Control of Organ Abscission and Other Cell Separation Processes by Evolutionary Conserved Peptide Signaling
Abstract
1. Introduction
2. The Role of IDA/IDL Peptides during Organ Abscission and Other Cell Separation Processes
2.1. Control of Cell Separation Processes by IDA and IDA-LIKE Peptides in Arabidospsis
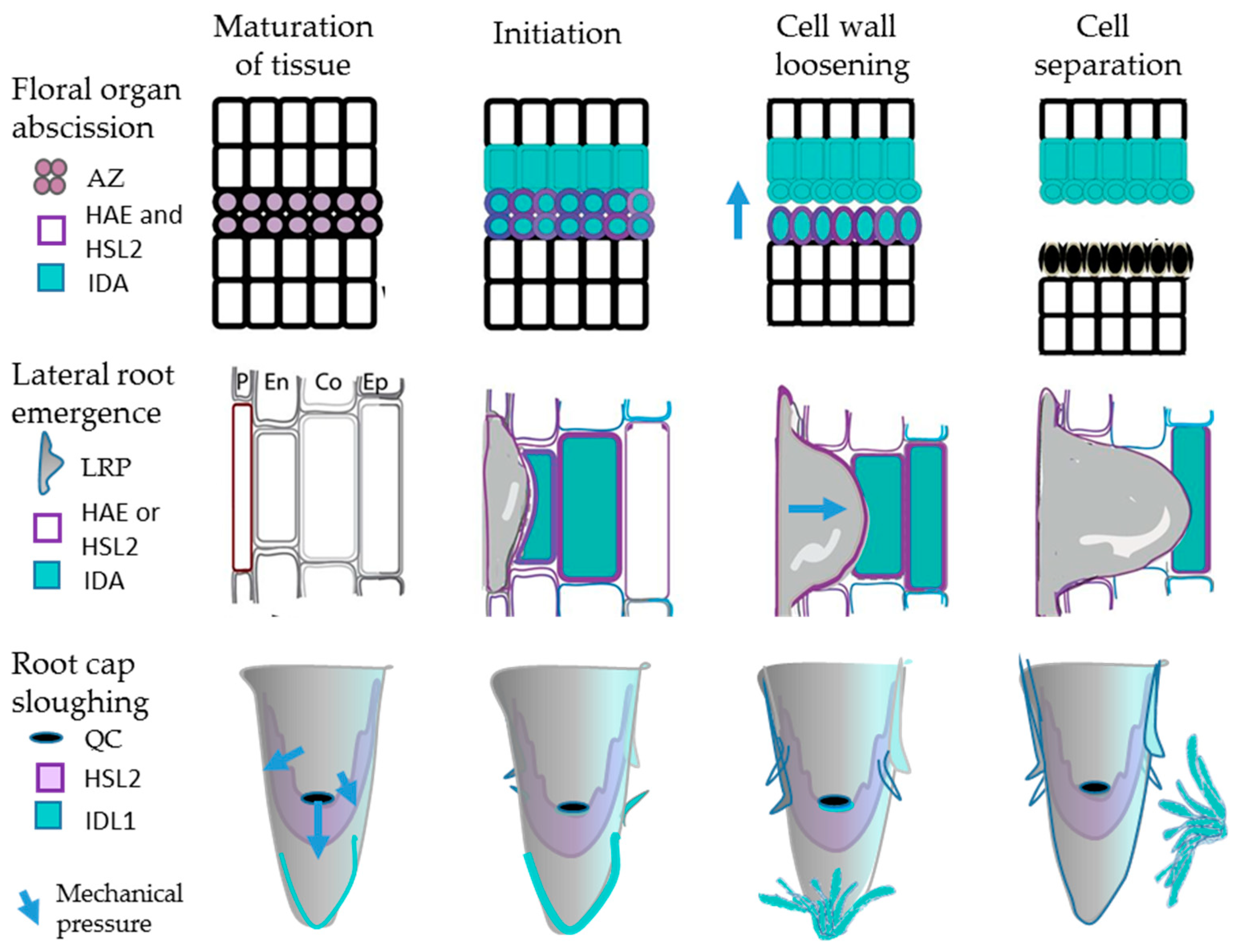
2.1.1. Mutants Deficient in Abscission
2.1.2. IDA and Induction of Abscission
2.1.3. IDA and IDL1 Are Involved in Cell Separation Events in the Root
2.2. Involvement of IDA/IDL1 in Abscission Processes in Other Flowering Plants
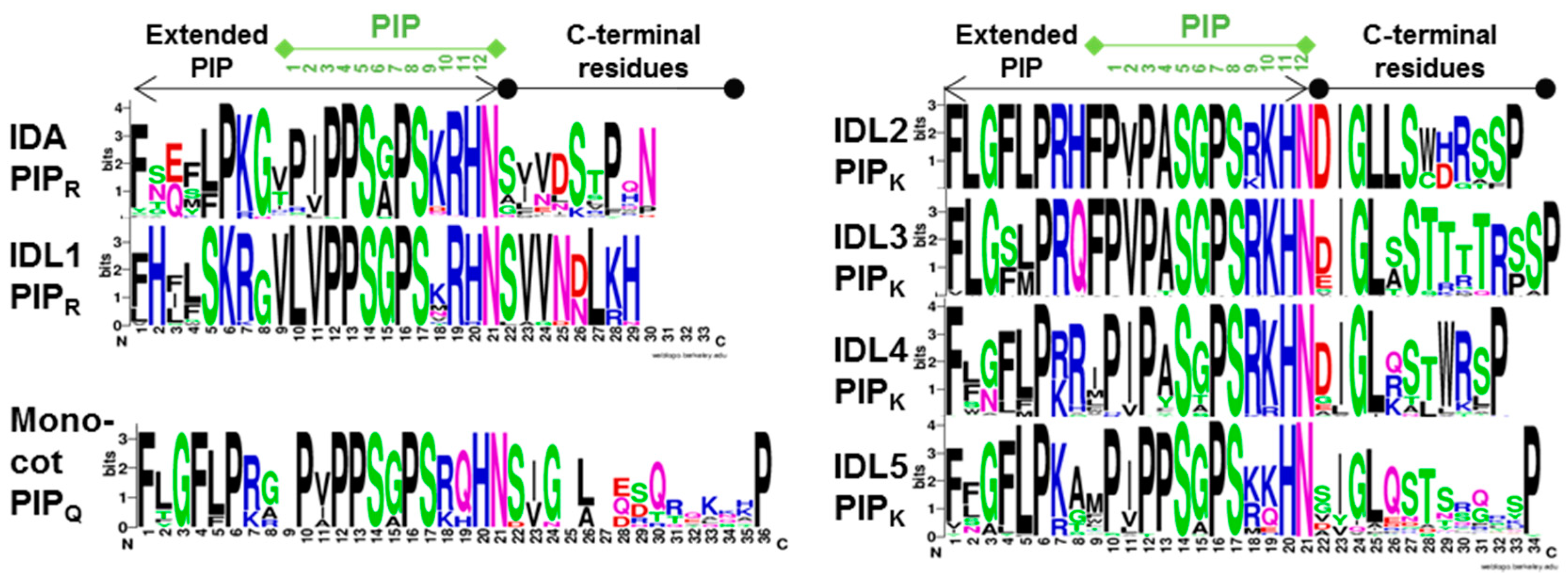
2.2.1. IDA/IDL1 and HAE/HSL Genes Are Present in All Angiosperm Orders
2.2.2. Expression Data and Functional Testing Suggest IDA/IDL Regulation of Abscission of Diverse Organs Both in Dicots and Monocots
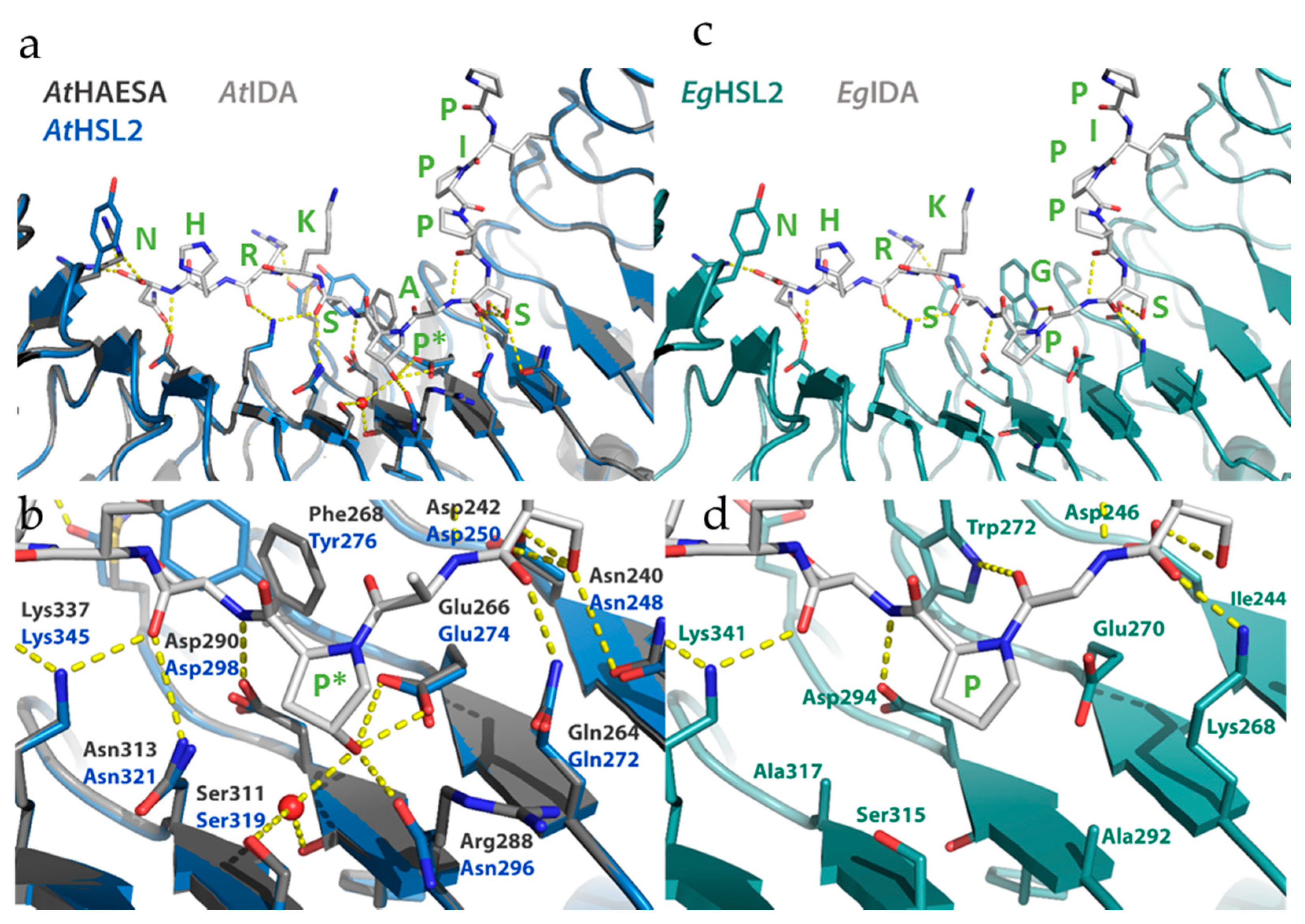
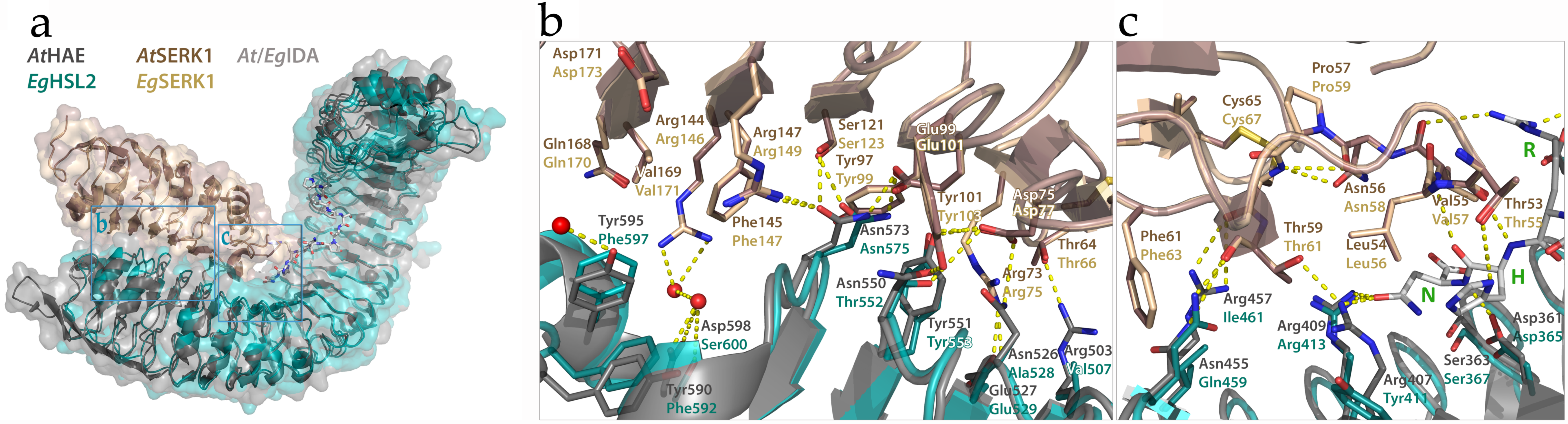
2.2.3. Matching Ligands and Receptors
3. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bleecker, A.; Patterson, S.E. Last exit: Senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 1997, 9, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Patterson, S.E. Cutting Loose. Abscission and Dehisence in Arabidopsis. Plant Physiol. 2001, 126, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Patharkar, O.R.; Walker, J.C. Connections between abscission, dehiscence, pathogen defense, drought tolerance, and senescence. Plant Sci. Int. J. Exp. Plant Boil. 2019, 284, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Patharkar, O.R.; Walker, J.C. Core Mechanisms Regulating Developmentally Timed and Environmentally Triggered Abscission. Plant Physiol. 2016, 172, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Kumpf, R.P.; Shi, C.L.; Larrieu, A.; Sto, I.M.; Butenko, M.A.; Peret, B.; Riiser, E.S.; Bennett, M.J.; Aalen, R.B. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. USA 2013, 110. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.-L.; von Wangenheim, D.; Herrmann, U.; Wildhagen, M.; Kulik, I.; Kopf, A.; Ishida, T.; Olsson, V.; Anker, M.K.; Albert, M.; et al. The dynamics of root cap sloughing in Arabidopsis is regulated by peptide signalling. Nat. Plants 2018, 4, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Begum, D.; Chuang, H.W.; Budiman, M.A.; Szymkowiak, E.J.; Irish, E.E.; Wing, R.A. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 2000, 406, 910–913. [Google Scholar] [CrossRef] [PubMed]
- McKim, S.M.; Stenvik, G.E.; Butenko, M.A.; Kristiansen, W.; Cho, S.K.; Hepworth, S.R.; Aalen, R.B.; Haughn, G.W. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 2008, 135, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Hepworth, S.R.; Zhang, Y.; McKim, S.; Li, X.; Haughn, G.W. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral pattering in Arabidpsis. Plant Cell 2005, 17, 1434–1448. [Google Scholar] [CrossRef]
- Butenko, M.A.; Patterson, S.E.; Grini, P.E.; Stenvik, G.-E.; Amundsen, S.S.; Mandal, A.; Aalen, R.B. INFLORESCENCE DEFICIENT IN ABSCISSION Controls Floral Organ Abscission in Arabidopsis and Identifies a Novel Family of Putative Ligands in Plants. Plant Cell 2003, 15, 2296–2307. [Google Scholar] [CrossRef]
- Liljegren, S.J.; Leslie, M.E.; Darnielle, L.; Lewis, M.W.; Taylor, S.M.; Luo, R.; Geldner, N.; Chory, J.; Randazzo, P.A.; Yanofsky, M.F.; et al. Regulation of membrane trafficking and organ separation by the NEVERSHED ARF-GAP protein. Development 2009, 136, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Butenko, M.A.; Shi, C.-L.; Bolivar, J.L.; Winge, P.; Stenvik, G.-E.; Vie, A.K.; Leslie, M.E.; Brembu, T.; Kristiansen, W.; et al. NEVERSHED and INFLORESCENCE DEFICIENT IN ABSCISSION are differentially required for cell expansion and cell separation during floral organ abscission in Arabidopsis thaliana. J. Exp. Bot. 2013, 64. [Google Scholar] [CrossRef] [PubMed]
- Stenvik, G.E.; Tandstad, N.M.; Guo, Y.; Shi, C.L.; Kristiansen, W.; Holmgren, A.; Clark, S.E.; Aalen, R.B.; Butenko, M.A. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 2008, 20, 1805–1817. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.K.; Larue, C.T.; Chevalier, D.; Wang, H.; Jinn, T.L.; Zhang, S.; Walker, J.C. Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 15629–15634. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.-L.; Butenko, M.A. Quantitative Analysis of Floral Organ Abscission in Arabidopsis Via a Petal Breakstrength Assay. In Plant Senescence: Methods and Protocols; Guo, Y., Ed.; Springer: New York, NY, USA, 2018; pp. 81–88. [Google Scholar] [CrossRef]
- Cai, S.; Lashbrook, C.C. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: Enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol. 2008, 146, 1305–1321. [Google Scholar] [CrossRef] [PubMed]
- Meir, S.; Philosoph-Hadas, S.; Riov, J.; Tucker, M.L.; Patterson, S.E.; Roberts, J.A. Re-evaluation of the ethylene-dependent and -independent pathways in the regulation of floral and organ abscission. J. Exp. Bot. 2019, 70, 1461–1467. [Google Scholar] [CrossRef]
- Stø, I.M.; Orr, R.J.S.; Fooyontphanich, K.; Jin, X.; Knutsen, J.M.B.; Fischer, U.; Tranbarger, T.J.; Nordal, I.; Aalen, R.B. Conservation of the abscission signaling peptide IDA during Angiosperm evolution: Withstanding genome duplications and gain and loss of the receptors HAE/HSL2. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Vie, A.K.; Najafi, J.; Winge, P.; Cattan, E.; Wrzaczek, M.; Kangasjarvi, J.; Miller, G.; Brembu, T.; Bones, A.M. The IDA-LIKE peptides IDL6 and IDL7 are negative modulators of stress responses in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 3557–3571. [Google Scholar] [CrossRef]
- Wang, X.; Hou, S.; Wu, Q.; Lin, M.; Acharya, B.R.; Wu, D.; Zhang, W. IDL6-HAE/HSL2 impacts pectin degradation and resistance to Pseudomonas syringae pv tomato DC3000 in Arabidopsis leaves. Plant J. Cell Mol. Boil. 2017, 89, 250–263. [Google Scholar] [CrossRef]
- Butenko, M.A.; Wildhagen, M.; Albert, M.; Jehle, A.; Kalbacher, H.; Aalen, R.B.; Felix, G. Tools and Strategies to Match Peptide-Ligand Receptor Pairs. Plant Cell 2014, 26, 1838–1847. [Google Scholar] [CrossRef]
- Santiago, J.; Brandt, B.; Wildhagen, M.; Hohmann, U.; Hothorn, L.A.; Butenko, M.A.; Hothorn, M. Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 2016, 5, e15075. [Google Scholar] [CrossRef]
- Schardon, K.; Hohl, M.; Graff, L.; Pfannstiel, J.; Schulze, W.; Stintzi, A.; Schaller, A. Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 2016, 354, 1594–1597. [Google Scholar] [CrossRef] [PubMed]
- Niederhuth, C.; Patharkar, O.R.; Walker, J. Transcriptional profiling of the Arabidopsis abscission mutant hae hsl2 by RNA-Seq. BMC Genom. 2013, 14, 37. [Google Scholar] [CrossRef]
- Shi, C.-L.; Stenvik, G.-E.; Vie, A.K.; Bones, A.M.; Pautot, V.; Proveniers, M.; Aalen, R.B.; Butenko, M.A. Arabidopsis Class I KNOTTED-Like Homeobox Proteins Act Downstream in the IDA-HAE/HSL2 Floral Abscission Signaling Pathway. Plant Cell Online 2011, 23, 2553–2567. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Shao, Y.; Ge, S.; Zhang, M.; Zhang, T.; Hu, X.; Liu, Y.; Walker, J.; Zhang, S.; Xu, J. A MAPK cascade downstream of IDA-HAE/HSL2 ligand-receptor pair in lateral root emergence. Nat. Plants 2019, 5, 414–423. [Google Scholar] [CrossRef]
- Kumpf, R.P.; Nowack, M.K. The root cap: A short story of life and death. J. Exp. Bot. 2015, 66, 5651–5662. [Google Scholar] [CrossRef]
- Tang, H.; Wang, X.; Bowers, J.E.; Ming, R.; Alam, M.; Paterson, A.H. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008, 18, 1944–1954. [Google Scholar] [CrossRef]
- Kim, J.; Yang, J.; Yang, R.; Sicher, R.C.; Chang, C.; Tucker, M.L. Transcriptome Analysis of Soybean Leaf Abscission Identifies Transcriptional Regulators of Organ Polarity and Cell Fate. Front. Plant Sci. 2016, 7, 125. [Google Scholar] [CrossRef]
- Lee, Y.; Yoon, T.H.; Lee, J.; Jeon, S.Y.; Lee, J.H.; Lee, M.K.; Chen, H.; Yun, J.; Oh, S.Y.; Wen, X.; et al. A Lignin Molecular Brace Controls Precision Processing of Cell Walls Critical for Surface Integrity in Arabidopsis. Cell 2018, 173, 1468–1480.e9. [Google Scholar] [CrossRef]
- Glazinska, P.; Wojciechowski, W.; Kulasek, M.; Glinkowski, W.; Marciniak, K.; Klajn, N.; Kesy, J.; Kopcewicz, J. De novo Transcriptome Profiling of Flowers, Flower Pedicels and Pods of Lupinus luteus (Yellow Lupine) Reveals Complex Expression Changes during Organ Abscission. Front. Plant Sci. 2017, 8, 641. [Google Scholar] [CrossRef]
- Kim, J.; Sundaresan, S.; Philosoph-Hadas, S.; Yang, R.; Meir, S.; Tucker, M.L. Examination of the Abscission-Associated Transcriptomes for Soybean, Tomato, and Arabidopsis Highlights the Conserved Biosynthesis of an Extensible Extracellular Matrix and Boundary Layer. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, S.; Philosoph-Hadas, S.; Riov, J.; Belausov, E.; Kochanek, B.; Tucker, M.L.; Meir, S. Abscission of flowers and floral organs is closely associated with alkalization of the cytosol in abscission zone cells. J. Exp. Bot. 2015, 66, 1355–1368. [Google Scholar] [CrossRef] [PubMed]
- Stenvik, G.E.; Butenko, M.A.; Urbanowicz, B.R.; Rose, J.K.; Aalen, R.B. Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 2006, 18, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Patharkar, O.R.; Gassmann, W.; Walker, J.C. Leaf shedding as an anti-bacterial defense in Arabidopsis cauline leaves. PLoS Genet. 2017, 13, e1007132. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.L.; Yang, R. IDA-like gene expression in soybean and tomato leaf abscission and requirement for a diffusible stelar abscission signal. AoB Plants 2012, 2012, pls035. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, C.; Fan, M.; Ma, W.; Chen, M.; Cai, F.; Liu, K.; Lin, F. GmIDL2a and GmIDL4a, Encoding the Inflorescence Deficient in Abscission-Like Protein, Are Involved in Soybean Cell Wall Degradation during Lateral Root Emergence. Int. J. Mol. Sci. 2018, 19, 2262. [Google Scholar] [CrossRef] [PubMed]
- Ying, P.; Li, C.; Liu, X.; Xia, R.; Zhao, M.; Li, J. Identification and molecular characterization of an IDA-like gene from litchi, LcIDL1, whose ectopic expression promotes floral organ abscission in Arabidopsis. Sci. Rep. 2016, 6, 37135. [Google Scholar] [CrossRef] [PubMed]
- Estornell, L.H.; Wildhagen, M.; Perez-Amador, M.A.; Talon, M.; Tadeo, F.R.; Butenko, M.A. The IDA Peptide Controls Abscission in Arabidopsis and Citrus. Front. Plant Sci. 2015, 6, 1003. [Google Scholar] [CrossRef]
- Wilmowicz, E.; Kućko, A.; Ostrowski, M.; Panek, K. INFLORESCENCE DEFICIENT IN ABSCISSION-like is an abscission-associated and phytohormone-regulated gene in flower separation of Lupinus luteus. Plant Growth Regul. 2018, 85, 91–100. [Google Scholar] [CrossRef]
- Tranbarger, T.J.; Domonhedo, H.; Cazemajor, M.; Dubreuil, C.; Fischer, U.; Morcillo, F. The PIP Peptide of INFLORESCENCE DEFICIENT IN ABSCISSION Enhances Populus Leaf and Elaeis guineensis Fruit Abscission. Plants 2019, 8, 143. [Google Scholar] [CrossRef]
- Jin, X.; Zimmermann, J.; Polle, A.; Fischer, U. Auxin is a long-range signal that acts independently of ethylene signaling on leaf abscission in Populus. Front. Plant Sci. 2015, 6, 634. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, W.G. Effect of ethylene on flower abscission: A survey. Ann. Bot. (Lond.) 2002, 89, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Butenko, M.A.; Stenvik, G.E.; Alm, V.; Saether, B.; Patterson, S.E.; Aalen, R.B. Ethylene-dependent and -independent pathways controlling floral abscission are revealed to converge using promoter:reporter gene constructs in the ida abscission mutant. J. Exp. Bot. 2006, 57, 3627–3637. [Google Scholar] [CrossRef] [PubMed]
- Aan den Toorn, M.; Albrecht, C.; de Vries, S. On the Origin of SERKs: Bioinformatics Analysis of the Somatic Embryogenesis Receptor Kinases. Mol. Plant 2015, 8, 762–782. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2005, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Olsson, V.; Joos, L.; Zhu, S.; Gevaert, K.; Butenko, M.A.; De Smet, I. Look Closely, the Beautiful May Be Small: Precursor-Derived Peptides in Plants. Annu. Rev. Plant Biol. 2019, 70, 153–186. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Sawa, S. Diverse function of plant peptide hormones in local signaling and development. Curr. Opin. Plant Biol. 2019, 51, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Bartels, S.; Boller, T. Quo vadis, Pep? Plant elicitor peptides at the crossroads of immunity, stress, and development. J. Exp. Bot. 2015, 66, 5183–5193. [Google Scholar] [CrossRef] [PubMed]
- Albert, M. Peptides as triggers of plant defence. J. Exp. Bot. 2013, 64, 5269–5279. [Google Scholar] [CrossRef] [PubMed][Green Version]

| Genes | Process | Methods 1 | Species |
|---|---|---|---|
| IDA, HAE, HSL2 | Floral abscission | Mutant; 35S-phenotype; Binding assay; Crystal structure | Arabidopsis [10,21,22,34] |
| IDA, HAE, HSL2 | LR emergence | Mutant | Arabidopsis [5] |
| IDA, HAE, HSL2 | Leaf abscission | Mutant | Arabidopsis [4,35] |
| IDL1, HSL2 | Root cap sloughing | Mutant; Enhanced-phenotype; Binding assay | Arabidopsis [6] |
| GmIDA2a/2b, GmHAE3b/5a/5b | Leaf abscission | Expressed in AZ | Soybean [36] |
| GmIDL2a/4b | LR emergence | 35S-phenotype | Soybean [37] |
| LcIDL1 | Flower/Fruit abscission | Expressed in AZ; 35S-phenotype; Rescue of ida mutant | Litchi [38] |
| CitIDA3 | Fruit abscission | Expressed in AZ 35S phenotype Rescue of ida mutant | Citrus [39] |
| LlIDA | Flower abscission | Expressed in AZ; Synthetic peptide | Lupin [40] |
| EgIDA2/5 | Fruit abscission | Expressed in AZ Synthetic peptide | Oil palm [18,41] |
| PpIDA, PpIDL1 | Leaf abscission | Expressed in AZ Synthetic peptide | Poplar [18,41] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, C.-L.; Alling, R.M.; Hammerstad, M.; Aalen, R.B. Control of Organ Abscission and Other Cell Separation Processes by Evolutionary Conserved Peptide Signaling. Plants 2019, 8, 225. https://doi.org/10.3390/plants8070225
Shi C-L, Alling RM, Hammerstad M, Aalen RB. Control of Organ Abscission and Other Cell Separation Processes by Evolutionary Conserved Peptide Signaling. Plants. 2019; 8(7):225. https://doi.org/10.3390/plants8070225
Chicago/Turabian StyleShi, Chun-Lin, Renate Marie Alling, Marta Hammerstad, and Reidunn B. Aalen. 2019. "Control of Organ Abscission and Other Cell Separation Processes by Evolutionary Conserved Peptide Signaling" Plants 8, no. 7: 225. https://doi.org/10.3390/plants8070225
APA StyleShi, C.-L., Alling, R. M., Hammerstad, M., & Aalen, R. B. (2019). Control of Organ Abscission and Other Cell Separation Processes by Evolutionary Conserved Peptide Signaling. Plants, 8(7), 225. https://doi.org/10.3390/plants8070225





