. Ecol. 2013, 86, 373–380. [Google Scholar] [CrossRef] [Green Version]
- Treves, H.; Raanan, H.; Kedem, I.; Murik, O.; Keren, N.; Zer, H.; Berkowicz, S.M.; Giordano, M.; Norici, A.; Shotland, Y.; et al. The mechanisms whereby the green alga Chlorella ohadii, isolated from desert soil crust, exhibits unparalleled photodamage resistance. New Phytol. 2016, 210, 1229–1243. [Google Scholar] [CrossRef] [Green Version]
- Whitney, S.M.; Houtz, R.L.; Alonso, H. Advancing Our Understanding and Capacity to Engineer Nature’s CO2-Sequestering Enzyme, Rubisco. Plant Physiol. 2011, 155, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Karkehabadi, S.; Peddi, S.R.; Anwaruzzaman, M.; Taylor, T.C.; Cederlund, A.; Genkov, T.; Andersson, I.; Spreitzer, R.J. Chimeric small subunits influence catalysis without causing global conformational changes in the crystal structure of ribulose-1,5-bisphosphate carboxylase/oxygenase. Biochemistry 2005, 44, 9851–9861. [Google Scholar] [CrossRef]
- Genkov, T.; Meyer, M.; Griffiths, H.; Spreitzer, R.J. Functional Hybrid Rubisco Enzymes with Plant Small Subunits and Algal Large Subunits: Engineered rbcS cDNA for expression in Chlamydomonas. J. Biol. Chem. 2010, 285, 19833–19841. [Google Scholar] [CrossRef] [Green Version]
- Larson, E.M.; O’Brien, C.M.; Zhu, G.; Spreitzer, R.J.; Portis, A.R. Specificity for activase is changed by a Pro-89 to Arg substitution in the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Biol. Chem. 1997, 272, 17033–17037. [Google Scholar] [CrossRef] [Green Version]
- Ott, C.M.; Smith, B.D.; Portis, A.R.; Spreitzer, R.J. Activase region on chloroplast ribulose-1,5-bisphosphate carboxylase/oxygenase: Nonconservative substitution in the large subunit alters species specificity of protein interaction. J. Biol. Chem. 2000, 275, 26241–26244. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Salvucci, M.E.; Portis, A.R. Two residues of Rubisco activase involved in recognition of the Rubisco substrate. J. Biol. Chem. 2005, 280, 24864–24869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; Wang, Q.; Xin, Y.; Lu, Y.; Xu, J. Enhancing photosynthetic biomass productivity of industrial oleaginous microalgae by overexpression of RuBisCO activase. Algal Res. 2017, 27, 366–375. [Google Scholar] [CrossRef]
- Liang, F.; Lindblad, P. Synechocystis PCC 6803 overexpressing RuBisCO grow faster with increased photosynthesis. Metab. Eng. Commun. 2017, 4, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Ruffing, A.M. Improved Free Fatty Acid Production in Cyanobacteria with Synechococcus sp. PCC 7002 as Host. Front. Bioeng. Biotechnol. 2014, 2, 17. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Kurek, I.; Liu, L. Chapter 20 Engineering Photosynthetic Enzymes Involved in CO2–Assimilation by Gene Shuffling BT-The Chloroplast: Basics and Applications; Rebeiz, C.A., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 307–322. [Google Scholar] [CrossRef]
- Pinto, T.S.; Malcata, F.X.; Arrabaça, J.D.; Silva, J.M.; Spreitzer, R.J.; Esquível, M.G. Rubisco mutants of Chlamydomonas reinhardtii enhance photosynthetic hydrogen production. Appl. Microbiol. Biotechnol. 2013, 97, 5635–5643. [Google Scholar] [CrossRef]
- Wunder, T.; Cheng, S.L.H.; Lai, S.-K.; Li, H.-Y.; Mueller-Cajar, O. The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nat. Commun. 2018, 9, 5076. [Google Scholar] [CrossRef]
- Fang, L.; Lin, H.X.; Low, C.S.; Wu, M.H.; Chow, Y.; Lee, Y.K. Expression of the Chlamydomonas reinhardtii Sedoheptulose-1,7-bisphosphatase in Dunaliella bardawil leads to enhanced photosynthesis and increased glycerol production. Plant Biotechnol. J. 2012, 10, 1129–1135. [Google Scholar] [CrossRef]
- Liang, F.; Lindblad, P. Effects of overexpressing photosynthetic carbon flux control enzymes in the cyanobacterium Synechocystis PCC 6803. Metab. Eng. 2016, 38, 56–64. [Google Scholar] [CrossRef]
- Liang, F.; Englund, E.; Lindberg, P.; Lindblad, P. Engineered cyanobacteria with enhanced growth show increased ethanol production and higher biofuel to biomass ratio. Metab. Eng. 2018, 46, 51–59. [Google Scholar] [CrossRef]
- Spalding, M.H. Modulation of Low Carbon Dioxide Inducible Proteins (lci) for Increased Biomass Production and Photosynthesis. U.S. Patent Application No. 13/535,842, 3 January 2013. [Google Scholar]
- Kao, P.H.; Ng, I.S. CRISPRi mediated phosphoenolpyruvate carboxylase regulation to enhance the production of lipid in Chlamydomonas reinhardtii. Bioresour. Technol. 2017, 245, 1527–1537. [Google Scholar] [CrossRef]
- Courchesne, N.M.D.; Parisien, A.; Wang, B.; Lan, C.Q. Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J. Biotechnol. 2009, 141, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Leu, S.; Boussiba, S. Advances in the Production of High-Value Products by Microalgae. Ind. Biotechnol. 2014, 10, 169–183. [Google Scholar] [CrossRef]
- Li, D.; Xie, W.; Hao, T.; Cai, J.; Zhou, T.; Balamurugan, S. Constitutive and Chloroplast Targeted Expression of Acetyl-CoA Carboxylase in Oleaginous Microalgae Elevates Fatty Acid Biosynthesis. Mar. Biotechnol. (NY) 2018, 20, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, W.; Hu, D.; Wang, X. Identification of a malonyl CoA-acyl carrier protein transacylase and its regulatory role in fatty acid biosynthesis in oleaginous microalga Nannochloropsis oceanica. Biotechnol. Appl. Biochem. 2017, 64, 620–626. [Google Scholar] [CrossRef]
- Li, Z.; Meng, T.; Ling, X.; Li, J.; Zheng, C.; Shi, Y.; Chen, Z.; Li, Z.; Li, Q.; Lu, Y.; et al. Overexpression of Malonyl-CoA: ACP Transacylase in Schizochytrium sp. to Improve Polyunsaturated Fatty Acid Production. J. Agric. Food Chem. 2018, 66, 5382–5391. [Google Scholar] [CrossRef]
- Wei, K.; Tan, M.; Lee, Y.K. Expression of the heterologous Dunaliella tertiolecta fatty acyl-ACP thioesterase leads to increased lipid production in Chlamydomonas reinhardtii. J. Biotechnol. 2017, 247, 60–67. [Google Scholar]
- Radakovits, R.; Eduafo, P.M.; Posewitz, M.C. Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab. Eng. 2011, 13, 89–95. [Google Scholar] [CrossRef]
- Lin, H.; Lee, Y.K. Genetic engineering of medium-chain-length fatty acid synthesis in Dunaliella tertiolecta for improved biodiesel production. J. Appl. Phycol. 2017, 29, 2811–2819. [Google Scholar] [CrossRef] [Green Version]
- Napier, J.A. Plumbing the depths of PUFA biosynthesis: A novel polyketide synthase-like pathway from marine organisms. Trends Plant Sci. 2002, 7, 51–54. [Google Scholar] [CrossRef]
- Kaye, Y.; Grundman, O.; Leu, S.; Zarka, A.; Zorin, B.; Didi-Cohen, S.; Khozin-Goldberg, I.; Boussiba, S. Metabolic engineering toward enhanced LC-PUFA biosynthesis in Nannochloropsis oceanica: Overexpression of endogenous Δ12 desaturase driven by stress-inducible promoter leads to enhanced deposition of polyunsaturated fatty acids in TAG. Algal Res. 2015, 11, 387–398. [Google Scholar] [CrossRef]
- Hamilton, M.L.; Haslam, R.P.; Napier, J.A.; Sayanova, O. Metabolic engineering of Phaeodactylum tricornutum for the enhanced accumulation of omega-3 long chain polyunsaturated fatty acids. Metab. Eng. 2014, 22, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.-T.; Zheng, C.-N.; Xue, J.; Chen, X.-Y.; Yang, W.-D.; Liu, J.-S.; Bai, W.; Li, H.-Y. Delta 5 Fatty Acid Desaturase Upregulates the Synthesis of Polyunsaturated Fatty Acids in the Marine Diatom Phaeodactylum tricornutum. J. Agric. Food Chem. 2014, 62, 8773–8776. [Google Scholar] [CrossRef]
- Zäuner, S.; Jochum, W.; Bigorowski, T.; Benning, C. A cytochrome b5-containing plastid-located fatty acid desaturase from Chlamydomonas reinhardtii. Eukaryot. Cell 2012, 11, 856–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, M.; Jiang, J. Progress in Lipid Research Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog. Lipid Res. 2013, 52, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Halim, R.; Danquah, M.K.; Webley, P.A. Extraction of oil from microalgae for biodiesel production: A review. Biotechnol. Adv. 2012, 30, 709–732. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.G.; Chen, J.W.; Zheng, D.L.; Balamurugan, S.; Li, D.W.; Yang, W.D.; Liu, J.S. High-efficiency promoter-driven coordinated regulation of multiple metabolic nodes elevates lipid accumulation in the model microalga Phaeodactylum tricornutum. Microb. Cell Fact. 2018, 17, 54. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.F.; Wang, X.; Hu, D.X.; Balamurugan, S.; Li, D.W. Molecular characterization of a glycerol-3-phosphate acyltransferase reveals key features essential for triacylglycerol production in Phaeodactylum tricornutum. Biotechnol. Biofuels 2016, 9, 60. [Google Scholar] [CrossRef] [Green Version]
- Yamaoka, Y.; Achard, D.; Jang, S.; Leg, B.; Kamisuki, S.; Ko, D.; Schulz-raffelt, M.; Kim, Y.; Song, W.; Nishida, I.; et al. Identification of a Chlamydomonas plastidial 2-lysophosphatidic acid acyltransferase and its use to engineer microalgae with increased oil content. Plant Biotechnol. J. 2016, 14, 2158–2167. [Google Scholar] [CrossRef]
- Balamurugan, S.; Wang, X.; Wang, H.-L.; An, C.-J.; Li, H.; Li, D.-W.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Occurrence of plastidial triacylglycerol synthesis and the potential regulatory role of AGPAT in the model diatom Phaeodactylum tricornutum. Biotechnol. Biofuels 2017, 10, 97. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Lu, J.; Deng, X.; Li, H.; Hu, Z. Effect of overexpression of LPAAT and GPD1 on lipid synthesis and composition in green microalga Chlamydomonas reinhardtii. J. Appl. Phycol. 2018, 30, 1711–1719. [Google Scholar] [CrossRef] [Green Version]
- Cases, S.; Smith, S.J.; Zheng, Y.W.; Myers, H.M.; Lear, S.R.; Sande, E.; Novak, S.; Collins, C.; Welch, C.B.; Lusis, A.J.; et al. Identification of a gene encoding an acyl CoA: Diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 1998, 95, 13018–13023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Zhao, J.; Wang, Y.; Qin, S.; Lu, Y. Characterization and engineering of a dual-function diacylglycerol acyltransferase in the oleaginous marine diatom Phaeodactylum tricornutum. Biotechnol. Biofuels 2018, 11, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, H.; Shi, Y.; Ma, X.; Pan, Y.; Hu, H.; Li, Y.; Luo, M.; Gerken, H.; Liu, J. A type-I diacylglycerol acyltransferase modulates triacylglycerol biosynthesis and fatty acid composition in the oleaginous microalga, Nannochloropsis oceanica. Biotechnol. Biofuels 2017, 10, 174. [Google Scholar] [CrossRef] [PubMed]
- Li, D.-W.; Cen, S.-Y.; Liu, Y.-H.; Balamurugan, S.; Zheng, X.-Y.; Alimujiang, A.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. A type 2 diacylglycerol acyltransferase accelerates the triacylglycerol biosynthesis in heterokont oleaginous microalga Nannochloropsis oceanica. J. Biotechnol. 2016, 229, 65–71. [Google Scholar] [CrossRef]
- Yan, J.; Cheng, R.; Lin, X.; You, S.; Li, K.; Rong, H.; Ma, Y. Overexpression of acetyl-CoA synthetase increased the biomass and fatty acid proportion in microalga Schizochytrium. Appl. Microbiol. Biotechnol. 2013, 97, 1933–1939. [Google Scholar] [CrossRef]
- Ma, Y.-H.; Wang, X.; Niu, Y.-F.; Yang, Z.-K.; Zhang, M.-H.; Wang, Z.-M.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Antisense knockdown of pyruvate dehydrogenase kinase promotes the neutral lipid accumulation in the diatom Phaeodactylum tricornutum. Microb. Cell Fact. 2014, 13, 100. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.; Niu, Y.-F.; Huang, T.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Genetic improvement of the microalga Phaeodactylum tricornutum for boosting neutral lipid accumulation. Metab. Eng. 2015, 27, 1–9. [Google Scholar] [CrossRef]
- Xue, J.; Balamurugan, S.; Li, D.-W.; Liu, Y.-H.; Zeng, H.; Wang, L.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Glucose-6-phosphate dehydrogenase as a target for highly efficient fatty acid biosynthesis in microalgae by enhancing NADPH supply. Metab. Eng. 2017, 41, 212–221. [Google Scholar] [CrossRef]
- Sun, X.-M.; Ren, L.-J.; Zhao, Q.-Y.; Ji, X.-J.; Huang, H. Enhancement of lipid accumulation in microalgae by metabolic engineering. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2019, 1864, 552–566. [Google Scholar] [CrossRef]
- Tian, Q.-L.; Shi, D.-J.; Jia, X.-H.; Mi, H.-L.; Huang, X.-W.; He, P.-M. Recombinant expression and functional analysis of a Chlamydomonas reinhardtii bacterial-type phosphoenolpyruvate carboxylase gene fragment. Biotechnol. Lett. 2014, 36, 821–827. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Chen, X.; Li, H.; Wang, J.; Hu, Z. Artificial miRNA inhibition of phosphoenolpyruvate carboxylase increases fatty acid production in a green microalga Chlamydomonas reinhardtii. Biotechnol. Biofuels 2017, 10, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Pan, Y.; Bowler, C.; Zhang, L.; Hu, H. Knockdown of phosphoenolpyruvate carboxykinase increases carbon flux to lipid synthesis in Phaeodactylum tricornutum. Algal Res. 2016, 15, 50–58. [Google Scholar] [CrossRef]
- Nobusawa, T.; Yamakawa-Ayukawa, K.; Saito, F.; Nomura, S.; Takami, A.; Ohta, H. A homolog of Arabidopsis SDP1 lipase in Nannochloropsis is involved in degradation of de novo-synthesized triacylglycerols in the endoplasmic reticulum. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2019, 1864, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez-Salazar, A.; Rosales-Mendoza, S.; Rocha-Uribe, A.; Ramírez-Alonso, J.I.; Lara-Hernández, I.; Hernández-Torres, A.; Paz-Maldonado, L.M.T.; Silva-Ramírez, A.S.; Bañuelos-Hernández, B.; Martínez-Salgado, J.L.; et al. Over-expression of Dof-type transcription factor increases lipid production in Chlamydomonas reinhardtii. J. Biotechnol. 2014, 184, 27–38. [Google Scholar] [CrossRef]
- Salas-Montantes, C.J.; González-Ortega, O.; Ochoa-Alfaro, A.E.; Camarena-Rangel, R.; Paz-Maldonado, L.M.T.; Rosales-Mendoza, S.; Rocha-Uribe, A.; Soria-Guerra, R.E. Lipid accumulation during nitrogen and sulfur starvation in Chlamydomonas reinhardtii overexpressing a transcription factor. J. Appl. Phycol. 2018, 30, 1721–1733. [Google Scholar] [CrossRef]
- Kwon, S.; Kang, N.K.; Koh, H.G.; Shin, S.-E.; Lee, B.; Jeong, B.; Chang, Y.K. Enhancement of biomass and lipid productivity by overexpression of a bZIP transcription factor in Nannochloropsis salina. Biotechnol. Bioeng. 2018, 115, 331–340. [Google Scholar] [CrossRef] [Green Version]
- Kang, N.K.; Jeon, S.; Kwon, S.; Koh, H.G.; Shin, S.-E.; Lee, B.; Choi, G.-G.; Yang, J.-W.; Jeong, B.; Chang, Y.K. Effects of overexpression of a bHLH transcription factor on biomass and lipid production in Nannochloropsis salina. Biotechnol. Biofuels 2015, 8, 200. [Google Scholar] [CrossRef] [Green Version]
- Kang, N.K.; Kim, E.K.; Kim, Y.U.; Lee, B.; Jeong, W.-J.; Jeong, B.-R.; Chang, Y.K. Increased lipid production by heterologous expression of AtWRI1 transcription factor in Nannochloropsis salina. Biotechnol. Biofuels 2017, 10, 231. [Google Scholar] [CrossRef]
- Li, D.-W.; Balamurugan, S.; Yang, Y.-F.; Zheng, J.-W.; Huang, D.; Zou, L.-G.; Yang, W.-D.; Liu, J.-S.; Guan, Y.; Li, H.-Y. Transcriptional regulation of microalgae for concurrent lipid overproduction and secretion. Sci. Adv. 2019, 5, 3795. [Google Scholar] [CrossRef] [Green Version]
- Ajjawi, I.; Verruto, J.; Aqui, M.; Soriaga, L.B.; Coppersmith, J.; Kwok, K.; Peach, L.; Orchard, E.; Kalb, R.; Xu, W.; et al. Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat. Publ. Gr. 2017, 35, 647–652. [Google Scholar] [CrossRef]
- Sharma, A.K.; Mühlroth, A.; Jouhet, J.; Maréchal, E.; Alipanah, L.; Kissen, R.; Brembu, T.; Bones, A.M.; Winge, P. The Myb-like transcription factor Phosphorus Starvation Response (PtPSR) controls conditional P acquisition and remodeling in marine microalgae. New Phytol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Torres-Romero, I.; Kong, F.; Légeret, B.; Beisson, F.; Peltier, G.; Li-Beisson, Y. Chlamydomonas cell cycle mutant crcdc5 over-accumulates starch and oil. Biochimie 2019. [Google Scholar] [CrossRef]
- Liu, J.; Sun, Z.; Mao, X.; Gerken, H.; Wang, X.; Yang, W. Multiomics analysis reveals a distinct mechanism of oleaginousness in the emerging model alga Chromochloris zofingiensis. Plant J. 2019, 98, 1060–1077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wen, F.; Xu, Z.; Sun, D.; Chew, W.; Liu, J. De novo transcriptomic analysis of the oleaginous alga Botryococcus braunii AC768 (Chlorophyta). J. Appl. Phycol. 2019, 31, 255–267. [Google Scholar] [CrossRef]
- D’Adamo, S.; Schiano di Visconte, G.; Lowe, G.; Szaub-Newton, J.; Beacham, T.; Landels, A.; Allen, M.J.; Spicer, A.; Matthijs, M. Engineering the unicellular alga Phaeodactylum tricornutum for high-value plant triterpenoid production. Plant Biotechnol. J. 2019, 17, 75–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, E.; Polle, J.E.W.; Lee, H.K.; Sang, M.; Man Chang, A. Xanthophylls in microalgae: From Biosynthesis to biotechnological mass production and application. J. Microbiol. Biotechnol. 2003, 13, 165–174. [Google Scholar]
- Jin, E.; Polle, J.E.W.; Melis, A. Involvement of zeaxanthin and of the Cbr protein in the repair of photosystem II from photoinhibition in the green alga Dunaliella salina. Biochim. Biophys. Acta-Bioenerg. 2001, 1506, 244–259. [Google Scholar] [CrossRef] [Green Version]
- Baek, K.L.; Yu, J.; Jeong, J.; Sim, S.J.; Bae, S.; Jin, E.S. Photoautotrophic production of macular pigment in a Chlamydomonas reinhardtii strain generated by using DNA-free CRISPR-Cas9 RNP-mediated mutagenesis. Biotechnol. Bioeng. 2018, 115, 719–728. [Google Scholar] [CrossRef]
- Steinbrenner, J.; Sandmann, G. Transformation of the Green Alga Haematococcus pluvialis with a Phytoene Desaturase for Accelerated Astaxanthin Biosynthesis. Appl. Environ. Microbiol. 2006, 72, 7477–7484. [Google Scholar] [CrossRef] [Green Version]
- Kathiresan, S.; Chandrashekar, A.; Ravishankar, G.A.; Sarada, R. Regulation of astaxanthin and its intermediates through cloning and genetic transformation of β-carotene ketolase in Haematococcus pluvialis. J. Biotechnol. 2015, 196–197, 33–41. [Google Scholar] [CrossRef]
- Galarza, J.I.; Gimpel, J.A.; Rojas, V.; Arredondo-vega, B.O. Over-accumulation of astaxanthin in Haematococcus pluvialis through chloroplast genetic engineering. Algal Res. 2018, 31, 291–297. [Google Scholar] [CrossRef]
- Couso, I.; Cordero, B.F.; Vargas, M.Á.; Rodríguez, H. Efficient heterologous transformation of Chlamydomonas reinhardtii npq2 mutant with the zeaxanthin epoxidase gene isolated and characterized from Chlorella zofingiensis. Mar. Drugs 2012, 10, 1955–1976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eilers, U.; Bikoulis, A.; Breitenbach, J.; Büchel, C.; Sandmann, G. Limitations in the biosynthesis of fucoxanthin as targets for genetic engineering in Phaeodactylum tricornutum. J. Appl. Phycol. 2016, 28, 123–129. [Google Scholar] [CrossRef]
- Lohr, M.; Schwender, J.; Polle, J.E.W. Isoprenoid biosynthesis in eukaryotic phototrophs: A spotlight on algae. Plant Sci. 2012, 185–186, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Lauersen, K.J.; Baier, T.; Wichmann, J.; Wördenweber, R.; Mussgnug, J.H.; Hübner, W.; Huser, T.; Kruse, O. Efficient phototrophic production of a high-value sesquiterpenoid from the eukaryotic microalga Chlamydomonas reinhardtii. Metab. Eng. 2016, 38, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Wichmann, J.; Baier, T.; Wentnagel, E.; Lauersen, K.J.; Kruse, O. Tailored carbon partitioning for phototrophic production of (E)-α-bisabolene from the green microalga Chlamydomonas reinhardtii. Metab. Eng. 2018, 45, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Lauersen, K.J.; Wichmann, J.; Baier, T.; Kampranis, S.C.; Pateraki, I.; Møller, B.L.; Kruse, O. Phototrophic production of heterologous diterpenoids and a hydroxy-functionalized derivative from Chlamydomonas reinhardtii. Metab. Eng. 2018, 49, 116–127. [Google Scholar] [CrossRef]
- Lin, P.-C.; Pakrasi, H.B. Engineering cyanobacteria for production of terpenoids. Planta 2019, 249, 145–154. [Google Scholar] [CrossRef]
- Nozzi, N.; Oliver, J.; Atsumi, S. Cyanobacteria as a Platform for Biofuel Production. Front. Bioeng. Biotechnol. 2013, 1, 7. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Gao, F.; Liu, D.; Zhang, H.; Nie, X.; Yang, C. Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2. Energy Environ. Sci. 2016, 9, 1400–1411. [Google Scholar] [CrossRef]
- Davies, F.K.; Work, V.H.; Beliaev, A.S.; Posewitz, M.C. Engineering Limonene and Bisabolene Production in Wild Type and a Glycogen-Deficient Mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2014, 2, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Liu, W.; Xin, C.; Zheng, Y.; Cheng, Y.; Sun, S.; Li, R.; Zhu, X.-G.; Dai, S.Y.; Rentzepis, P.M.; et al. Enhanced limonene production in cyanobacteria reveals photosynthesis limitations. Proc. Natl. Acad. Sci. USA 2016, 113, 14225–14230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, P.-C.; Saha, R.; Zhang, F.; Pakrasi, H.B. Metabolic engineering of the pentose phosphate pathway for enhanced limonene production in the cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2017, 7, 17503. [Google Scholar] [CrossRef] [PubMed]
- Lauersen, K.J. Eukaryotic microalgae as hosts for light-driven heterologous isoprenoid production. Planta 2018. [Google Scholar] [CrossRef]
- Baier, T.; Wichmann, J.; Kruse, O.; Lauersen, K.J. Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii. Nucleic Acids Res. 2018, 46, 6909–6919. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vecchi, V.; Barera, S.; Bassi, R.; Dall’Osto, L. Potential and Challenges of Improving Photosynthesis in Algae. Plants 2020, 9, 67. https://doi.org/10.3390/plants9010067
Vecchi V, Barera S, Bassi R, Dall’Osto L. Potential and Challenges of Improving Photosynthesis in Algae. Plants. 2020; 9(1):67. https://doi.org/10.3390/plants9010067
Chicago/Turabian StyleVecchi, Valeria, Simone Barera, Roberto Bassi, and Luca Dall’Osto. 2020. "Potential and Challenges of Improving Photosynthesis in Algae" Plants 9, no. 1: 67. https://doi.org/10.3390/plants9010067




