Plant Mitochondrial Carriers: Molecular Gatekeepers That Help to Regulate Plant Central Carbon Metabolism
Abstract
1. Introduction
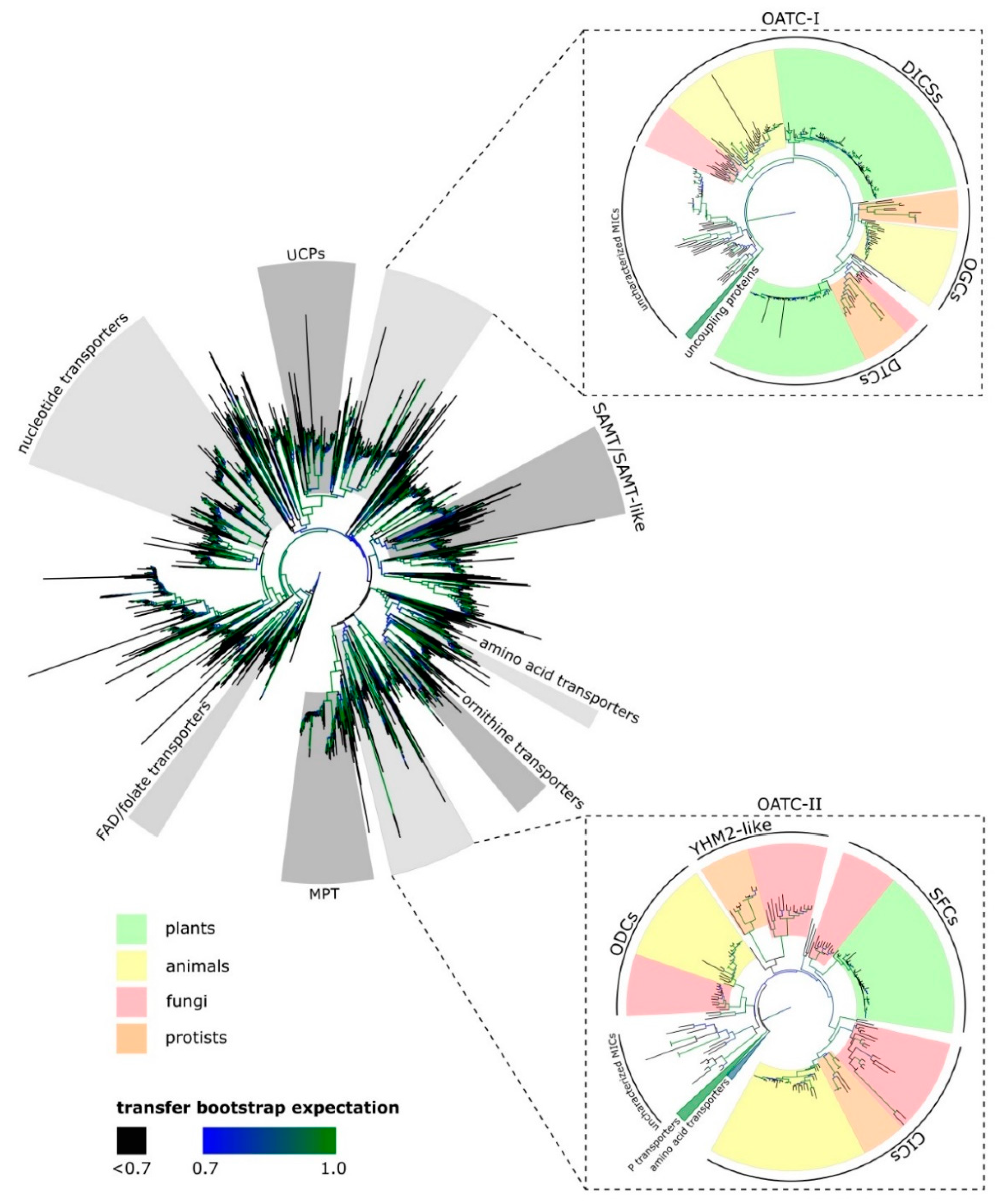
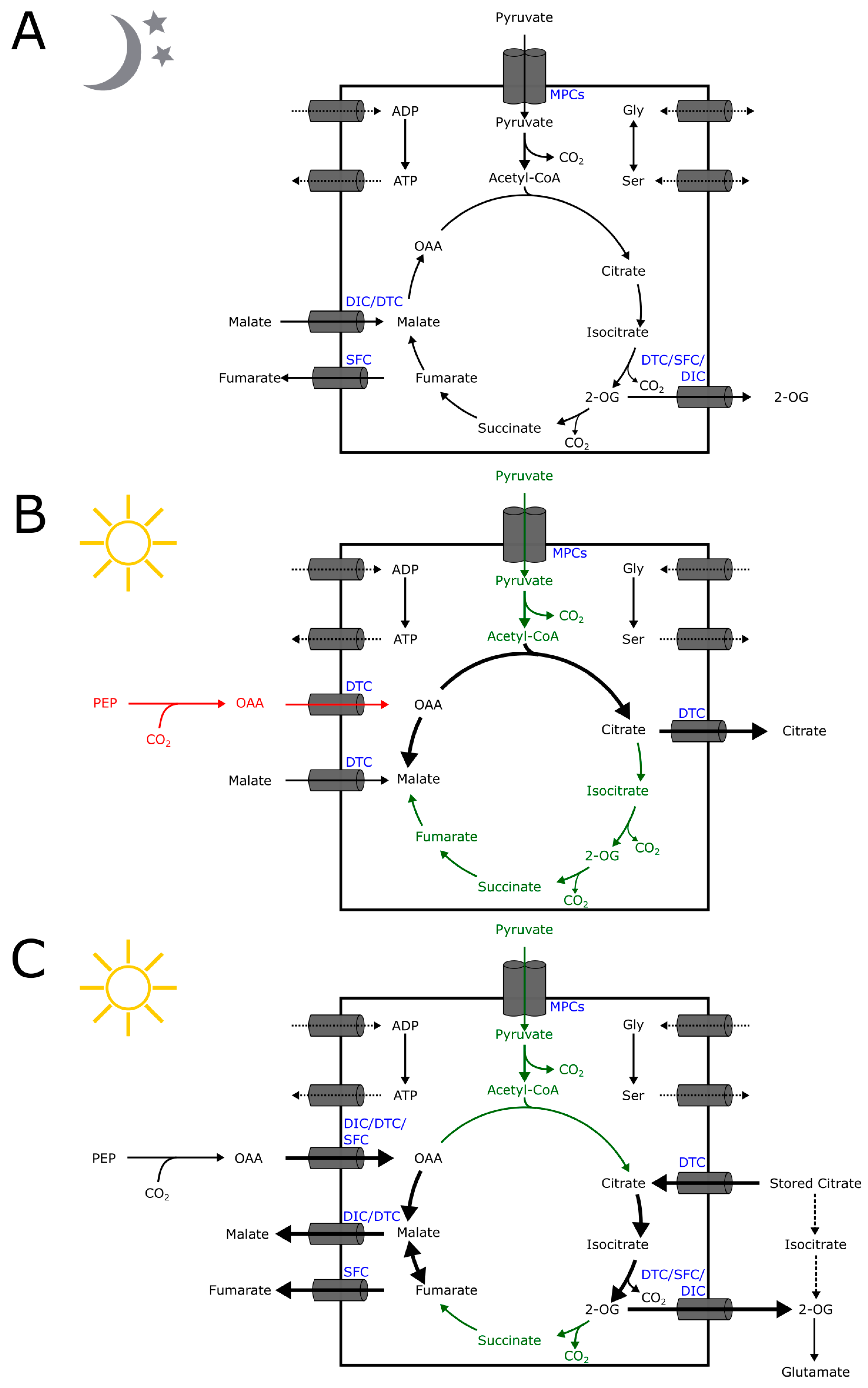
2. Mitochondrial Organic Acid Transporters Are Important to Central Carbon Metabolism
2.1. Dicarboxylate Carriers (DICs)
2.2. Dicarboxylate/Tricarboxylate Carrier (DTC)
2.3. Succinate/Fumarate Carriers (SFC)
3. Mitochondrial Carriers Relevant to Mitochondrial Oxidative Phosphorylation
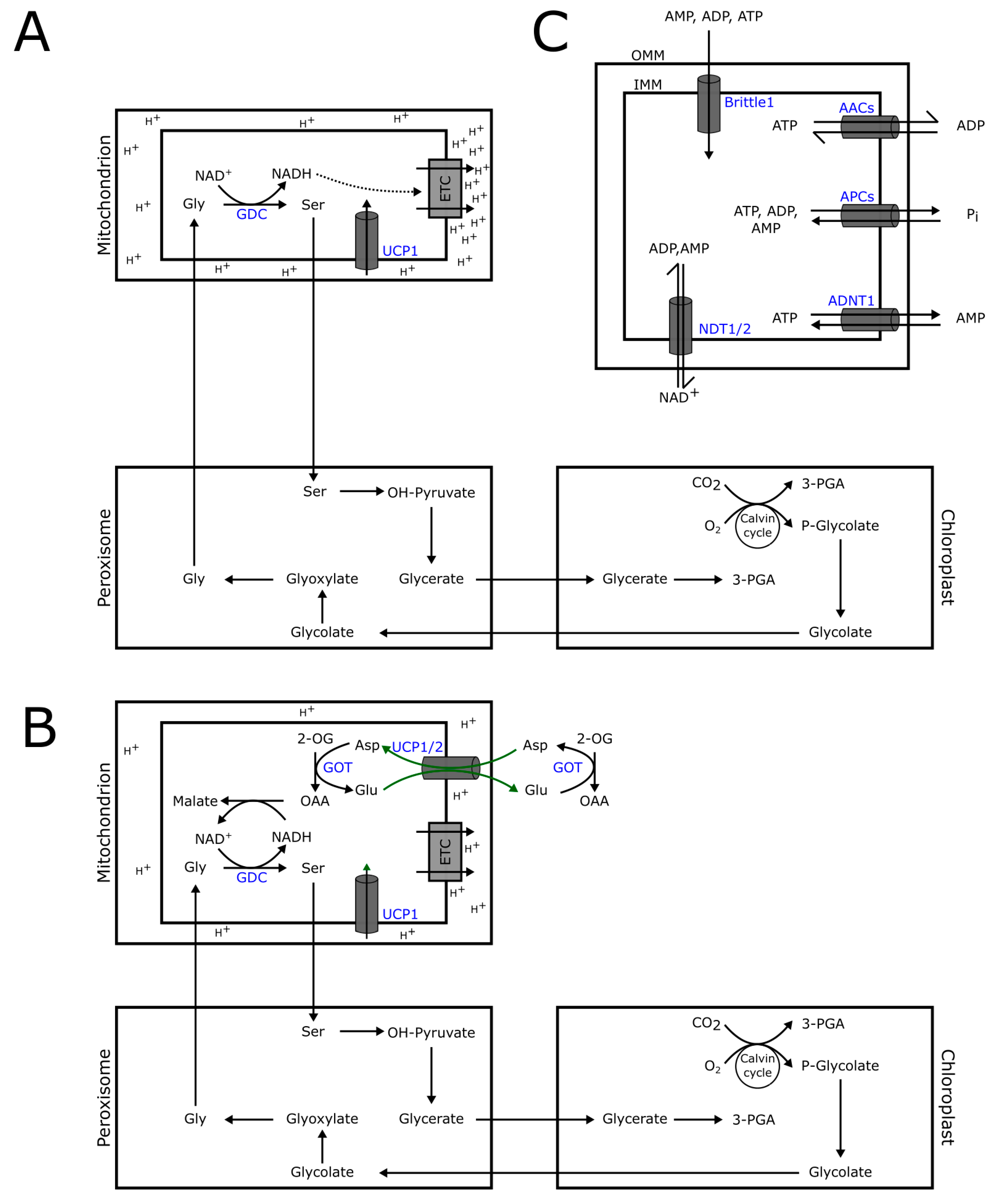
3.1. NAD+ Transport
3.2. Uncoupling Proteins (UCPs)
3.3. Adenylate Transporters
4. Novel Approaches to Investigate MCFs
5. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Gray, M.W.; Burger, G.; Lang, B.F. The origin and early evolution of mitochondria. Genome Biol. 2001, 2, reviews1018. [Google Scholar] [CrossRef] [PubMed]
- Sweetlove, L.J.; Fernie, A.R. The spatial organization of metabolism within the plant cell. Annu. Rev. Plant Biol. 2013, 64, 723–746. [Google Scholar] [CrossRef] [PubMed]
- Monné, M.; Vozza, A.; Lasorsa, F.M.; Porcelli, V.; Palmieri, F. Mitochondrial carriers for aspartate, glutamate and other amino acids: A review. Int. J. Mol. Sci. 2019, 20, 4456. [Google Scholar] [CrossRef] [PubMed]
- Gueguen, V.; Macherel, D.; Jaquinod, M.; Douce, R.; Bourguignon, J. Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J. Biol. Chem. 2000, 275, 5016–5025. [Google Scholar] [CrossRef] [PubMed]
- Douce, R.; Neuburger, M. Biochemical dissection of photorespiration. Curr. Opin. Plant Biol. 1999, 2, 214–222. [Google Scholar] [CrossRef]
- Edwards, G.E.; Franceschi, V.R.; Ku, M.S.B.; Voznesenskaya, E.V.; Pyankov, V.I.; Andreo, C.S. Compartmentation of photosynthesis in cells and tissues of C4 plants. J. Exp. Bot. 2001, 52, 577–590. [Google Scholar] [CrossRef]
- Demine, S.; Reddy, N.; Renard, P.; Raes, M.; Arnould, T. Unraveling biochemical pathways affected by mitochondrial dysfunctions using metabolomic approaches. Metabolites 2014, 4, 831–878. [Google Scholar] [CrossRef]
- Bayrhuber, M.; Meins, T.; Habeck, M.; Becker, S.; Giller, K.; Villinger, S.; Vonrhein, C.; Griesinger, C.; Zweckstetter, M.; Zeth, K. Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. USA 2008, 105, 15370–15375. [Google Scholar] [CrossRef]
- Nury, H.; Dahout-Gonzalez, C.; Trézéguet, V.; Lauquin, G.J.M.; Brandolin, G.; Pebay-Peyroula, E. Relations between structure and function of the mitochondrial ADP/ATP carrier. Annu. Rev. Biochem. 2006, 75, 713–741. [Google Scholar] [CrossRef]
- Kühlbrandt, W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015, 13, 89. [Google Scholar] [CrossRef]
- Palmieri, F.; Pierri, C.L.; De Grassi, A.; Nunes-Nesi, A.; Fernie, A.R. Evolution, structure and function of mitochondrial carriers: A review with new insights. Plant J. 2011, 66, 161–181. [Google Scholar] [CrossRef] [PubMed]
- Haferkamp, I.; Schmitz-Esser, S. The plant mitochondrial carrier family: Functional and evolutionary aspects. Front. Plant Sci. 2012, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; Millar, A.H. The plant mitochondrial transportome: Balancing metabolic demands with energetic constraints. Trends Plant Sci. 2016, 21, 662–676. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. The mitochondrial transporter family SLC25: Identification, properties and physiopathology. Mol. Asp. Med. 2013, 34, 465–484. [Google Scholar] [CrossRef]
- Saraste, M.; Walker, J.E. Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett. 1982, 144, 250–254. [Google Scholar] [CrossRef]
- Capobianco, L.; Bisaccia, F.; Michel, A.; Sluse, F.E.; Palmieri, F. The N- and C-termini of the tricarboxylate carrier are exposed to the cytoplasmic side of the inner mitochondrial membrane. FEBS Lett. 1995, 357, 297–300. [Google Scholar] [CrossRef]
- Pebay-Peyroula, E.; Dahout-Gonzalez, C.; Kahn, R.; Trézéguet, V.; Lauquin, G.J.M.; Brandolin, G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 2003, 426, 39–44. [Google Scholar] [CrossRef]
- Palmieri, F.; Monné, M. Discoveries, metabolic roles and diseases of mitochondrial carriers: A review. BBA-Mol. Cell Res. 2016, 1863, 2362–2378. [Google Scholar] [CrossRef]
- Robinson, A.J.; Overy, C.; Kunji, E.R.S. The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc. Natl. Acad. Sci. USA 2008, 105, 17766–17771. [Google Scholar] [CrossRef]
- Ruprecht, J.J.; King, M.S.; Zögg, T.; Aleksandrova, A.A.; Pardon, E.; Crichton, P.G.; Steyaert, J.; Kunji, E.R.S. The molecular mechanism of transport by the mitochondrial ADP/ATP carrier. Cell 2019, 176, 435–447.e415. [Google Scholar] [CrossRef]
- Ruprecht, J.J.; Kunji, E.R.S. The SLC25 mitochondrial carrier family: Structure and mechanism. Trends Biochem. Sci. 2019. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, M. The ADP and ATP transport in mitochondria and its carrier. BBA-Biomembranes 2008, 1778, 1978–2021. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, M. The ADP,ATP shuttle of the mitochondrion. Trends Biochem. Sci. 1979, 4, 249–252. [Google Scholar] [CrossRef]
- Indiveri, C.; Tonazzi, A.; Palmieri, F. The reconstituted carnitine carrier from rat liver mitochondria: Evidence for a transport mechanism different from that of the other mitochondrial translocators. BBA-Biomembranes 1994, 1189, 65–73. [Google Scholar] [CrossRef]
- Ruprecht, J.J.; Hellawell, A.M.; Harding, M.; Crichton, P.G.; McCoy, A.J.; Kunji, E.R. Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc. Natl. Acad. Sci. USA 2014, 111, E426–E434. [Google Scholar] [CrossRef]
- Pietropaolo, A.; Pierri, C.L.; Palmieri, F.; Klingenberg, M. The switching mechanism of the mitochondrial ADP/ATP carrier explored by free-energy landscapes. BBA-Bioenergetics 2016, 1857, 772–781. [Google Scholar] [CrossRef]
- Springett, R.; King, M.S.; Crichton, P.G.; Kunji, E.R.S. Modelling the free energy profile of the mitochondrial ADP/ATP carrier. BBA-Bioenergetics 2017, 1858, 906–914. [Google Scholar] [CrossRef]
- Ruprecht, J.J.; Kunji, E.R. Structural changes in the transport cycle of the mitochondrial ADP/ATP carrier. Curr. Opin. Struc. Biol. 2019, 57, 135–144. [Google Scholar] [CrossRef]
- Leroch, M.; Neuhaus, H.E.; Kirchberger, S.; Zimmermann, S.; Melzer, M.; Gerhold, J.; Tjaden, J. Identification of a novel adenine nucleotide transporter in the endoplasmic reticulum of Arabidopsis. Plant Cell 2008, 20, 438–451. [Google Scholar] [CrossRef]
- Bahaji, A.; Ovecka, M.; Bárány, I.; Risueño, M.C.; Muñoz, F.J.; Baroja-Fernández, E.; Montero, M.; Li, J.; Hidalgo, M.; Sesma, M.T.; et al. Dual targeting to mitochondria and plastids of AtBT1 and ZmBT1, two members of the mitochondrial carrier family. Plant Cell Physiol. 2011, 52, 597–609. [Google Scholar] [CrossRef]
- Zhang, Y.; Swart, C.; Alseekh, S.; Scossa, F.; Jiang, L.; Obata, T.; Graf, A.; Fernie, A.R. The extra-pathway interactome of the TCA cycle: Expected and unexpected metabolic interactions. Plant Physiol. 2018, 177, 966–979. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Beard, K.F.M.; Swart, C.; Bergmann, S.; Krahnert, I.; Nikoloski, Z.; Graf, A.; George Ratcliffe, R.; Sweetlove, L.J.; Fernie, A.R.; et al. Protein-protein interactions and metabolite channelling in the plant tricarboxylic acid cycle. Nat. Commun. 2017, 8, 15212. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, F.; Domelevo Entfellner, J.B.; Wilkinson, E.; Correia, D.; Dávila Felipe, M.; De Oliveira, T.; Gascuel, O. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 2018, 556, 452–456. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Beard, K.F.M.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef]
- Bricker, D.K.; Taylor, E.B.; Schell, J.C.; Orsak, T.; Boutron, A.; Chen, Y.-C.; Cox, J.E.; Cardon, C.M.; Vraken, J.G.V.; Dephoure, N.; et al. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila and humans. Science 2012, 337, 96–100. [Google Scholar] [CrossRef]
- Zhang, Y.; Fernie, A.R. On the role of the tricarboxylic acid cycle in plant productivity. J. Integr. Plant Biol. 2018, 60, 1199–1216. [Google Scholar] [CrossRef]
- Fernie, A.R.; Zhang, Y.; Sweetlove, L.J. Passing the baton: Substrate channelling in respiratory metabolism. Research 2018, 2018, 1–16. [Google Scholar] [CrossRef]
- Steuer, R.; Nesi, A.N.; Fernie, A.R.; Gross, T.; Blasius, B.; Selbig, J. From structure to dynamics of metabolic pathways: Application to the plant mitochondrial TCA cycle. Bioinformatics 2007, 23, 1378–1385. [Google Scholar] [CrossRef] [PubMed]
- Tcherkez, G. In vivo respiratory metabolism of illuminated leaves. Plant Physiol. 2005, 138, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, P.; Rugen, N.; Carrie, C.; Elsässer, M.; Finkemeier, I.; Giese, J.; Hildebrandt, T.M.; Kühn, K.; Maurino, V.G.; Ruberti, C.; et al. Single organelle function and organization as estimated from Arabidopsis mitochondrial proteomics. Plant J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Picault, N.; Arrigoni, R.; Besin, E.; Palmieri, F.; Hodges, M. Molecular identification of three arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: Organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem. J. 2008, 410, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Haferkamp, I.; Hackstein, J.H.P.; Voncken, F.G.J.; Schmit, G.; Tjaden, J. Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur. J. Biochem. 2002, 269, 3172–3181. [Google Scholar] [CrossRef]
- Iacopetta, D.; Madeo, M.; Tasco, G.; Carrisi, C.; Curcio, R.; Martello, E.; Casadio, R.; Capobianco, L.; Dolce, V. A novel subfamily of mitochondrial dicarboxylate carriers from Drosophila melanogaster: Biochemical and computational studies. BBA-Bioenergetics 2011, 1807, 251–261. [Google Scholar] [CrossRef]
- Fiermonte, G.; Palmieri, L.; Dolce, V.; Lasorsa, F.M.; Palmieri, F.; Runswick, M.J.; Walker, J.E. The sequence, bacterial expression, and functional reconstitution of the rat mitochondrial dicarboxylate transporter cloned via distant homologs in yeast and Caenorhabditis elegans. J. Biol. Chem. 1998, 273, 24754–24759. [Google Scholar] [CrossRef]
- Picault, N.; Palmieri, L.; Pisano, I.; Hodges, M.; Palmieri, F. Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria: Bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 2002, 277, 24204–24211. [Google Scholar] [CrossRef]
- Regalado, A.; Pierri, C.L.; Bitetto, M.; Laera, V.L.; Pimentel, C.; Francisco, R.; Passarinho, J.; Chaves, M.M.; Agrimi, G. Characterization of mitochondrial dicarboxylate/tricarboxylate transporters from grape berries. Planta 2013, 237, 693–703. [Google Scholar] [CrossRef]
- Spagnoletta, A.; Santis, A.D.; Tampieri, E.; Baraldi, E.; Bachi, A.; Genchi, G. Identification and kinetic characterization of HtDTC, the mitochondrial dicarboxylate-tricarboxylate carrier of Jerusalem artichoke tubers. J. Bioenerg. Biomembr. 2006, 38, 57–65. [Google Scholar] [CrossRef]
- Deng, W.; Luo, K.; Li, Z.; Yang, Y. Molecular cloning and characterization of a mitochondrial dicarboxylate/tricarboxylate transporter gene in Citrus junos response to aluminum stress. Mitochondrial DNA 2008, 19, 376–384. [Google Scholar] [CrossRef]
- Genchi, G.; Spagnoletta, A.; De Santis, A.; Stefanizzi, L.; Palmieri, F. Purification and characterization of the reconstitutively active citrate carrier from maize mitochondria. Plant Physiol. 1999, 120, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zeng, Y.; Zhuang, X.; Sun, L.; Yao, X.; Pimpl, P.; Jiang, L. Organelle pH in the Arabidopsis endomembrane system. Mol. Plant 2013, 6, 1419–1437. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Lasorsa, F.M.; De Palma, A.; Palmieri, F.; Runswick, M.J.; Walker, J.E. Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 1997, 417, 114–118. [Google Scholar] [CrossRef]
- Fernández, M.; Fernández, E.; Rodicio, R. ACR1, a gene encoding a protein related to mitochondrial carriers, is essential for acetyl-CoA synthetase activity in Saccharomyces cerevisiae. Mol. Gen. Genet. 1994, 242, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Catoni, E.; Schwab, R.; Hilpert, M.; Desimone, M.; Schwacke, R.; Flügge, U.I.; Schumacher, K.; Frommer, W.B. Identification of an Arabidopsis mitochondrial succinate-fumarate translocator. FEBS Lett. 2003, 534, 87–92. [Google Scholar] [CrossRef]
- Kunze, M.; Pracharoenwattana, I.; Smith, S.M.; Hartig, A. A central role for the peroxisomal membrane in glyoxylate cycle function. BBA-Mol. Cell Res. 2006, 1763, 1441–1452. [Google Scholar] [CrossRef]
- Lee, Y.J.; Jang, J.W.; Kim, K.J.; Maeng, P.J. TCA cycle-independent acetate metabolism via the glyoxylate cycle in Saccharomyces cerevisiae. Yeast 2011, 28, 153–166. [Google Scholar] [CrossRef]
- Meyer, E.H.; Welchen, E.; Carrie, C. Assembly of the complexes of the oxidative phosphorylation system in land plant mitochondria. Annu. Rev. Plant Biol. 2019, 70, 23–50. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Lytovchenko, A.; Morgan, M.; Nunes-Nesi, A.; Taylor, N.L.; Baxter, C.J.; Eickmeier, I.; Fernie, A.R. Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19587–19592. [Google Scholar] [CrossRef]
- Monné, M.; Daddabbo, L.; Gagneul, D.; Obata, T.; Hielscher, B.; Palmieri, L.; Miniero, D.V.; Fernie, A.R.; Weber, A.P.M.; Palmieri, F. Uncoupling proteins 1 and 2 (UCP1 and UCP2) from Arabidopsis thaliana are mitochondrial transporters of aspartate, glutamate, and dicarboxylates. J. Biol. Chem. 2018, 293, 4213–4227. [Google Scholar] [CrossRef]
- Geigenberger, P.; Fernie, A.R. Metabolic control of redox and redox control of metabolism in plants. Antioxid. Redox Signal. 2014, 21, 1389–1421. [Google Scholar] [CrossRef]
- Gakière, B.; Hao, J.; de Bont, L.; Pétriacq, P.; Nunes-Nesi, A.; Fernie, A.R. NAD+ biosynthesis and signaling in plants. Crit. Rev. Plant Sci. 2018, 37, 259–307. [Google Scholar] [CrossRef]
- De Souza Chaves, I.; Araújo, E.F.; Florian, A.; Medeiros, D.B.; da Fonseca-Pereira, P.; Charton, L.; Heyneke, E.; Apfata, J.A.C.; Pires, M.V.; Mettler-Altmann, T. Mitochondrial NAD+ transporter (NDT1) plays important roles in cellular NAD+ homeostasis in Arabidopsis thaliana. Plant J. 2019, 100, 487–504. [Google Scholar] [CrossRef]
- Palmieri, F.; Rieder, B.; Ventrella, A.; Blanco, E.; Do, P.T.; Nunes-Nesi, A.; Trauth, A.U.; Fiermonte, G.; Tjaden, J.; Agrimi, G.; et al. Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J. Biol. Chem. 2009, 284, 31249–31259. [Google Scholar] [CrossRef] [PubMed]
- Van Roermund, C.W.; Schroers, M.G.; Wiese, J.; Facchinelli, F.; Kurz, S.; Wilkinson, S.; Charton, L.; Wanders, R.J.; Waterham, H.R.; Weber, A.P. The peroxisomal NAD carrier from Arabidopsis imports NAD in exchange with AMP. Plant Physiol. 2016, 171, 2127–2139. [Google Scholar] [CrossRef] [PubMed]
- Agrimi, G.; Russo, A.; Pierri, C.L.; Palmieri, F. The peroxisomal NAD+ carrier of Arabidopsis thaliana transports coenzyme A and its derivatives. J. Bioenerg. Biomembr. 2012, 44, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, K.; Wilkinson, S.; Weber, A.P.; Linka, N. A peroxisomal carrier delivers NAD+ and contributes to optimal fatty acid degradation during storage oil mobilization. Plant J. 2012, 69, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Daloso, D.M.; Müller, K.; Obata, T.; Florian, A.; Tohge, T.; Bottcher, A.; Riondet, C.; Bariat, L.; Carrari, F.; Nunes-Nesi, A.; et al. Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc. Natl. Acad. Sci. USA 2015, 112, E1392–E1400. [Google Scholar] [CrossRef]
- Ježek, P.; Holendová, B.; Garlid, K.D.; Jabůrek, M. Mitochondrial uncoupling proteins: Subtle regulators of cellular redox signaling. Antioxid. Redox Signal. 2018, 29, 667–714. [Google Scholar] [CrossRef]
- Ježek, P.; Jabůrek, M.; Porter, R.K. Uncoupling mechanism and redox regulation of mitochondrial uncoupling protein 1 (UCP1). BBA-Bioenergetics 2019, 1860, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Echtay, K.S. Mitochondrial uncoupling proteins-What is their physiological role? Free Radic. Biol. Med. 2007, 43, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Vercesi, A.E.; Borecky, J.; Maia Ide, G.; Arruda, P.; Cuccovia, I.M.; Chaimovich, H. Plant uncoupling mitochondrial proteins. Annu. Rev. Plant Biol. 2006, 57, 383–404. [Google Scholar] [CrossRef] [PubMed]
- Jabůrek, M.; Vařecha, M.; Gimeno, R.E.; Dembski, M.; Ježek, P.; Zhang, M.; Burn, P.; Tartaglia, L.A.; Garlid, K.D. Transport function and regulation of mitochondrial uncoupling proteins 2 and 3. J. Biol. Chem. 1999, 274, 26003–26007. [Google Scholar] [CrossRef]
- Kunji, E.R.S.; Aleksandrova, A.; King, M.S.; Majd, H.; Ashton, V.L.; Cerson, E.; Springett, R.; Kibalchenko, M.; Tavoulari, S.; Crichton, P.G.; et al. The transport mechanism of the mitochondrial ADP/ATP carrier. BBA-Mol. Cell Res. 2016, 1863, 2379–2393. [Google Scholar] [CrossRef]
- Da Fonseca-Pereira, P.; Neri-Silva, R.; Cavalcanti, J.H.F.; Brito, D.S.; Weber, A.P.M.; Araújo, W.L.; Nunes-Nesi, A. Data-mining bioinformatics: Connecting adenylate transport and metabolic responses to stress. Trends Plant Sci. 2018, 23, 961–974. [Google Scholar] [CrossRef]
- Monné, M.; Miniero, D.V.; Obata, T.; Daddabbo, L.; Palmieri, L.; Vozza, A.; Nicolardi, M.C.; Fernie, A.R.; Palmieri, F. Functional characterization and organ distribution of three mitochondrial ATP–Mg/Pi carriers in Arabidopsis thaliana. BBA-Bioenergetics 2015, 1847, 1220–1230. [Google Scholar] [CrossRef]
- Lorenz, A.; Lorenz, M.; Vothknecht, U.C.; Niopek-Witz, S.; Neuhaus, H.E.; Haferkamp, I. In vitro analyses of mitochondrial ATP/phosphate carriers from Arabidopsis thaliana revealed unexpected Ca2+-effects. BMC Plant Biol. 2015, 15, 238. [Google Scholar] [CrossRef]
- Monne, M.; Daddabbo, L.; Giannossa, L.C.; Nicolardi, M.C.; Palmieri, L.; Miniero, D.V.; Mangone, A.; Palmieri, F. Mitochondrial ATP-Mg/phosphate carriers transport divalent inorganic cations in complex with ATP. J. Bioenerg. Biomembr. 2017, 49, 369–380. [Google Scholar] [CrossRef]
- Iacobazzi, V.; Infantino, V.; Palmieri, F. Epigenetic mechanisms and Sp1 regulate mitochondrial citrate carrier gene expression. Biochem. Biophys. Res. Commun. 2008, 376, 15–20. [Google Scholar] [CrossRef]
- Inan, G.; Goto, F.; Jin, J.B.; Rosado, A.; Koiwa, H.; Shi, H.; Hasegawa, P.M.; Bressan, R.A.; Maggio, A.; Li, X. Isolation and characterization of shs1, a sugar-hypersensitive and ABA-insensitive mutant with multiple stress responses. Plant Mol. Biol. 2007, 65, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Kirchberger, S.; Tjaden, J.; Ekkehard Neuhaus, H. Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J. 2008, 56, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Bahaji, A.; Muñoz, F.J.; Ovecka, M.; Baroja-Fernández, E.; Montero, M.; Li, J.; Hidalgo, M.; Almagro, G.; Sesma, M.T.; Ezquer, I.; et al. Specific delivery of AtBT1 to mitochondria complements the aberrant growth and sterility phenotype of homozygous AtBT1 Arabidopsis mutants. Plant J. 2011, 68, 1115–1121. [Google Scholar] [CrossRef] [PubMed]
- Kadooka, C.; Izumitsu, K.; Onoue, M.; Okutsu, K.; Yoshizaki, Y.; Takamine, K.; Goto, M.; Tamaki, H.; Futagami, T. Mitochondrial citrate transporters CtpA and YhmA are required for extracellular citric acid accumulation and contribute to cytosolic acetyl coenzyme A generation in Aspergillus luchuensis mut. kawachii. Appl. Environ. Microbiol. 2019, 85, e03136-18. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Geier, M.; Ashykhmina, N.; Frerigmann, H.; Wulfert, S.; Krueger, S.; Mugfor, S.G.; Kopriv, S.; Haferkamp, I.; Flügge, U.I. The Arabidopsis thylakoid ADP/ATP carrier TAAC Has an additional role in supplying plastidic phosphoadenosine 5′-phosphosulfate to the cytosol. Plant Cell 2012, 24, 4187–4204. [Google Scholar] [CrossRef]
- Ashykhmina, N.; Lorenz, M.; Frerigmann, H.; Koprivova, A.; Hofsetz, E.; Stührwohldt, N.; Flügge, U.I.; Haferkamp, I.; Kopriva, S.; Gigolashvilia, T. PAPST2 plays critical roles in removing the stress signaling molecule 3′-phosphoadenosine 5′-phosphate from the cytosol and its subsequent degradation in plastids and mitochondria. Plant Cell 2019, 31, 231–249. [Google Scholar] [CrossRef]
- Boulet, A.; Vest, K.E.; Maynard, M.K.; Gammon, M.G.; Russell, A.C.; Mathews, A.T.; Cole, S.E.; Zhu, X.; Phillips, C.B.; Kwong, J.Q.; et al. The mammalian phosphate carrier SLC25A3 is a mitochondrial copper transporter required for cytochrome c oxidase biogenesis. J. Biol. Chem. 2018, 293, 1887–1896. [Google Scholar] [CrossRef]
- Rautengarten, C.; Ebert, B.; Moreno, I.; Temple, H.; Herter, T.; Link, B.; Donas-Cofre, D.; Moreno, A.; Saez-Aguayo, S.; Blanco, F.; et al. The Golgi localized bifunctional UDP-rhamnose/UDP-galactose transporter family of Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 11563–11568. [Google Scholar] [CrossRef]
- Crichton, P.G.; Lee, Y.; Ruprecht, J.J.; Cerson, E.; Thangaratnarajah, C.; King, M.S.; Kunji, E.R.S. Trends in thermostability provide information on the nature of substrate, inhibitor, and lipid interactions with mitochondrial carriers. J. Biol. Chem. 2015, 290, 8206–8217. [Google Scholar] [CrossRef]
- Alexandrov, A.I.; Mileni, M.; Chien, E.Y.T.; Hanson, M.A.; Stevens, R.C. Microscale fluorescent thermal stability assay for membrane proteins. Structure 2008, 16, 351–359. [Google Scholar] [CrossRef]
- Majd, H.; King, M.S.; Palmer, S.M.; Smith, A.C.; Elbourne, L.D.H.; Paulsen, I.T.; Sharples, D.; Henderson, P.J.F.; Kunji, E.R.S. Screening of candidate substrates and coupling ions of transporters by thermostability shift assays. eLife 2018, 7, e38821. [Google Scholar] [CrossRef]
- Tavoulari, S.; Thangaratnarajah, C.; Mavridou, V.; Harbour, M.E.; Martinou, J.C.; Kunji, E.R. The yeast mitochondrial pyruvate carrier is a hetero-dimer in its functional state. EMBO J. 2019, 38, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, H.M.; Marquardt, K.; Lash, L.H.; Linseman, D.A. Bcl-2 is a novel interacting partner for the 2-oxoglutarate carrier and a key regulator of mitochondrial glutathione. Free Radic. Biol. Med. 2012, 52, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, E.M.; Spera, I.; Menga, A.; Infantino, V.; Porcelli, V.; Iacobazzi, V.; Pierri, C.L.; Hooper, D.C.; Palmieri, F.; Castegna, A. Acetylation of human mitochondrial citrate carrier modulates mitochondrial citrate/malate exchange activity to sustain NADPH production during macrophage activation. BBA-Bioenergetics 2015, 1847, 729–738. [Google Scholar] [CrossRef]
- Giangregorio, N.; Palmieri, F.; Indiveri, C. Glutathione controls the redox state of the mitochondrial carnitine/acylcarnitine carrier Cys residues by glutathionylation. BBA-Gen. Subj. 2013, 1830, 5299–5304. [Google Scholar] [CrossRef]
- Cai, T.; Hua, B.; Luo, D.; Xu, L.; Cheng, Q.; Yuan, G.; Yan, Z.; Sun, N.; Hua, L.; Lu, C. The circadian protein CLOCK regulates cell metabolism via the mitochondrial carrier SLC25A10. BBA-Mol. Cell Res. 2019, 1866, 1310–1321. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Sew, Y.S.; Stroher, E.; Holzmann, C.; Huang, S.; Taylor, N.L.; Jordana, X.; Millar, A.H. Multiplex micro-respiratory measurements of Arabidopsis tissues. New Phytol. 2013, 200, 922–932. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toleco, M.R.; Naake, T.; Zhang, Y.; Heazlewood, J.L.; Fernie, A.R. Plant Mitochondrial Carriers: Molecular Gatekeepers That Help to Regulate Plant Central Carbon Metabolism. Plants 2020, 9, 117. https://doi.org/10.3390/plants9010117
Toleco MR, Naake T, Zhang Y, Heazlewood JL, Fernie AR. Plant Mitochondrial Carriers: Molecular Gatekeepers That Help to Regulate Plant Central Carbon Metabolism. Plants. 2020; 9(1):117. https://doi.org/10.3390/plants9010117
Chicago/Turabian StyleToleco, M. Rey, Thomas Naake, Youjun Zhang, Joshua L. Heazlewood, and Alisdair R. Fernie. 2020. "Plant Mitochondrial Carriers: Molecular Gatekeepers That Help to Regulate Plant Central Carbon Metabolism" Plants 9, no. 1: 117. https://doi.org/10.3390/plants9010117
APA StyleToleco, M. R., Naake, T., Zhang, Y., Heazlewood, J. L., & Fernie, A. R. (2020). Plant Mitochondrial Carriers: Molecular Gatekeepers That Help to Regulate Plant Central Carbon Metabolism. Plants, 9(1), 117. https://doi.org/10.3390/plants9010117




