Fast Track to Discover Novel Promoters in Rice
Abstract
1. Introduction
2. Results and Discussion
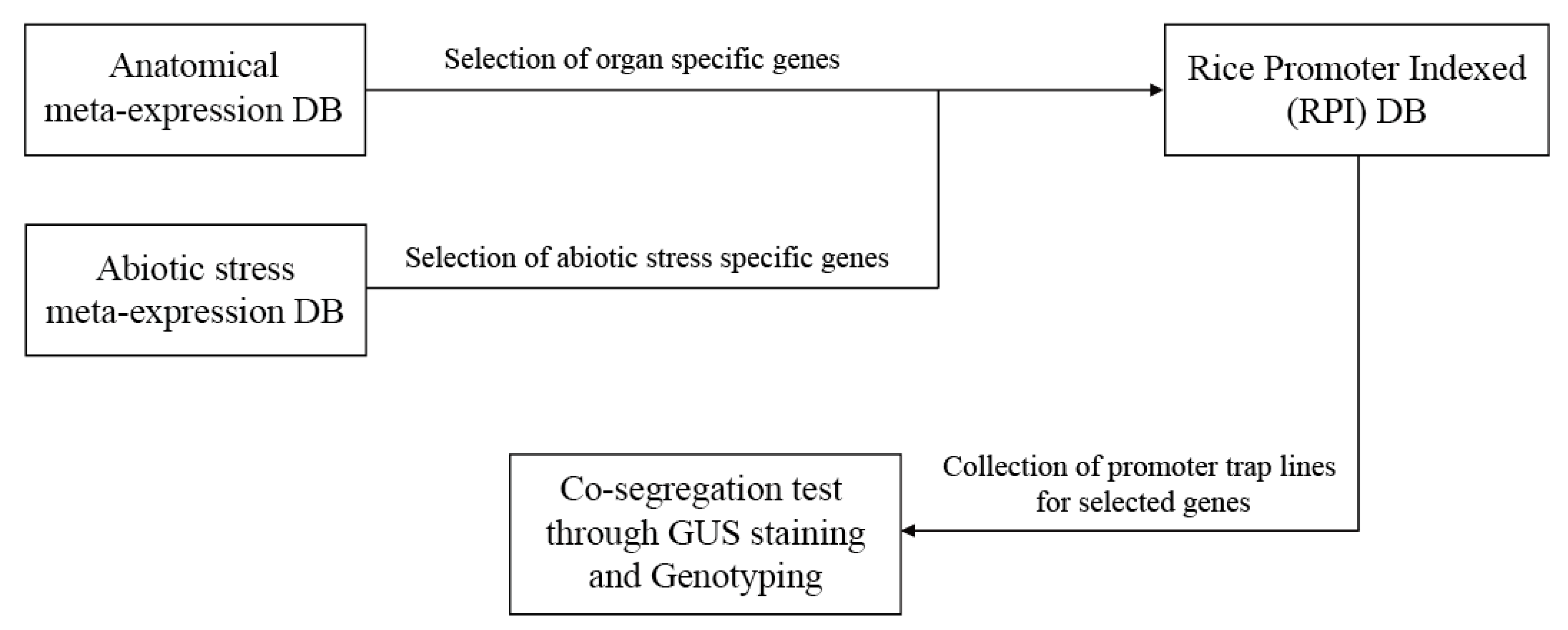
2.1. Summary of the Promoter Trap Line Analysis Process
2.2. Integration of Annotated Rice Genes from the Rice Genome Annotation Project
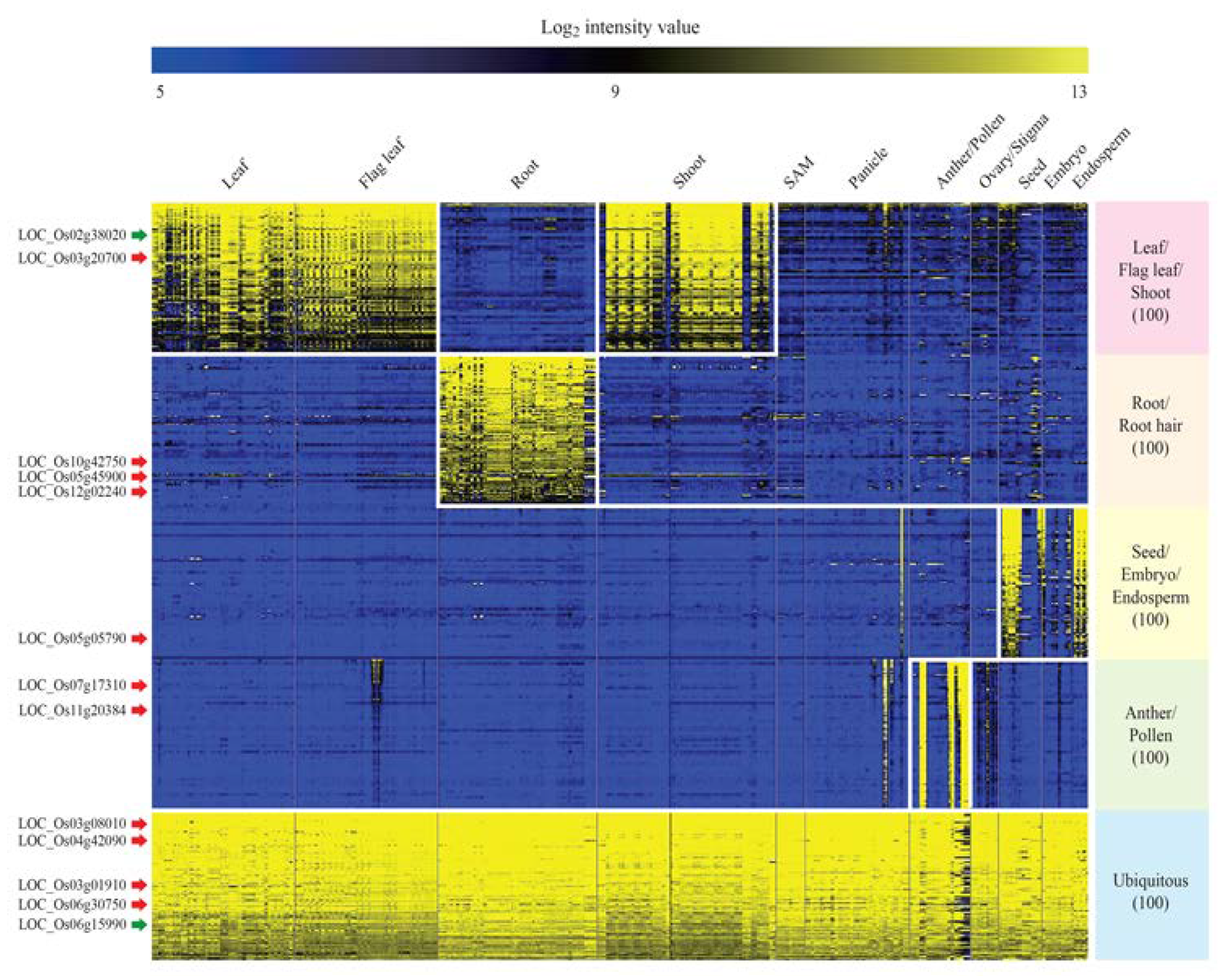
2.3. Identification of Tissue/Organ-Preferred Genes in Rice Using Meta-Expression Data
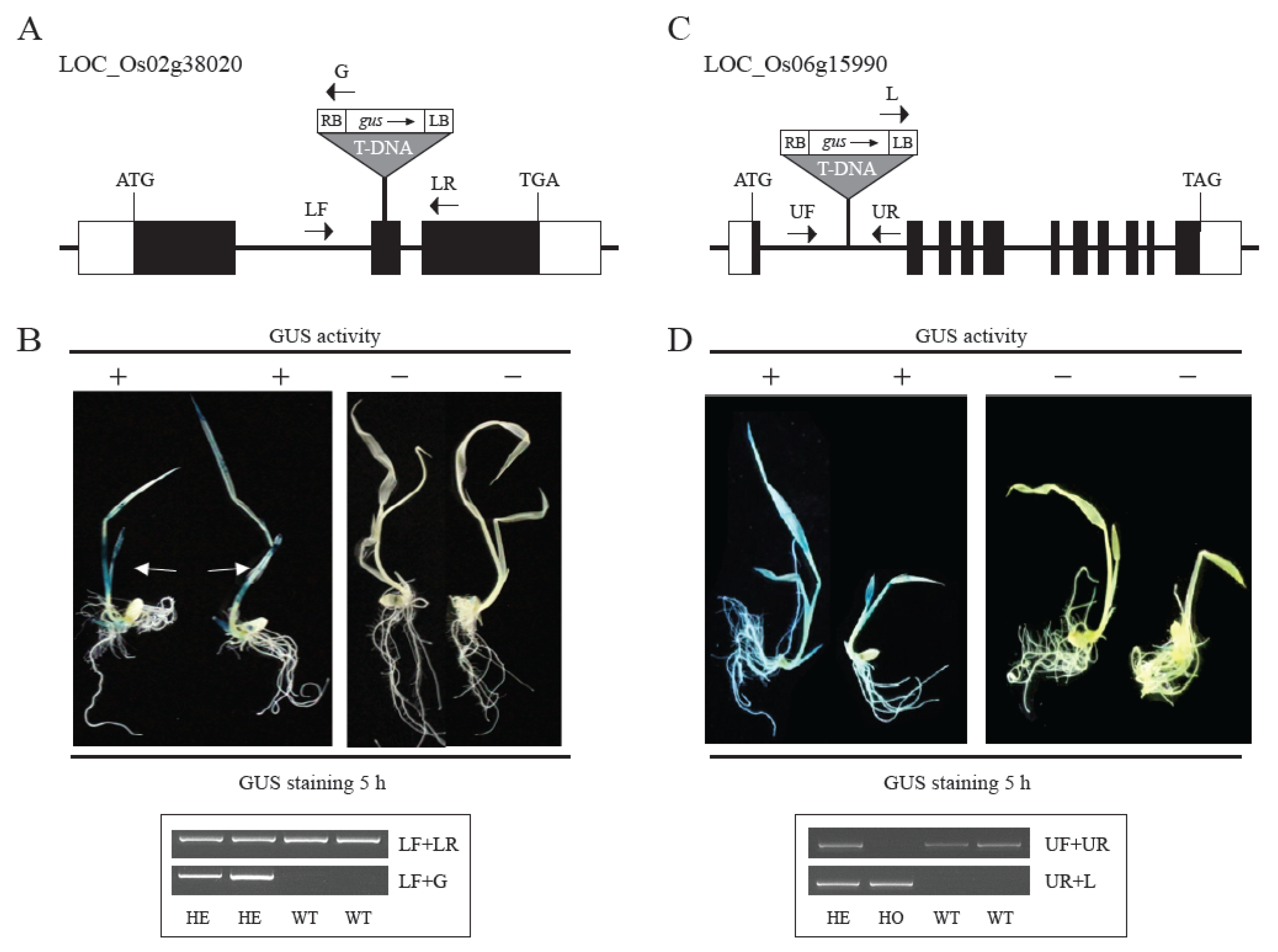
2.4. Validation of Promoters of Tissue/Organ-Preferential Genes Using the Promoter Trap System and Genotyping
2.5. Abiotic Stress Analyses of Rice Genes via Meta-Expression Data
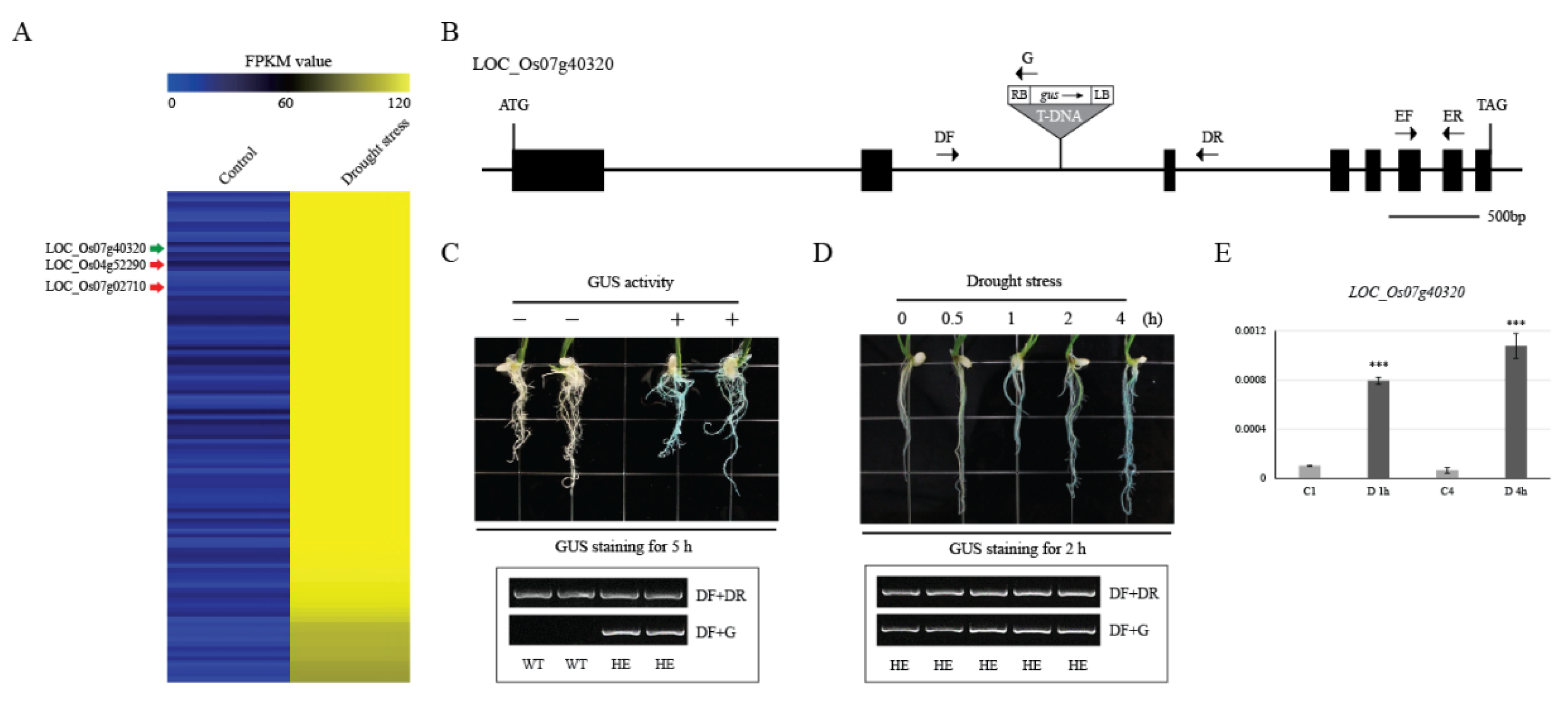
2.6. Evaluation of Promoter Trap Lines through a Literature Search
2.7. Validation of Drought-Inducible Genes Using the GUS Reporter System and qRT-PCR
3. Materials and Methods
3.1. Integration of Whole Rice Genes from Public Data Source
3.2. Collection of Transcriptome Data
3.3. Classification of Organ-Preferential or Abiotic Stress-Responsive Gene Groups
3.4. Histochemical GUS Assay
3.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
3.6. Analysis of Promoter Trap Lines via Literature Search
4. Conclusions and Future Prospects
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jung, K.H.; Hur, J.; Ryu, C.H.; Choi, Y.; Chung, Y.Y.; Miyao, A.; Hirochika, H.; An, G. Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol. 2003, 44, 463–472. [Google Scholar] [CrossRef]
- Moon, S.; Chandran, A.K.N.; An, G.; Lee, C.; Jung, K.H. Genome-wide analysis of root hair-preferential genes in rice. Rice 2018, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Oo, M.M.; Kim, B.; Koh, H.-J.; Oh, S.A.; Yi, G.; An, G.; Park, S.K.; Jung, K.-H. Genome-wide analyses of late pollen-preferred genes conserved in various rice cultivars and functional identification of a gene involved in the key processes of late pollen development. Rice 2018, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.-H.; Kim, S.-R.; Giong, H.-K.; Nguyen, M.X.; Koh, H.-J.; An, G. Genome-wide identification and functional analysis of genes expressed ubiquitously in rice. Mol. Plant 2015, 8, 276–289. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoo, Y.-H.; Nalini Chandran, A.K.; Park, J.-C.; Gho, Y.-S.; Lee, S.-W.; An, G.; Jung, K.-H. OsPhyB-Mediating Novel Regulatory Pathway for Drought Tolerance in Rice Root Identified by a Global RNA-Seq Transcriptome Analysis of Rice Genes in Response to Water Deficiencies. Front. Plant Sci. 2017, 8, 580. [Google Scholar] [CrossRef]
- Kumar, M.; Gho, Y.S.; Jung, K.H.; Kim, S.R. Genome-wide identification and analysis of genes, conserved between japonica and indica rice cultivars, that respond to low-temperature stress at the vegetative growth stage. Front. Plant Sci. 2017, 8, 1120. [Google Scholar] [CrossRef]
- Hong, W.J.; Kim, Y.J.; Chandran, A.K.N.; Jung, K.H. Infrastructures of systems biology that facilitate functional genomic study in rice. Rice 2019, 12, 15. [Google Scholar] [CrossRef]
- Hong, W.J.; Jung, K.H. Comparative Analysis of Flanking Sequence Tags of T-DNA/Transposon Insertional Mutants and Genetic Variations of Fast-neutron Treated Mutants in Rice. J. Plant Biol. 2018, 61, 80–84. [Google Scholar] [CrossRef]
- Chandran, A.; Jung, K.-H. Resources for systems biology in rice. J. Plant Biol. 2014, 57, 80–92. [Google Scholar] [CrossRef]
- Jeong, D.H.; An, S.; Kang, H.-G.; Moon, S.; Han, J.-J.; Park, S.; Lee, H.S.; An, K.; An, G. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002, 130, 1636–1644. [Google Scholar] [CrossRef]
- Zimmermann, P.; Laule, O.; Schmitz, J.; Hruz, T.; Bleuler, S.; Gruissem, W. Genevestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Mol. Plant 2008, 1, 851–857. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jeong, H.J.; Jung, K.H. Rice tissue-specific promoters and condition-dependent promoters for effective translational application. J. Integr. Plant Biol. 2015, 57, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.J.; Yoo, Y.H.; Park, S.A.; Moon, S.; Kim, S.R.; An, G.; Jung, K.H. Genome-wide identification and extensive analysis of rice-endosperm preferred genes using reference expression database. J. Plant Biol. 2017, 60, 249–258. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Chandran, A.K.N.; Yoo, Y.H.; Cao, P.; Sharma, R.; Sharma, M.; Dardick, C.; Ronald, P.C.; Jung, K.H. Updated Rice Kinase Database RKD 2.0: Enabling transcriptome and functional analysis of rice kinase genes. Rice 2016, 9, 40. [Google Scholar] [CrossRef][Green Version]
- Yoo, Y.H.; Hong, W.J.; Jung, K.H. A systematic view exploring the role of chloroplasts in plant abiotic stress responses. BioMed Res. Int. 2019, 2019, 1–14. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Yao, W.; Li, G.; Yu, Y.; Ouyang, Y. funRiceGenes dataset for comprehensive understanding and application of rice functional genes. Gigascience 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 2007, 35, D883–D887. [Google Scholar] [CrossRef]
- Cao, P.; Jung, K.H.; Choi, D.; Hwang, D.; Zhu, J.; Ronald, P.C. The rice Oligonucleotide array database: An atlas of rice gene expression. Rice 2012, 5, 17. [Google Scholar] [CrossRef]
- Hoang, T.V.; Vo, K.T.X.; Hong, W.J.; Jung, K.H.; Jeon, J.S. Defense Response to Pathogens Through Epigenetic Regulation in Rice. J. Plant Biol. 2018, 61, 1–10. [Google Scholar] [CrossRef]
- Yu, G.H.; Huang, S.C.; He, R.; Li, Y.Z.; Cheng, X.G. Transgenic Rice Overexperessing a Tomato Mitochondrial Phosphate Transporter, SlMPT3;1, Promotes Phosphate Uptake and Increases Grain Yield. J. Plant Biol. 2018, 61, 383–400. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef] [PubMed]
| Category | Expression Pattern | Locus_ID | T-DNA Line No. | Putative Function | DOI References a |
|---|---|---|---|---|---|
| Anatomy | Leaf/Flag leaf/Shoot | LOC_Os03g20700 | 9-07117 | Magnesium chelatase | 10.1093/pcp/pcg064 |
| Root (root hair) | LOC_Os05g45900 | 3A-00457 | endonuclease/exonuclease/phosphatase family domain-containing protein | 10.1186/s12284-018-0241-2 | |
| LOC_Os10g42750 | 2B-60199 | CSLD1, cellulose synthase-like family D | |||
| LOC_Os12g02240 | 4A-50567 | expressed protein | |||
| Seed/Embryo/Endosperm | LOC_Os05g05790 | 1A-10540 | double-stranded RNA binding motif-containing protein | 10.1104/pp.014357 | |
| Anther/Pollen | LOC_Os11g20384 | 1A-13819 | SacI homology domain-containing protein | 10.1186/s12284-018-0219-0 | |
| LOC_Os07g17310 | 2D-41188 | B12D protein | |||
| Ubiquitous | LOC_Os03g01910 | 4A-04197 | transcription factor BTF3 | 10.1016/j.molp.2014.10.013 | |
| LOC_Os03g08010 | 5A-00191 | elongation factor Tu | |||
| LOC_Os04g42090 | 3A-05916 | CPuORF7, conserved peptide uORF-containing transcript | |||
| LOC_Os06g30750 | 2D-00098 | reticulon domain-containing protein | |||
| Abiotic stress | Drought | LOC_Os04g52290 | 3A-03417 | PPR repeat domain-containing protein | 10.3389/fpls.2017.00580 |
| LOC_Os07g02710 | 3A-13738 | expressed protein | |||
| Cold | LOC_Os01g31370 | 3A-50649 | glycosyltransferase | 10.3389/fpls.2017.01120 | |
| LOC_Os03g49830 | 1C-08613 | expressed protein |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoo, Y.-H.; Kim, Y.-J.; Moon, S.; Gho, Y.-S.; Hong, W.-J.; Kim, E.-J.; Jiang, X.; Jung, K.-H. Fast Track to Discover Novel Promoters in Rice. Plants 2020, 9, 125. https://doi.org/10.3390/plants9010125
Yoo Y-H, Kim Y-J, Moon S, Gho Y-S, Hong W-J, Kim E-J, Jiang X, Jung K-H. Fast Track to Discover Novel Promoters in Rice. Plants. 2020; 9(1):125. https://doi.org/10.3390/plants9010125
Chicago/Turabian StyleYoo, Yo-Han, Yu-Jin Kim, Sunok Moon, Yun-Shil Gho, Woo-Jong Hong, Eui-Jung Kim, Xu Jiang, and Ki-Hong Jung. 2020. "Fast Track to Discover Novel Promoters in Rice" Plants 9, no. 1: 125. https://doi.org/10.3390/plants9010125
APA StyleYoo, Y.-H., Kim, Y.-J., Moon, S., Gho, Y.-S., Hong, W.-J., Kim, E.-J., Jiang, X., & Jung, K.-H. (2020). Fast Track to Discover Novel Promoters in Rice. Plants, 9(1), 125. https://doi.org/10.3390/plants9010125





