Understanding the Role of the WRKY Gene Family under Stress Conditions in Pigeonpea (Cajanus Cajan L.)
Abstract
1. Introduction
2. Results
2.1. Identification of WRKY Gene Family in Pigeonpea
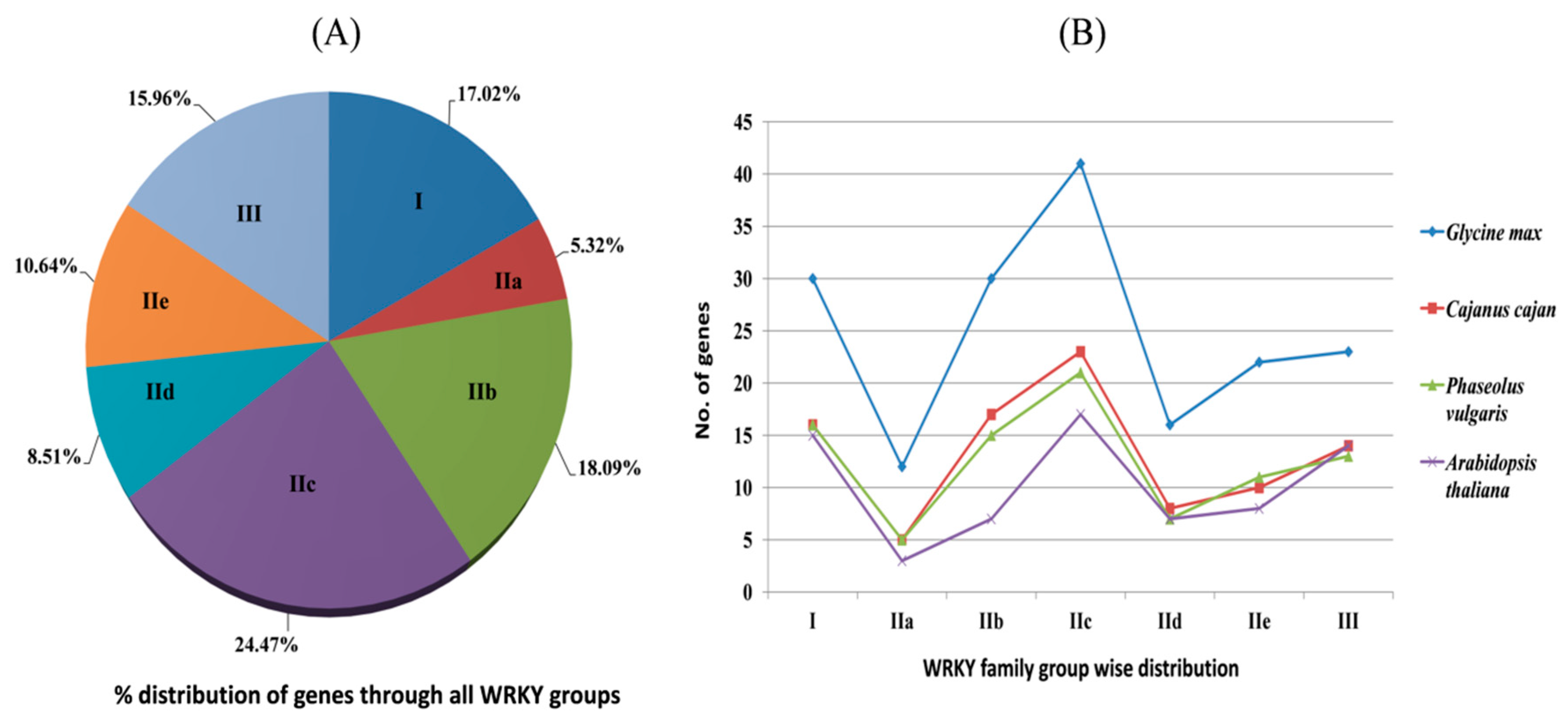
2.2. Phylogenetic Distribution of CcWRKY Proteins
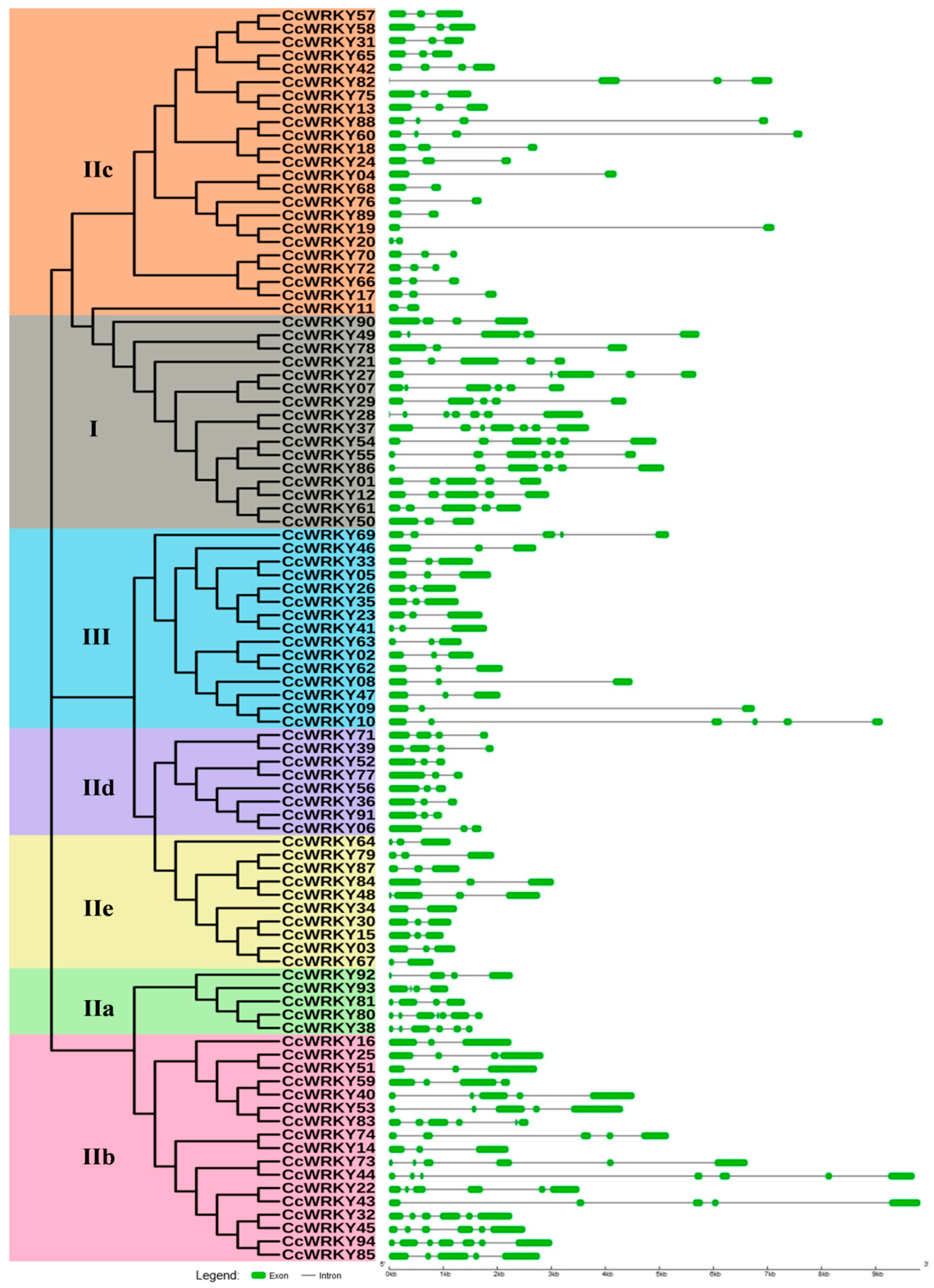
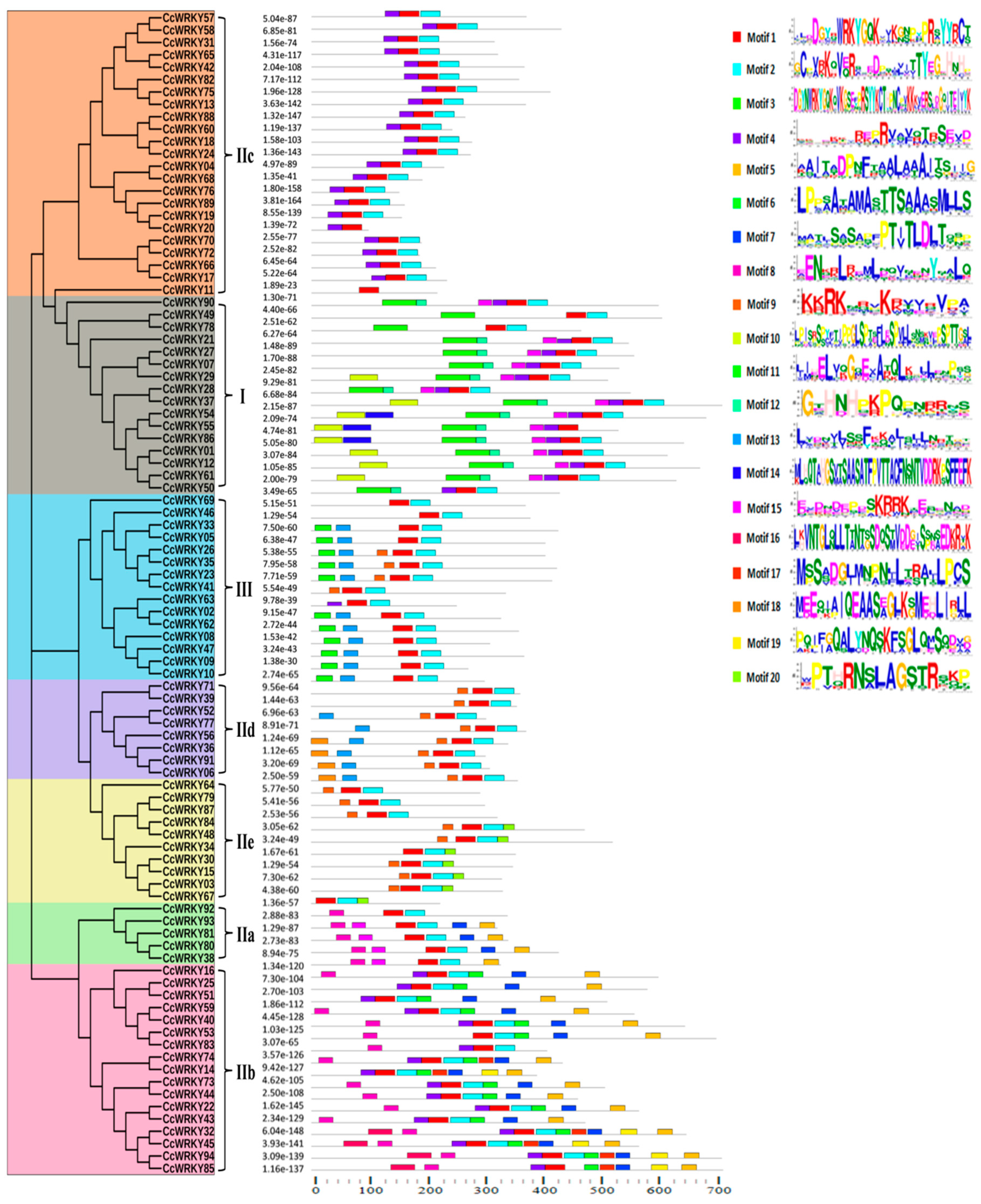
2.3. Exon-Intron Organization and Motif Composition of WRKY Proteins
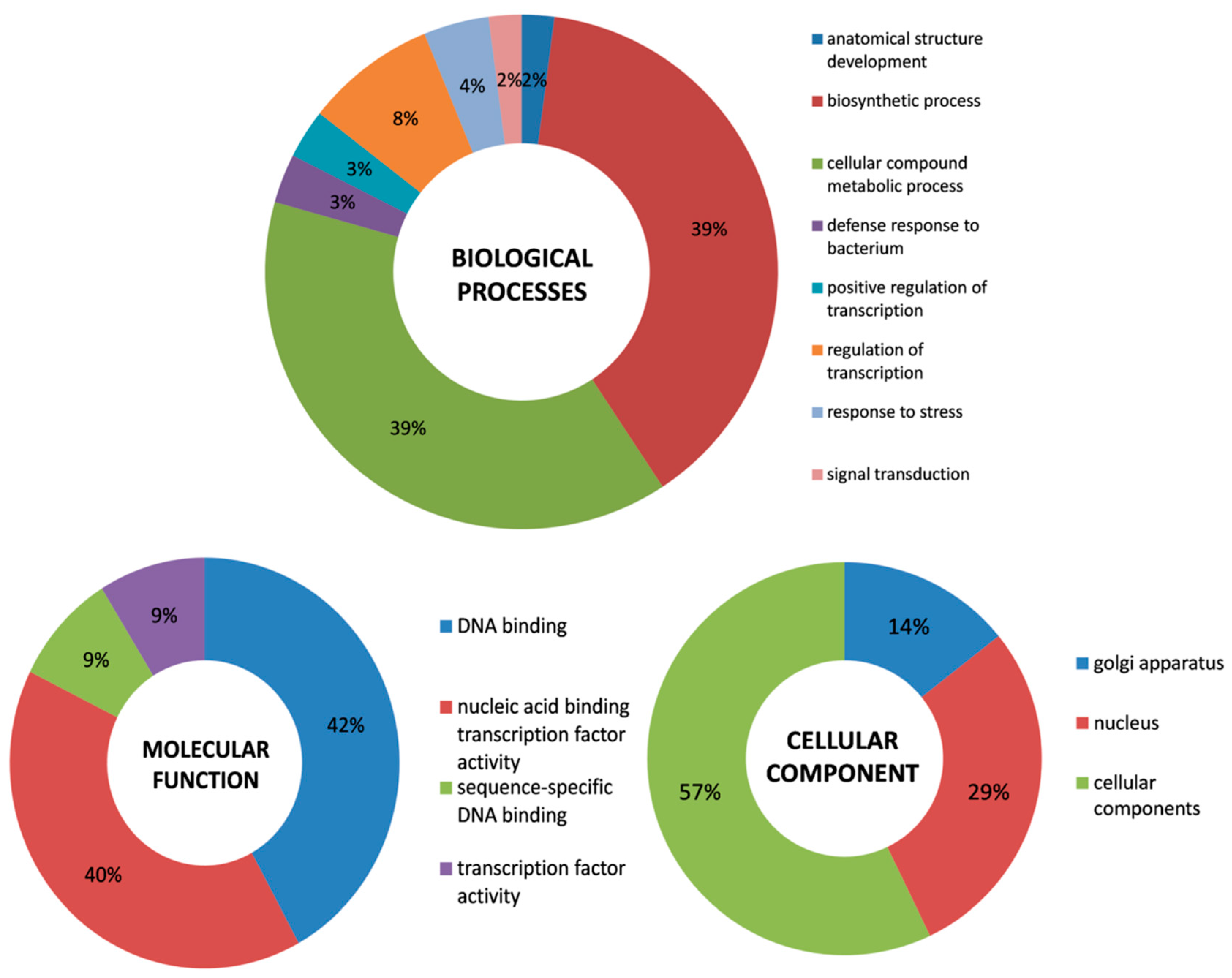
2.4. Stress-related Regulatory Elements in the Upstream Promoter Region and Functional Annotation of WRKY Genes
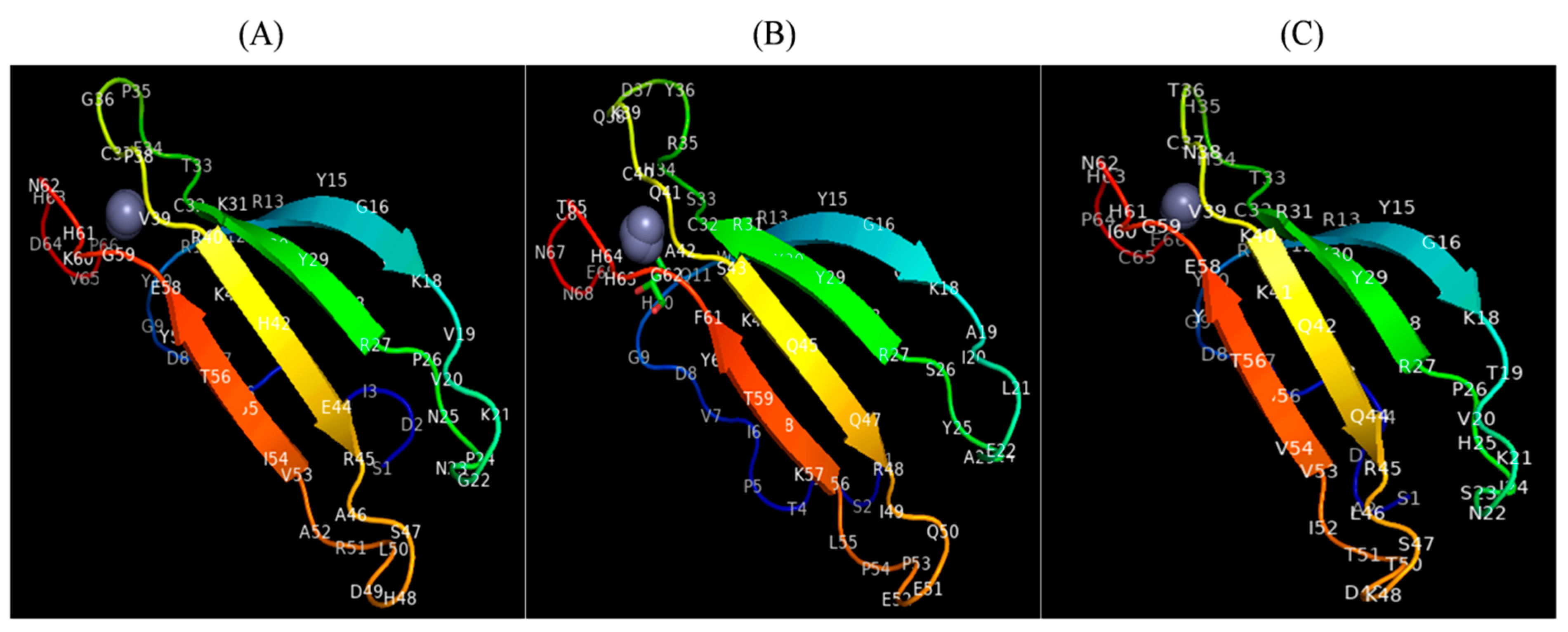
2.5. Tertiary Structures of CcWRKY Proteins
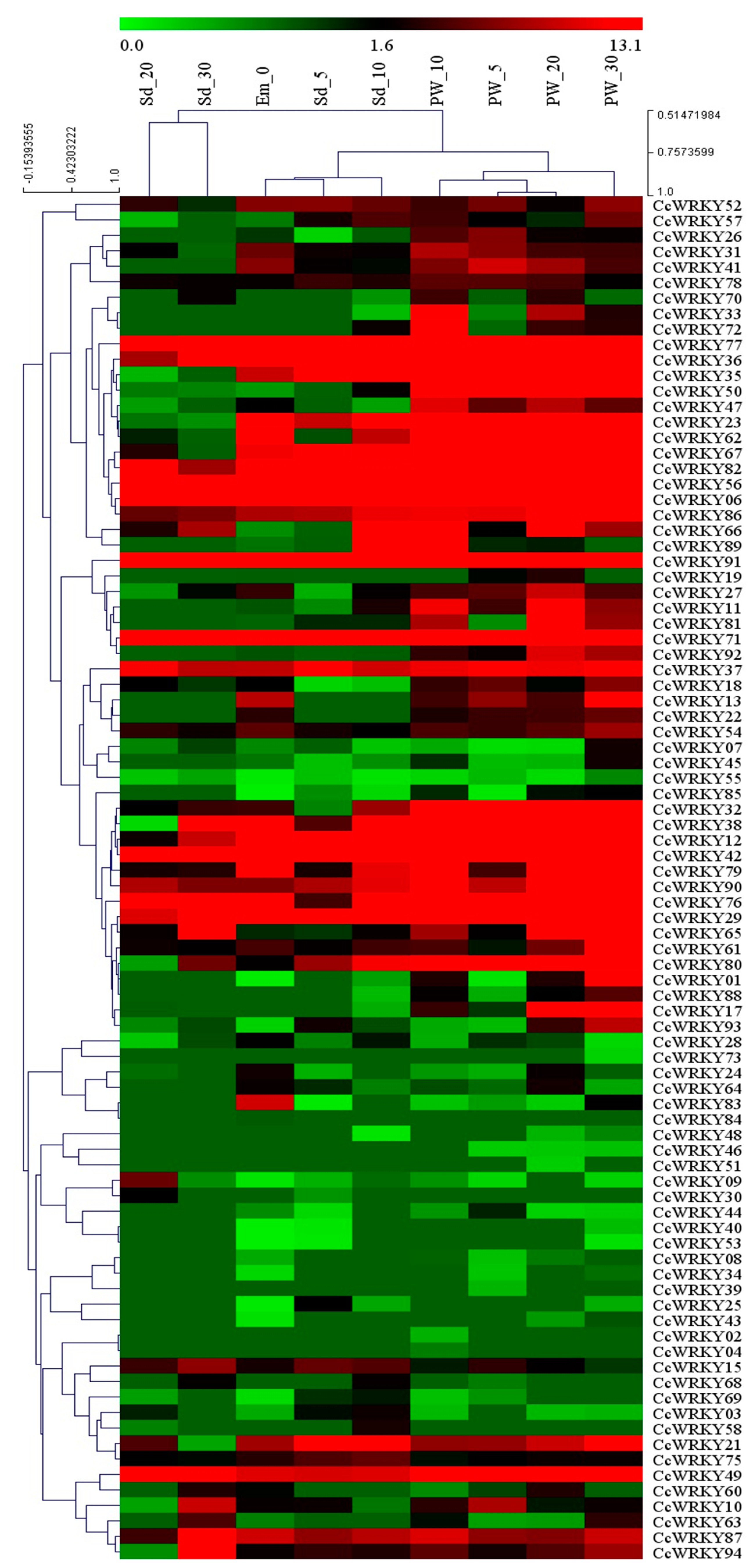
2.6. Expression of CcWRKY Genes During Different Developmental Stages
2.7. Gene Co-expression Network of CcWRKY Proteins in Pigeonpea
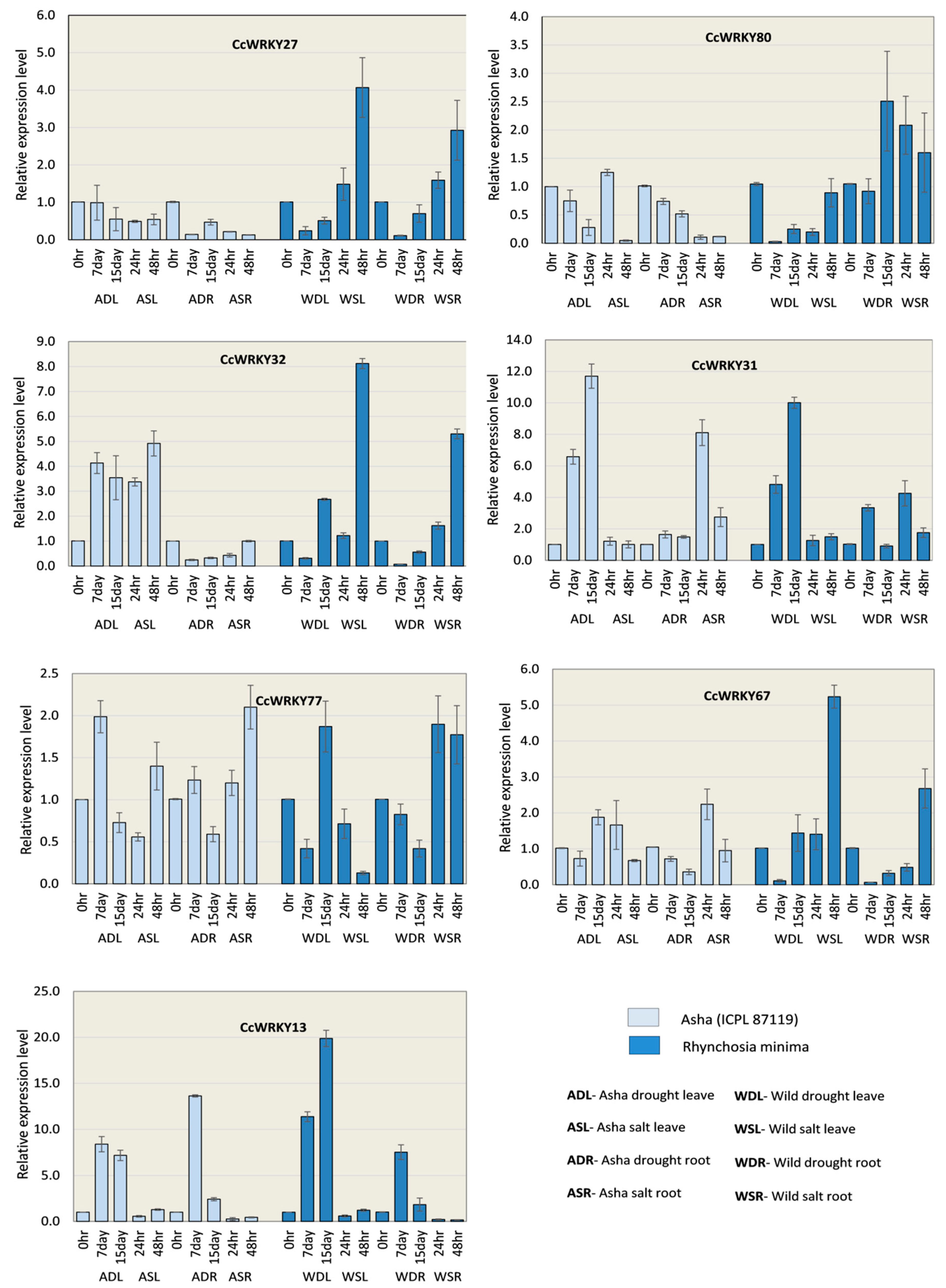
2.8. Expression of CcWRKYs Under Drought and Salt Conditions
3. Discussion
4. Materials and Methods
4.1. Identification of Genes Encoding WRKY Transcription Factors (TFs) in Pigeonpea Genome
4.2. Sequence Analysis and Phylogeny
4.3. Gene Structure, Motif Conservation, and Regulatory Element Analysis
4.4. Gene Annotation and Co-expression Network Analysis
4.5. Homology-Based CcWRKYs Protein Structure Prediction
4.6. Transcript Abundance Analysis
4.7. Plant Materials and Stress Treatments
4.8. RNA Isolation, cDNA Synthesis, and qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Singh, A.; Sharma, A.K.; Singh, N.K.; Sharma, T.R. PpTFDB: A pigeonpea transcription factor database for exploring functional genomics in legumes. PLoS ONE 2017, 12, e0179736. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Chen, W.; Li, Y.; Bharti, A.K.; Saxena, R.K.; Schlueter, J.A.; Donoghue, M.T.A.; Azam, S.; Fan, G.; Whaley, A.M.; et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 2011, 30, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Gowda, C.L.; Saxena, K.; Srivastava, R.; Upadhyaya, H.; Silim, S. Pigeonpea: From an Orphan to a Leader in Food Legumes. Biodivers. Agric. 2013, 361–374. [Google Scholar] [CrossRef]
- FAOSTAT. Available online: http://faostat.fao.org/ (accessed on 16 November 2018).
- Singh, N.K.; Gupta, D.K.; Jayaswal, P.K.; Mahato, A.K.; Dutta, S.; Singh, S.; Bhutani, S.; Dogra, V.; Singh, B.P.; Kumawat, G.; et al. The first draft of the pigeonpea genome sequence. J. Plant Biochem. Biotechnol. 2011, 21, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Penmetsa, R.V.; Dutta, S.; Kulwal, P.L.; Saxena, R.K.; Datta, S.; Sharma, T.R.; Rosen, B.; Carrasquilla-Garcia, N.; Farmer, A.D.; et al. Pigeonpea genomics initiative (PGI): An international effort to improve crop productivity of pigeonpea (Cajanus cajan L.). Mol. Breed. 2009, 26, 393–408. [Google Scholar] [CrossRef] [PubMed]
- Kingwell-Banham, E.; Fuller, D.Q. Pigeon Pea: Origins and Development. Encycl. Glob. Archaeol. 2014, 5941–5944. [Google Scholar] [CrossRef]
- Riechmann, J.L. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Encycl. Plant Crop Sci. 2004, 290, 2105–2110. [Google Scholar]
- Karkute, S.G.; Gujjar, R.S.; Rai, A.; Akhtar, M.; Singh, M.; Singh, B. Genome wide expression analysis of WRKY genes in tomato (Solanum lycopersicum) under drought stress. Plant Gene 2018, 13, 8–17. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Bai, Y.; Sunarti, S.; Kissoudis, C.; Visser, R.G.F.; Linden, C.G.V.D. The Role of Tomato WRKY Genes in Plant Responses to Combined Abiotic and Biotic Stresses. Front. Plant Sci. 2018, 9, 801. [Google Scholar] [CrossRef]
- Song, H.; Wang, P.; Hou, L.; Zhao, S.; Zhao, C.; Xia, H.; Li, P.; Zhang, Y.; Bian, X.; Wang, X. Global Analysis of WRKY Genes and Their Response to Dehydration and Salt Stress in Soybean. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Somssich, I.E. The Role of WRKY Transcription Factors in Plant Immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochimica et Biophysica Acta (BBA)-Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2011, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Muthamilarasan, M.; Bonthala, V.S.; Khandelwal, R.; Jaishankar, J.; Shweta, S.; Nawaz, K.; Prasad, M. Global analysis of WRKY transcription factor superfamily in Setaria identifies potential candidates involved in abiotic stress signaling. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.-Y.; Choi, C.-H.; Hwang, D.-J. The WRKY Superfamily of Rice Transcription Factors. Plant Pathol. J. 2010, 26, 110–114. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and amylase from sweet potato. MGG Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y. Cloning and analysis of expression profile of 13 WRKY genes in rice. Chin. Sci. Bull. 2004, 49, 2159–2168. [Google Scholar] [CrossRef]
- Wu, H.; Ni, Z.; Yao, Y.; Guo, G.; Sun, Q. Cloning and expression profiles of 15 genes encoding WRKY transcription factor in wheat (Triticum aestivem L.). Prog. Nat. Sci. 2008, 18, 697–705. [Google Scholar] [CrossRef]
- Sun, J.; An, H.; Shi, W.; Guo, X.; Li, H. Molecular cloning and characterization of GhWRKY11, a gene implicated in pathogen responses from cotton. S. Afr. J. Bot. 2012, 81, 113–123. [Google Scholar] [CrossRef]
- Zhou, Q.-Y.; Tian, A.-G.; Zou, H.-F.; Xie, Z.-M.; Lei, G.; Huang, J.; Wang, C.-M.; Wang, H.-W.; Zhang, J.-S.; Chen, S.-Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.G.; Sinha, A.K.; Proels, R.K.; Roitsch, T. Cloning and characterization of a novel LpWRKY1 transcription factor in tomato. Plant Physiol. Biochem. 2008, 46, 533–540. [Google Scholar] [CrossRef]
- Shekhawat, U.K.S.; Ganapathi, T.R.; Srinivas, L. Cloning and characterization of a novel stress-responsive WRKY transcription factor gene (MusaWRKY71) from Musa spp. cv. Karibale Monthan (ABB group) using transformed banana cells. Mol. Biol. Rep. 2010, 38, 4023–4035. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Dabi, M.; Agarwal, P.K. Molecular Cloning and Characterization of a Group II WRKY Transcription Factor from Jatropha curcas, an Important Biofuel Crop. DNA Cell Biol. 2014, 33, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY Gene Family in Rice (Oryza sativa). J. Integr. Plant Biol. 2007, 49, 827–842. [Google Scholar] [CrossRef]
- Wang, N.; Xia, E.-H.; Gao, L.-Z. Genome-wide analysis of WRKY family of transcription factors in common bean, Phaseolus vulgaris: Chromosomal localization, structure, evolution and expression divergence. Plant Gene 2016, 5, 22–30. [Google Scholar] [CrossRef]
- Kumar, K.; Srivastava, V.; Purayannur, S.; Kaladhar, V.C.; Cheruvu, P.J.; Verma, P.K. WRKY domain-encoding genes of a crop legume chickpea (Cicer arietinum): Comparative analysis with Medicago truncatula WRKY family and characterization of group-III gene(s). DNA Res. 2016, 23, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Yu, D. Arabidopsis WRKY2 transcription factor mediates seed germination and post germination arrest of development by abscisic acid. BMC Plant Biol. 2009, 9, 96. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54 and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Response. Plant Cell 2017, 1425–1439. [Google Scholar] [CrossRef]
- Wang, H.; Avci, U.; Nakashima, J.; Hahn, M.G.; Chen, F.; Dixon, R.A. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. USA 2010, 107, 22338–22343. [Google Scholar] [CrossRef]
- Johnson, C.S. TRANSPARENT TESTA GLABRA2, a Trichome and Seed Coat Development Gene of Arabidopsis, Encodes a WRKY Transcription Factor. Plant Cell Online 2002, 14, 1359–1375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-Q.; Xu, Y.; Lu, Y.; Yu, H.-X.; Gu, M.-H.; Liu, Q.-Q. The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 2011, 234, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Tang, S.; Zhi, H.; Jia, G.; Wang, H.; Diao, X. Loose Panicle1 encoding a novel WRKY transcription factor, regulates panicle development, stem elongation, and seed size in foxtail millet [Setaria italica (L.) P. Beauv.]. PLoS ONE 2017, 12, e0178730. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Li, D.; Wang, F.; Yu, D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, E1963–E1971. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef]
- Liu, H.; Sun, M.; Du, D.; Pan, H.; Cheng, T.; Wang, J.; Zhang, Q. Whole-Transcriptome Analysis of Differentially Expressed Genes in the Vegetative Buds, Floral Buds and Buds of Chrysanthemum morifolium. PLoS ONE 2015, 10, e0128009. [Google Scholar] [CrossRef]
- Chen, L.; Yang, Y.; Liu, C.; Zheng, Y.; Xu, M.; Wu, N.; Sheng, J.; Shen, L. Characterization of WRKY transcription factors in Solanum lycopersicum reveals collinearity and their expression patterns under cold treatment. Biochem. Biophys. Res. Commun. 2015, 464, 962–968. [Google Scholar] [CrossRef]
- Wu, Z.-J.; Li, X.-H.; Liu, Z.-W.; Li, H.; Wang, Y.-X.; Zhuang, J. Transcriptome-wide identification of Camellia sinensis WRKY transcription factors in response to temperature stress. Mol. Genet. Genom. 2015, 291, 255–269. [Google Scholar] [CrossRef]
- Wu, J.; Chen, J.; Wang, L.; Wang, S. Genome-Wide Investigation of WRKY Transcription Factors Involved in Terminal Drought Stress Response in Common Bean. Front. Plant Sci. 2017, 8, 380. [Google Scholar] [CrossRef]
- Chu, X.; Wang, C.; Chen, X.; Lu, W.; Li, H.; Wang, X.; Hao, L.; Guo, X. The Cotton WRKY Gene GhWRKY41 Positively Regulates Salt and Drought Stress Tolerance in Transgenic Nicotiana benthamiana. PLoS ONE 2015, 10, e0143022. [Google Scholar] [CrossRef]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Deslandes, L.; Olivier, J.; Theulieres, F.; Hirsch, J.; Feng, D.X.; Bittner-Eddy, P.; Beynon, J.; Marco, Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 2002, 99, 2404–2409. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, Y.; Chi, Y.; Fan, B.; Chen, Z. Characterization of Soybean WRKY Gene Family and Identification of Soybean WRKY Genes that Promote Resistance to Soybean Cyst Nematode. Sci. Rep. 2017, 7, 17804. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.U.; Choi, L.M.; Lee, G.-J.; Kim, Y.J.; Paek, K.-H. Capsicum annuum WRKY transcription factor d (CaWRKYd) regulates hypersensitive response and defense response upon Tobacco mosaic virus infection. Plant Sci. 2012, 197, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Yagi, M.; Kusano, T.; Sano, H. Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. MGG-Mol. Gen. Genet. 2000, 263, 30–37. [Google Scholar] [CrossRef]
- Li, P.; Song, A.; Gao, C.; Jiang, J.; Chen, S.; Fang, W.; Zhang, F.; Chen, F. The over-expression of a chrysanthemum WRKY transcription factor enhances aphid resistance. Plant Physiol. Biochem. 2015, 95, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, R.; Sonah, H.; Patil, G.; Chen, W.; Prince, S.; Mutava, R.; Vuong, T.; Valliyodan, B.; Nguyen, H.T. Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 2014, 5, 244. [Google Scholar] [CrossRef]
- Tuan, V.P.; Narith, D.; Tshibangu-Kabamba, E.; Dung, H.D.Q.; Viet, P.T.; Sokomoth, S.; Binh, T.T.; Sokhem, S.; Tri, T.D.; Ngov, S.; et al. A Next-Generation Sequencing-Based Approach to Identify Genetic Determinants of Antibiotic Resistance in Cambodian Helicobacter pylori Clinical Isolates. J. Clin. Med. 2019, 8, 858. [Google Scholar] [CrossRef]
- Zargar, S.M.; Raatz, B.; Sonah, H.; Bhat, J.A.; Dar, Z.A.; Agrawal, G.K.; Rakwal, R. Recent advances in molecular marker techniques: Insight into QTL mapping, GWAS and genomic selection in plants. J. Crop Sci. Biotechnol. 2015, 18, 293–308. [Google Scholar] [CrossRef]
- Sonah, H.; Bastien, M.; Iquira, E.; Tardivel, A.; Légaré, G.; Boyle, B.; Normandeau, É.; Laroche, J.; Larose, S.; Jean, M.; et al. An Improved Genotyping by Sequencing (GBS) Approach Offering Increased Versatility and Efficiency of SNP Discovery and Genotyping. PLoS ONE 2013, 8, e54603. [Google Scholar] [CrossRef]
- Satheesh, V.; Jagannadham, P.T.K.; Chidambaranathan, P.; Jain, P.K.; Srinivasan, R. NAC transcription factor genes: Genome-wide identification, phylogenetic, motif and cis-regulatory element analysis in pigeonpea (Cajanus cajan (L.) Millsp.). Mol. Biol. Rep. 2014, 41, 7763–7773. [Google Scholar] [CrossRef] [PubMed]
- Sirhindi, G.; Sharma, P.; Arya, P.; Goel, P.; Kumar, G.; Acharya, V.; Singh, A.K. Genome-wide characterization and expression profiling of TIFY gene family in pigeonpea (Cajanus cajan (L.) Millsp.) under copper stress. J. Plant Biochem. Biotechnol. 2016, 25, 301–310. [Google Scholar] [CrossRef]
- Pazhamala, L.T.; Agarwal, G.; Bajaj, P.; Kumar, V.; Kulshreshtha, A.; Saxena, R.K.; Varshney, R.K. Deciphering Transcriptional Programming during Pod and Seed Development Using RNA-Seq in Pigeonpea (Cajanus cajan). PLoS ONE 2016, 11, e0164959. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pazhamala, L.T.; Purohit, S.; Saxena, R.K.; Garg, V.; Krishnamurthy, L.; Verdier, J.; Varshney, R.K. Gene expression atlas of pigeonpea and its application to gain insights into genes associated with pollen fertility implicated in seed formation. J. Exp. Bot. 2017, 68, 2037–2054. [Google Scholar] [CrossRef] [PubMed]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Chapman, J.; Hellsten, U.; Rokhsar, D. Sequence and Assembly of the Soybean Genome. Genetics and Genomics of Soybean Plant; Genetics and Genomics: Crops and Models101–112; Springer: New York, NY, USA, 2008. [Google Scholar]
- Mingyu, Z. WRKY transcription factor superfamily: Structure, origin and functions. Afr. J. Biotechnol. 2012, 11. [Google Scholar] [CrossRef]
- Wu, K.-L. The WRKY Family of Transcription Factors in Rice and Arabidopsis and Their Origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Brand, L.H.; Fischer, N.M.; Harter, K.; Kohlbacher, O.; Wanke, D. Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 2013, 41, 9764–9778. [Google Scholar] [CrossRef] [PubMed]
- Verk, M.C.V.; Pappaioannou, D.; Neeleman, L.; Bol, J.F.; Linthorst, H.J. A Novel WRKY Transcription Factor Is Required for Induction of PR-1a Gene Expression by Salicylic Acid and Bacterial Elicitors. Plant Physiol. 2008, 146, 1983–1995. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Silva, J.A.T.D.; Tan, J.; Zhang, J.; Pan, X.; Li, M.; Luo, J.; Duan, J. A Genome-Wide Identification of the WRKY Family Genes and a Survey of Potential WRKY Target Genes in Dendrobium officinale. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Watanabe, K.A.; Zhang, L.; Shen, Q.J. WRKY transcription factor genes in wild rice Oryza nivara. DNA Res. 2016, 23, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Xu, H.; Xiao, S.; Qin, Y.; Li, Y.; Yan, Y.; Hu, Y. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 2013, 13, 148. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Rabara, R.C.; Langum, T.J.; Boken, A.K.; Rushton, D.L.; Boomsma, D.D.; Rinerson, C.I.; Rabara, J.; Reese, R.; Chen, X.; et al. The WRKY transcription factor family in Brachypodium distachyon. BMC Genom. 2012, 13, 270. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, R.; Jiang, S.-Y.; Kumar, N.; Venkatesh, P.N.; Ramachandran, S. A Comprehensive Transcriptional Profiling of the WRKY Gene Family in Rice Under Various Abiotic and Phytohormone Treatments. Plant Cell Physiol. 2008, 49, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Li, W.; Zhao, J.; Liu, Y.; Guo, C. Transcriptome sequencing and expression profiling of genes involved in the response to abiotic stress in Medicago ruthenica. Genet. Mol. Biol. 2018, 41, 638–648. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016, 45, D200–D203. [Google Scholar] [CrossRef]
- Larkin, M.; Blackshields, G.; Brown, N.; Chenna, R.; Mcgettigan, P.; Mcwilliam, H.; Valentin, F.; Wallace, I.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Higo, K. PLACE: A database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998, 26, 358–359. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S. Blast2GO: A Comprehensive Suite for Functional Analysis in Plant Genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef]
- Tzfadia, O.; Diels, T.; Meyer, S.D.; Vandepoele, K.; Aharoni, A.; Peer, Y.V.D. CoExpNetViz: Comparative Co-Expression Networks Construction and Visualization Tool. Front. Plant Sci. 2016, 6, 1194. [Google Scholar] [CrossRef]
- Shannon, P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2017, 27, 129–134. [Google Scholar] [CrossRef]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B.; Bakker, P.I.W.D.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins Struct. Funct. Bioinform. 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Seo, C.H.; Alver, B.H.; Lee, S.; Park, P.J. EMSAR: Estimation of transcript abundance from RNA-seq data by mappability-based segmentation and reclustering. BMC Bioinform. 2015, 16, 278. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sinha, P.; Singh, V.K.; Suryanarayana, V.; Krishnamurthy, L.; Saxena, R.K.; Varshney, R.K. Evaluation and Validation of Housekeeping Genes as Reference for Gene Expression Studies in Pigeonpea (Cajanus cajan) Under Drought Stress Conditions. PLoS ONE 2015, 10, e0122847. [Google Scholar] [CrossRef] [PubMed]
- Schonfeld, M.A.; Johnson, R.C.; Carver, B.F.; Mornhinweg, D.W. Water Relations in Winter Wheat as Drought Resistance Indicators. Crop Sci. 1988, 28, 526–531. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.; Singh, P.K.; Sharma, A.K.; Singh, N.K.; Sonah, H.; Deshmukh, R.; Sharma, T.R. Understanding the Role of the WRKY Gene Family under Stress Conditions in Pigeonpea (Cajanus Cajan L.). Plants 2019, 8, 214. https://doi.org/10.3390/plants8070214
Singh A, Singh PK, Sharma AK, Singh NK, Sonah H, Deshmukh R, Sharma TR. Understanding the Role of the WRKY Gene Family under Stress Conditions in Pigeonpea (Cajanus Cajan L.). Plants. 2019; 8(7):214. https://doi.org/10.3390/plants8070214
Chicago/Turabian StyleSingh, Akshay, Pankaj Kumar Singh, Ajay Kumar Sharma, Nagendra Kumar Singh, Humira Sonah, Rupesh Deshmukh, and Tilak Raj Sharma. 2019. "Understanding the Role of the WRKY Gene Family under Stress Conditions in Pigeonpea (Cajanus Cajan L.)" Plants 8, no. 7: 214. https://doi.org/10.3390/plants8070214
APA StyleSingh, A., Singh, P. K., Sharma, A. K., Singh, N. K., Sonah, H., Deshmukh, R., & Sharma, T. R. (2019). Understanding the Role of the WRKY Gene Family under Stress Conditions in Pigeonpea (Cajanus Cajan L.). Plants, 8(7), 214. https://doi.org/10.3390/plants8070214







