Pollination Type Recognition from a Distance by the Ovary Is Revealed Through a Global Transcriptomic Analysis
Abstract
1. Introduction
2. Results and Discussion
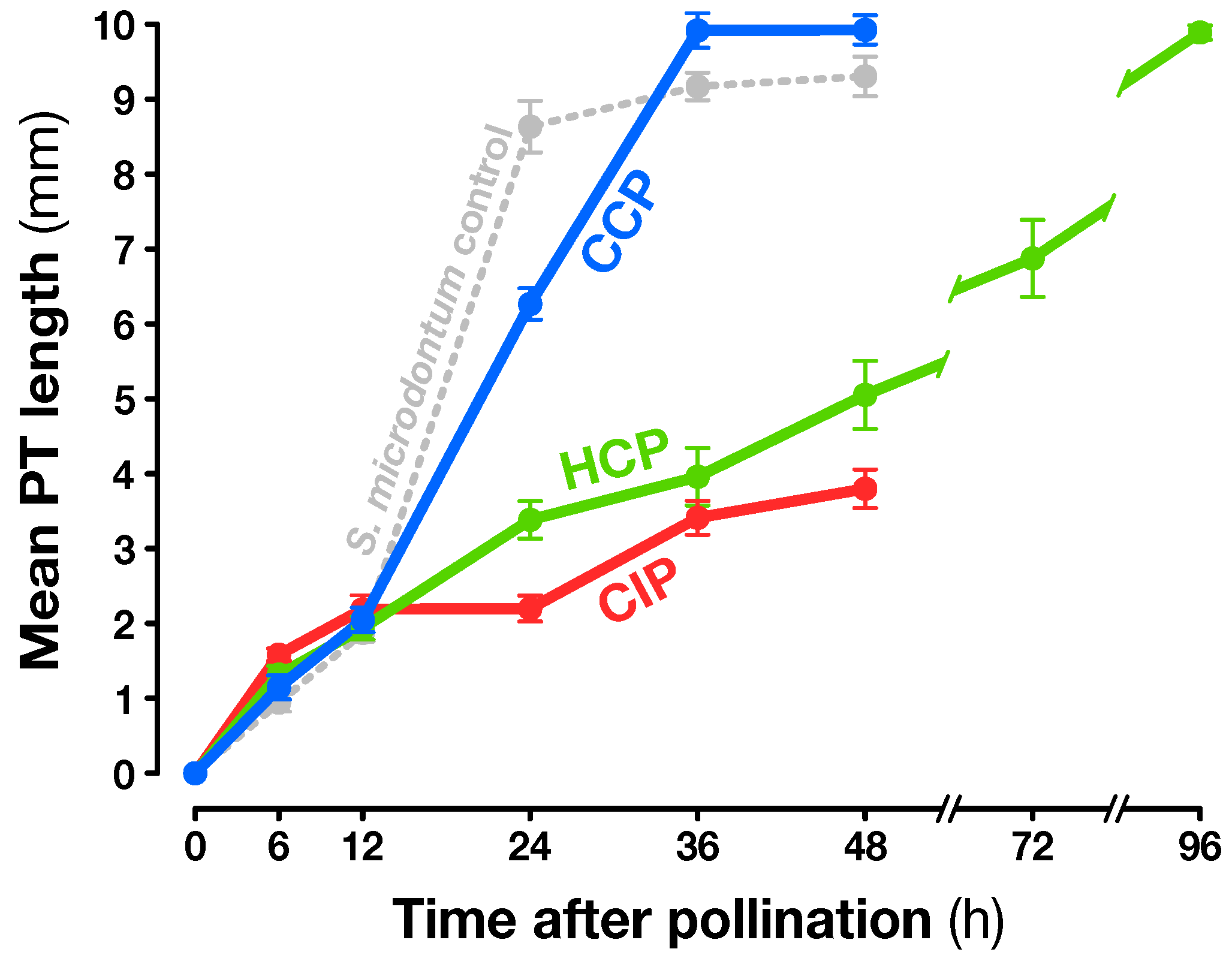
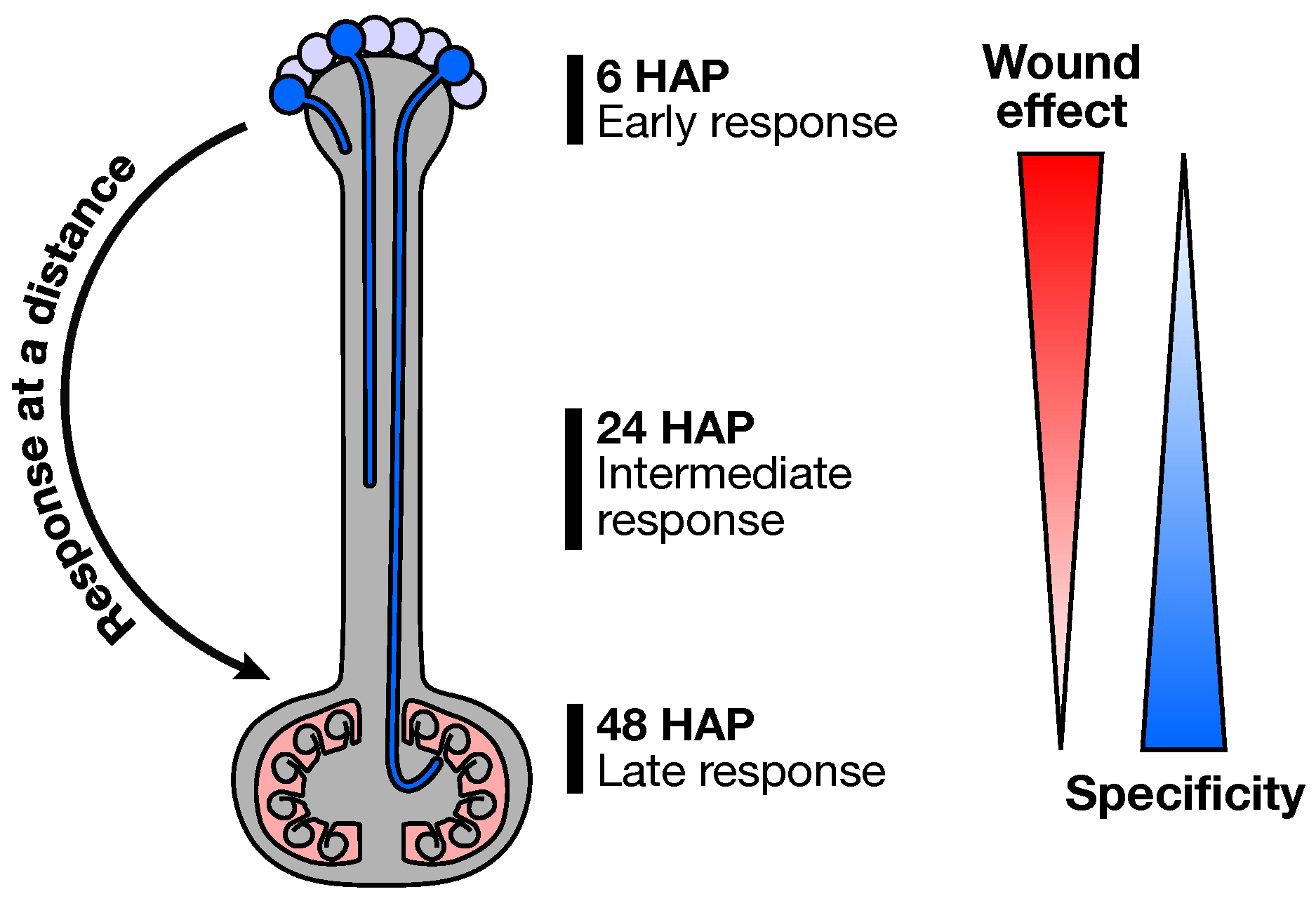
2.1. Experimental Design
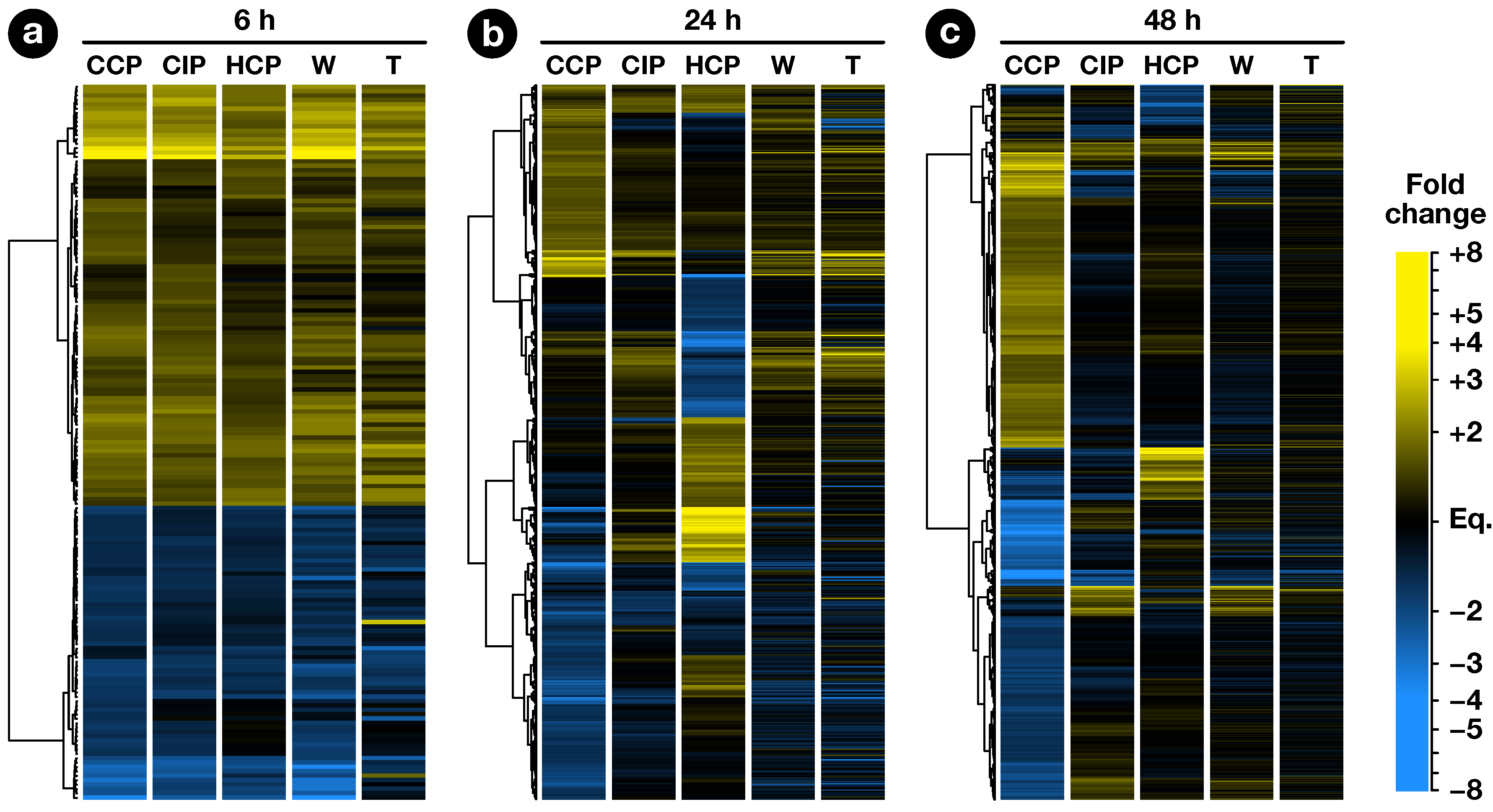
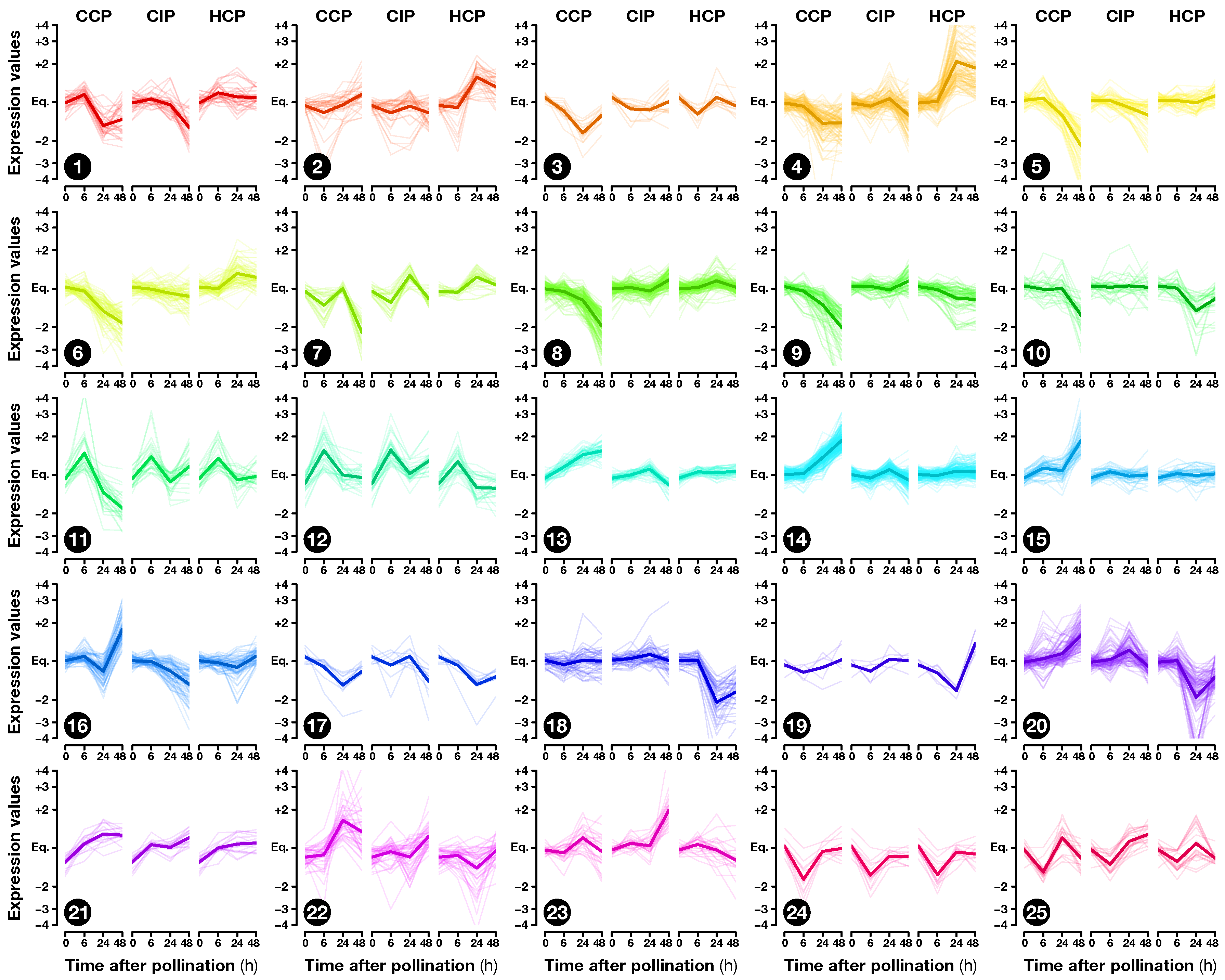
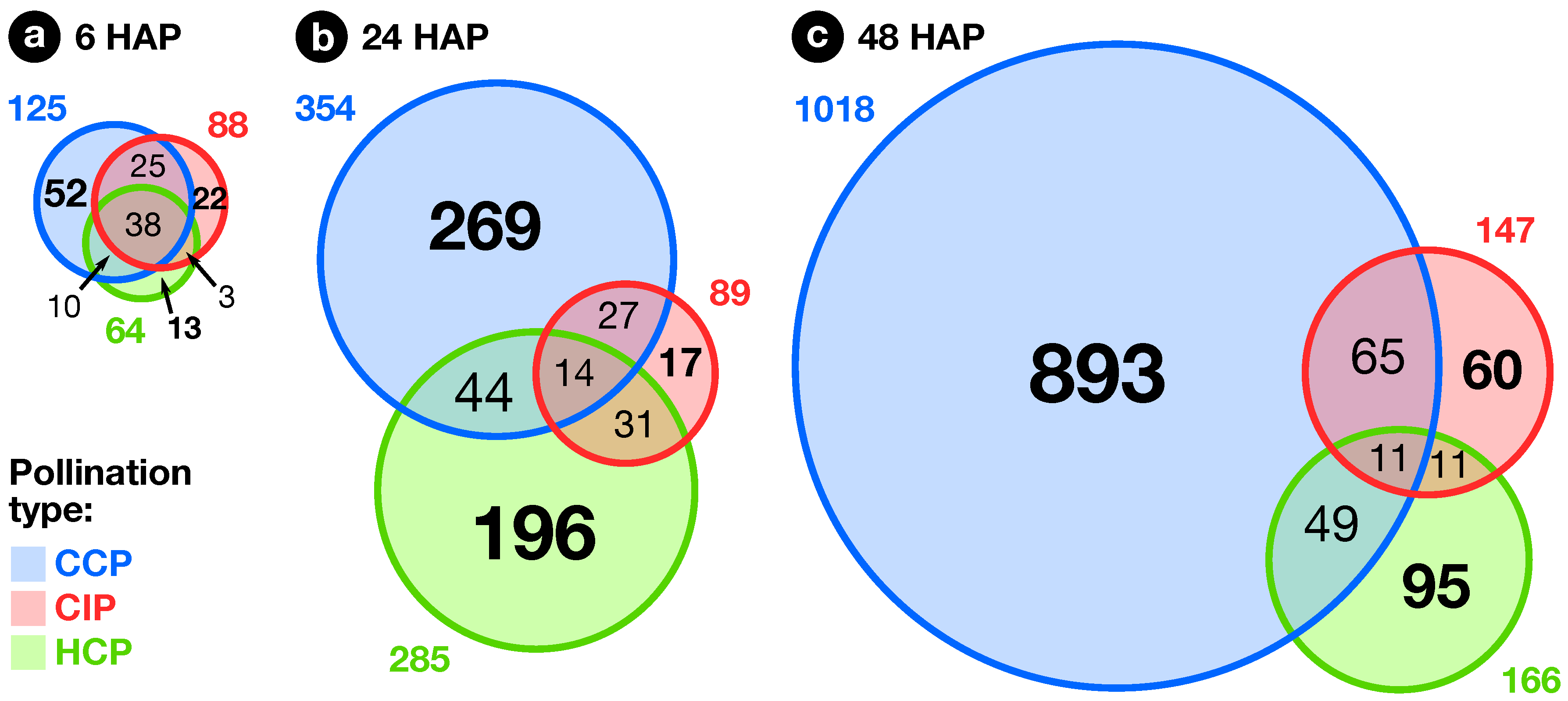
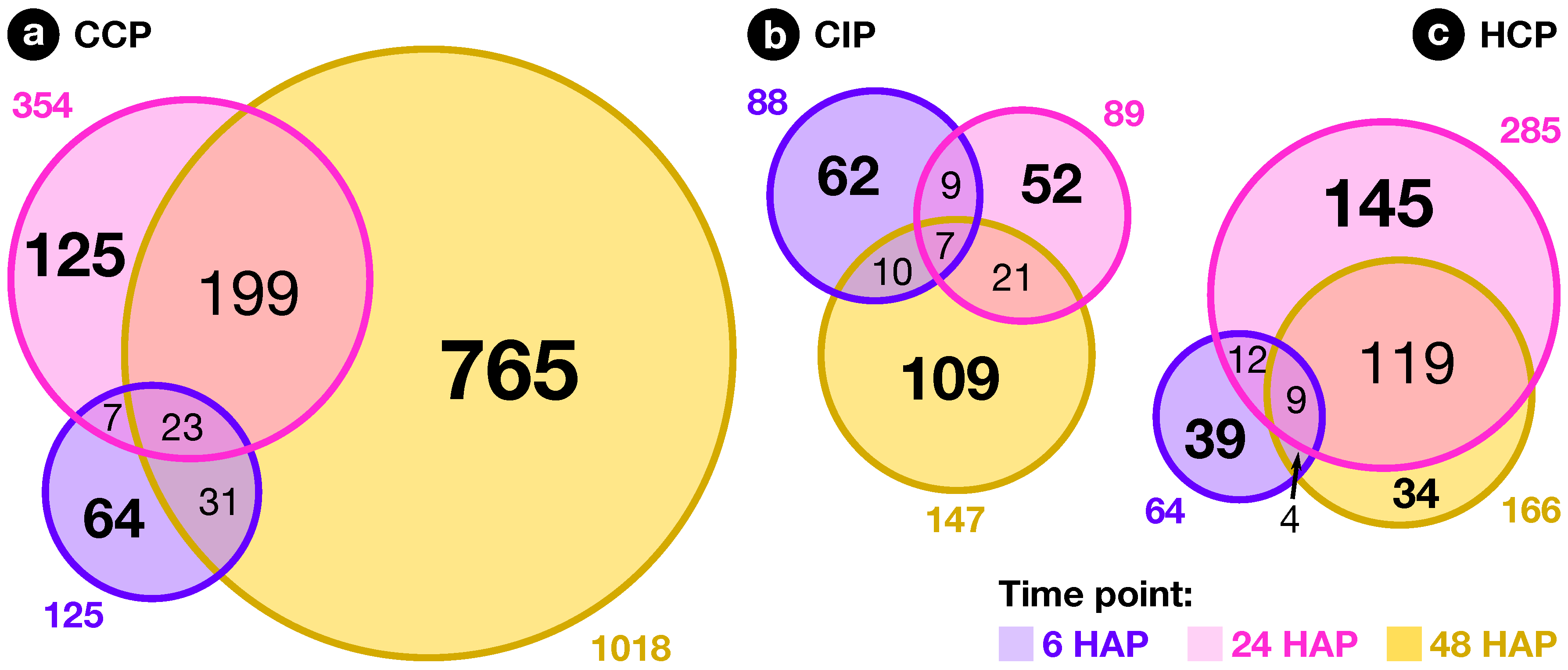
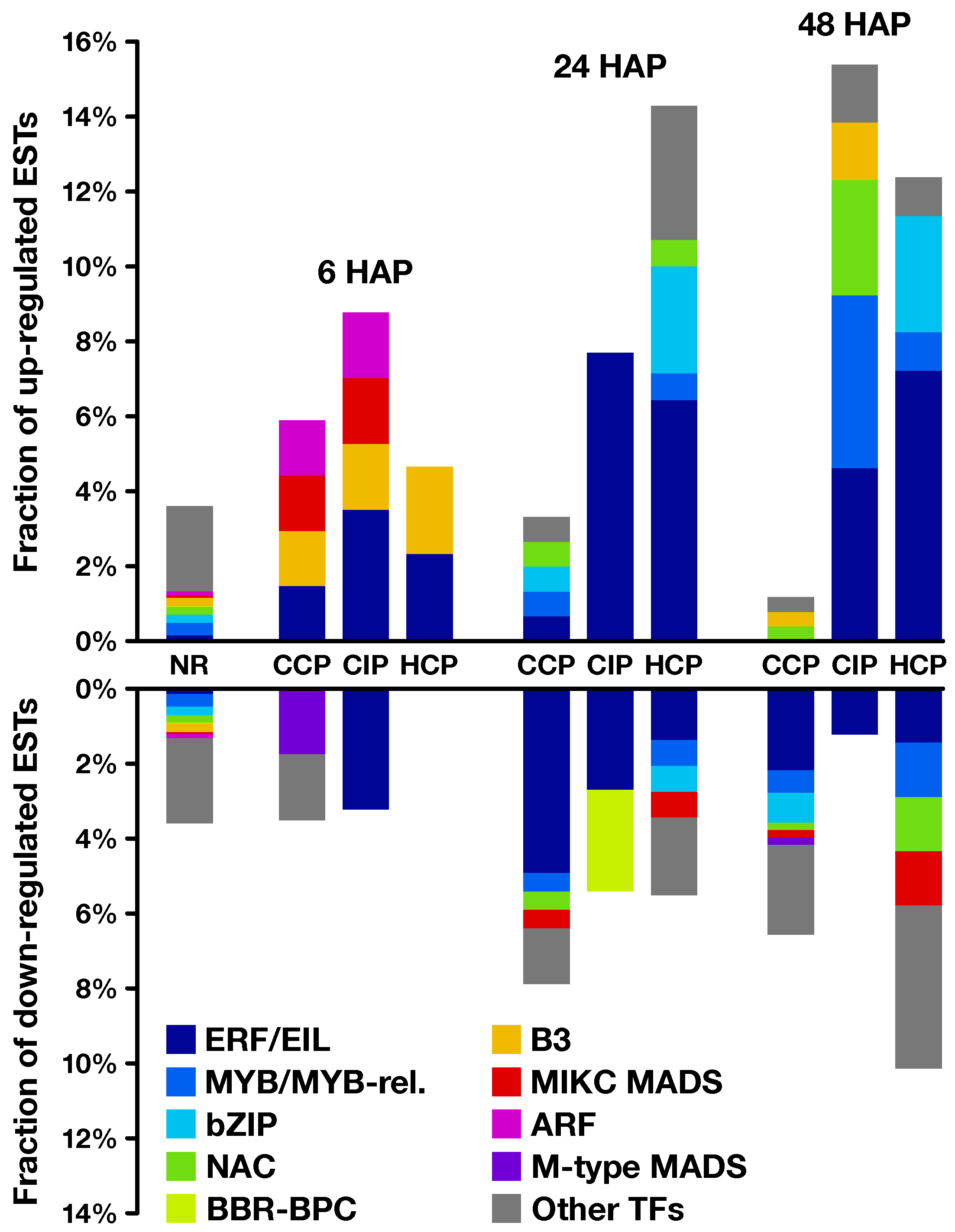
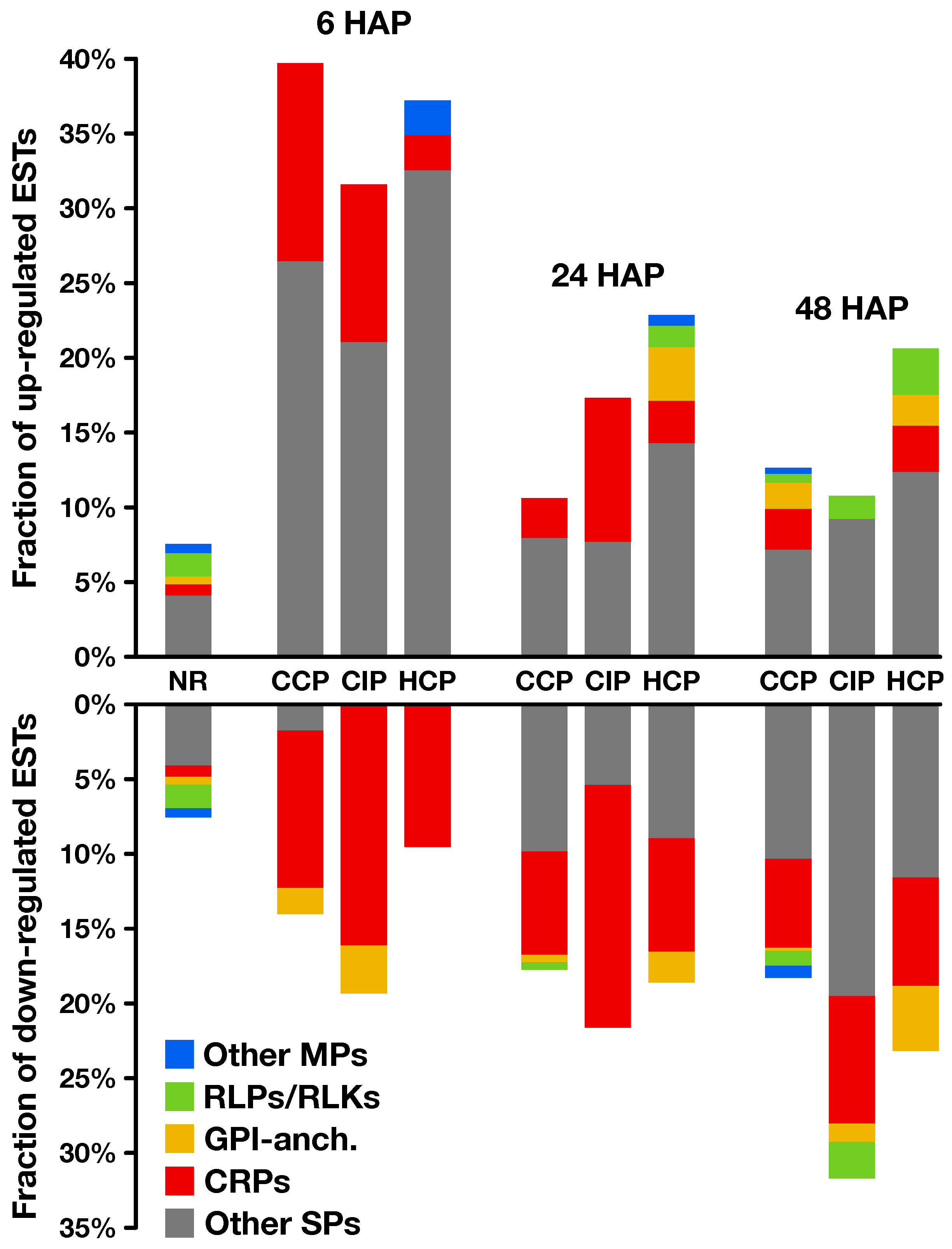
2.2. Expression Profiling of Pollination-Responsive Genes
2.3. Early Response to Pollination
2.4. Pollination Response after Completion of the SI Reaction
2.5. Fertilization and Late Pollination Responses
3. Conclusions
4. Materials and Methods
4.1. Plant Material and Pollination Conditions
4.2. Pollen Tube Growth Assay and Aniline Blue Staining
4.3. RNA Isolation and Microarray Experimental Design
4.4. Differential Expression Analaysis
4.5. Sequence Annotation
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mizuta, Y.; Higashiyama, T. Chemical signaling for pollen tube guidance at a glance. J. Cell Sci. 2018, 131, jcs208447. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.A.; Harper, J.F.; Palanivelu, R. A fruitful journey: Pollen tube navigation from germination to fertilization. Annu. Rev. Plant Biol. 2019, 70. [Google Scholar] [CrossRef] [PubMed]
- Doucet, J.; Lee, H.K.; Goring, D.R. Pollen acceptance or rejection: A tale of two pathways. Trends Plant Sci. 2016, 21, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lin, Z.; Triviño, M.; Nowack, M.K.; Franklin-Tong, V.E.; Bosch, M. Self-incompatibility in Papaver pollen: Programmed cell death in an acidic environment. J. Exp. Bot. 2018, 70, 2113–2123. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.S.; Wu, L.; Li, S.; Sun, P.; Kao, T.H. Insight into S-RNase-based self-incompatibility in Petunia: Recent findings and future directions. Front. Plant Sci. 2015, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.D.; Boavida, L.C.; Carneiro, J.; Haury, M.; Feijó, J.A. Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 2003, 133, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Honys, D.; Twell, D. Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 2003, 132, 640–652. [Google Scholar] [CrossRef]
- Honys, D.; Twell, D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 2004, 5, R85. [Google Scholar] [CrossRef]
- Pina, C.; Pinto, F.; Feijó, J.A.; Becker, J.D. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 2005, 138, 744–756. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.Z.; Song, L.F.; Zou, J.J.; Su, Z.; Wu, W.H. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 2008, 148, 1201–1211. [Google Scholar] [CrossRef]
- Swanson, R.; Clark, T.; Preuss, D. Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sex. Plant Reprod. 2005, 18, 163–171. [Google Scholar] [CrossRef]
- Tung, C.W.; Dwyer, K.G.; Nasrallah, M.E.; Nasrallah, J.B. Genome-wide identification of genes expressed in Arabidopsis pistils specifically along the path of pollen tube growth. Plant Physiol. 2005, 138, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xu, W.; Yang, W.; Kong, Z.; Xue, Y. Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol. 2007, 144, 1797–1812. [Google Scholar] [CrossRef] [PubMed]
- Quiapim, A.C.; Brito, M.S.; Bernardes, L.A.S.; Dasilva, I.; Malavazi, I.; DePaoli, H.C.; Molfetta-Machado, J.B.; Giuliatti, S.; Goldman, G.H.; Goldman, M.H.S. Analysis of the Nicotiana tabacum stigma/style transcriptome reveals gene expression differences between wet and dry stigma species. Plant Physiol. 2009, 149, 1211–1230. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Sun, N.; Liu, X.; Jiao, Y.; Zhao, H.; Deng, X.W. Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 2005, 138, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.J.; Hogan, P.; Sundaresan, V. Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol. 2005, 139, 1853–1869. [Google Scholar] [CrossRef] [PubMed]
- Peiffer, J.A.; Kaushik, S.; Sakai, H.; Arteaga-Vazquez, M.; Sanchez-Leon, N.; Ghazal, H.; Vielle-Calzada, J.P.; Meyers, B.C. A spatial dissection of the Arabidopsis floral transcriptome by MPSS. BMC Plant Biol. 2008, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Vriezen, W.H.; Feron, R.; Maretto, F.; Keijman, J.; Mariani, C. Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol. 2008, 177, 60–76. [Google Scholar] [CrossRef]
- Tebbji, F.; Nantel, A.; Matton, D.P. Transcription profiling of fertilization and early seed development events in a solanaceous species using a 7.7 K cDNA microarray from Solanum chacoense ovules. BMC Plant Biol. 2010, 10, 174. [Google Scholar] [CrossRef]
- Dresselhaus, T.; Lörz, H.; Kranz, E. Representative cDNA libraries from few plant cells. Plant J. 1994, 5, 605–610. [Google Scholar] [CrossRef]
- Wuest, S.E.; Vijverberg, K.; Schmidt, A.; Weiss, M.; Gheyselinck, J.; Lohr, M.; Wellmer, F.; Rahnenführer, J.; von Mering, C.; Grossniklaus, U. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 2010, 20, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Leydon, A.R.; Manziello, A.; Pandey, R.; Mount, D.; Denic, S.; Vasic, B.; Johnson, M.A.; Palanivelu, R. Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 2009, 5, e1000621. [Google Scholar] [CrossRef] [PubMed]
- Boavida, L.C.; Borges, F.; Becker, J.D.; Feijó, J.A. Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol. 2011, 155, 2066–2080. [Google Scholar] [CrossRef] [PubMed]
- Leydon, A.R.; Weinreb, C.; Venable, E.; Reinders, A.; Ward, J.M.; Johnson, M.A. The molecular dialog between flowering plant reproductive partners defined by SNP-informed RNA-sequencing. Plant Cell 2017, 29, 984–1006. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Zhang, J.; Qi, X.; Ye, W.; Wang, X.; Xiang, X. Integrated metabolite profiling and transcriptome analysis reveals a dynamic metabolic exchange between pollen tubes and the style during fertilization of Brassica napus. Plant Mol. Biol. 2018, 97, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Zhang, L.; Zhao, L. Dissection of the style’s response to pollination using transcriptome profiling in self-compatible (Solanum pimpinellifolium) and self-incompatible (Solanum chilense) tomato species. BMC Plant Biol. 2015, 15, 119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Gao, C.; Yue, Y.; Liu, Z.; Ma, C.; Zhou, G.; Yang, Y.; Duan, Z.; Li, B.; Wen, J.; et al. Time-course transcriptome analysis of compatible and incompatible pollen-stigma interactions in Brassica napus L. Front. Plant Sci. 2017, 8, 682. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Chen, C.; Zeng, Z.; Zou, Z.; Li, H.; Zhou, Q.; Chen, X.; Sun, K.; Li, X. Transcriptomic analysis between self- and cross-pollinated pistils of tea plants (Camellia sinensis). BMC Genom. 2018, 19, 289. [Google Scholar] [CrossRef]
- Pease, J.B.; Guerrero, R.F.; Sherman, N.A.; Hahn, M.W.; Moyle, L.C. Molecular mechanisms of postmating prezygotic reproductive isolation uncovered by transcriptome analysis. Mol. Ecol. 2016, 25, 2592–2608. [Google Scholar] [CrossRef]
- Broz, A.K.; Guerrero, R.F.; Randle, A.M.; Baek, Y.S.; Hahn, M.W.; Bedinger, P.A. Transcriptomic analysis links gene expression to unilateral pollen-pistil reproductive barriers. BMC Plant Biol. 2017, 17, 81. [Google Scholar] [CrossRef]
- Wang, M.; Chen, Z.; Zhang, H.; Chen, H.; Gao, X. Transcriptome analysis provides insight into the molecular mechanisms underlying gametophyte factor 2-mediated cross-incompatibility in maize. Int. J. Mol. Sci. 2018, 19, 1757. [Google Scholar] [CrossRef] [PubMed]
- Mondragón-Palomino, M.; John-Arputharaj, A.; Pallmann, M.; Dresselhaus, T. Similarities between reproductive and immune pistil transcriptomes of Arabidopsis species. Plant Physiol. 2017, 174, 1559–1575. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.; Chen, Z.; Yang, X.; Gao, K.; Yang, X.; Zhao, T.; Li, S.; Wu, B.; An, X. Dynamic transcriptomic analysis of the early response of female flowers of Populus alba × P. glandulosa to pollination. Sci. Rep. 2017, 7, 6048. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, F. Die Fruchtbildung der Orchideen, ein Beweis für die doppelte Wirkung des pollen. Bot. Zeitung 1863, 21, 329–333. [Google Scholar]
- Treub, M.M. Notes sur l’embryon, le sac embryonnaire et l’ovule. 4. L’action des tubes polliniques sur le développement des ovules chez les orchidées. Ann. Jard. Bot. Buitenzorg 1883, 3, 122–128. [Google Scholar]
- Guignard, L. Sur la pollinisation et ses effets chez les orchidées. Ann. Sci. Nat. Sér. 7 Bot. 1886, 4, 202–240. [Google Scholar]
- Zhang, X.S.; O’Neill, S.D. Ovary and gametophyte development are coordinately regulated by auxin and ethylene following pollination. Plant Cell 1993, 5, 403–418. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, S.D.; Bui, A.Q.; Potter, D.; Zhang, X.S. Pollination of orchid flowers: Quantitative and domain-specific analysis of ethylene biosynthetic and hormone-induced gene expression. Int. J. Plant Sci. 2017, 178, 188–210. [Google Scholar] [CrossRef]
- Pimienta, E.; Polito, V.S. Embryo sac development in almond [Prunus dulcis (Mill.) D. A. Webb] as affected by cross-, self- and non-pollination. Ann. Bot. 1983, 51, 469–479. [Google Scholar] [CrossRef]
- Mól, R.; Filek, M.; Machackova, I.; Matthys-Rochon, E. Ethylene synthesis and auxin augmentation in pistil tissues are important for egg cell differentiation after pollination in maize. Plant Cell Physiol. 2004, 45, 1396–1405. [Google Scholar] [CrossRef]
- Brito, M.S.; Bertolino, L.T.; Cossalter, V.; Quiapim, A.C.; DePaoli, H.C.; Goldman, G.H.; Teixeira, S.P.; Goldman, M.H.S. Pollination triggers female gametophyte development in immature Nicotiana tabacum flowers. Front. Plant Sci. 2015, 6, 561. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, S.D. Pollination regulation of flower development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 547–574. [Google Scholar] [CrossRef] [PubMed]
- Huda, M.K.; Wilcock, C.C. Rapid floral senescence following male function and breeding systems of some tropical orchids. Plant Biol. 2012, 14, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, K.; Niki, T.; Ichimura, K. Pollination induces autophagy in petunia petals via ethylene. J. Exp. Bot. 2013, 64, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Theis, N.; Raguso, R.A. The effect of pollination on floral fragrance in thistles. J. Chem. Ecol. 2005, 31, 2581–2600. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, S.D.; Nadeau, J.A.; Zhang, X.S.; Bui, A.Q.; Halevy, A.H. Interorgan regulation of ethylene biosynthetic genes by pollination. Plant Cell 1993, 5, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Llop-Tous, I.; Barry, C.S.; Grierson, D. Regulation of ethylene biosynthesis in response to pollination in tomato flowers. Plant Physiol. 2000, 123, 971–978. [Google Scholar] [CrossRef]
- Lantin, S.; O’Brien, M.; Matton, D.P. Pollination, wounding and jasmonate treatments induce the expression of a developmentally regulated pistil dioxygenase at a distance, in the ovary, in the wild potato Solanum chacoense Bitt. Plant Mol. Biol. 1999, 41, 371–386. [Google Scholar] [CrossRef]
- Broderick, S.R.; Wijeratne, S.; Wijeratn, A.J.; Chapin, L.J.; Meulia, T.; Jones, M.L. RNA-sequencing reveals early, dynamic transcriptome changes in the corollas of pollinated petunias. BMC Plant Biol. 2014, 14, 307. [Google Scholar] [CrossRef]
- Chen, C.; Zeng, L.; Ye, Q. Proteomic and biochemical changes during senescence of Phalaenopsis ‘Red Dragon’ petals. Int. J. Mol. Sci. 2018, 19, 1317. [Google Scholar] [CrossRef]
- Germain, H.; Rudd, S.; Zotti, C.; Caron, S.; O’Brien, M.; Chantha, S.C.; Lagacé, M.; Major, F.; Matton, D.P. A 6374 unigene set corresponding to low abundance transcripts expressed following fertilization in Solanum chacoense Bitt, and characterization of 30 receptor-like kinases. Plant Mol. Biol. 2005, 59, 515–532. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, M.; Kapfer, C.; Major, G.; Laurin, M.; Bertrand, C.; Kondo, K.; Kowyama, Y.; Matton, D.P. Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J. 2002, 32, 985–996. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Chen, X.; Wu, Y.; Liu, W.; Liang, Q.; Zhang, L. Detection and transcript expression of S-RNase gene associated with self-incompatibility in apricot (Prunus armeniaca L.). Mol. Biol. Rep. 2006, 33, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Morse, D.; Cappadocia, M. Compatible pollinations in Solanum chacoense decrease both S-RNase and S-RNase mRNA. PLoS ONE 2009, 4, e5774. [Google Scholar] [CrossRef] [PubMed]
- Maune, J.F.; Camadro, E.L.; Erazzú, L.E. Cross-incompatibility and self-incompatibility: Unrelated phenomena in wild and cultivated potatoes? Botany 2018, 96, 33–45. [Google Scholar] [CrossRef]
- Brücher, H. Über das natürliche Vorkommen von Hybriden zwischen Solanum simplicifolium und Solanum subtilius im Aconquija Gebirge. Z. Indukt. Abstamm. Vererbungsl. 1953, 85, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, A.G.; Travers, S.E.; Mena-Ali, J.I.; Winsor, J.A. Pollen performance before and during the autotrophic-heterotrophic transition of pollen tube growth. Philos. Trans. R. Soc. B 2003, 358, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Chantha, S.C.; Emerald, B.S.; Matton, D.P. Characterization of the plant Notchless homolog, a WD repeat protein involved in seed development. Plant Mol. Biol. 2006, 62, 897–912. [Google Scholar] [CrossRef] [PubMed]
- Hogenboom, N.G.; Mather, K. Incompatibility and incongruity: Two different mechanisms for the non-functioning of intimate partner relationships [and comment]. Proc. R. Soc. Lond. Ser. B 1975, 188, 361–375. [Google Scholar] [CrossRef]
- Liu, Y.; Joly, V.; Dorion, S.; Rivoal, J.; Matton, D.P. The plant ovule secretome: A different view toward pollen–pistil interactions. J. Proteome Res. 2015, 14, 4763–4775. [Google Scholar] [CrossRef]
- Olsen, S.; Krause, K. Activity of xyloglucan endotransglucosylases/hydrolases suggests a role during host invasion by the parasitic plant Cuscuta reflexa. PLoS ONE 2017, 12, e0176754. [Google Scholar] [CrossRef] [PubMed]
- Bircheneder, S.; Dresselhaus, T. Why cellular communication during plant reproduction is particularly mediated by CRP signalling. J. Exp. Bot. 2016, 67, 4849–4861. [Google Scholar] [CrossRef] [PubMed]
- Chae, K.; Gonong, B.J.; Kim, S.C.; Kieslich, C.A.; Morikis, D.; Balasubramanian, S.; Lord, E.M. A multifaceted study of stigma/style cysteine-rich adhesin (SCA)-like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction. J. Exp. Bot. 2010, 61, 4277–4290. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Borevitz, J.O.; Preuss, D. Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet. 2007, 3, 1848–1861. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Rotino, G.L.; Dusi, V.; Chignola, R.; Sala, T.; Mennella, G.; Francese, G.; Pandolfini, T. Two metallocarboxypeptidase inhibitors are implicated in tomato fruit development and regulated by the inner no outer transcription factor. Plant Sci. 2018, 266, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Lagana, E.; Rodríguez-Leal, D.; Lúa, J.; Vielle-Calzada, J.P. A multigenic network of ARGONAUTE4 clade members controls early megaspore formation in Arabidopsis. Genetics 2016, 204, 1045–1056. [Google Scholar] [CrossRef] [PubMed]
- Ocarez, N.; Mejía, N. Suppression of the D-class MADS-box AGL11 gene triggers seedlessness in fleshy fruits. Plant Cell Rep. 2016, 35, 239–254. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, Y.; Feng, Q.; Qin, L.; Pan, C.; Lamin-Samu, A.T.; Lu, G. Tomato AUXIN RESPONSE FACTOR 5 regulates fruit set and development via the mediation of auxin and gibberellin signaling. Sci. Rep. 2018, 8, 2971. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Evensen, K.B.; Kao, T.H. Ethylene synthesis and floral senescence following compatible and incompatible pollinations in Petunia inflata. Plant Physiol. 1992, 99, 38–45. [Google Scholar] [CrossRef]
- De Martinis, D.; Cotti, G.; te Lintel Hekker, S.; Harren, F.J.M.; Mariani, C. Ethylene response to pollen tube growth in Nicotiana tabacum flowers. Planta 2002, 214, 806–812. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Baldwin, I.T. The post-pollination ethylene burst and the continuation of floral advertisement are harbingers of non-random mate selection in Nicotiana attenuata. Plant J. 2012, 71, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Yang, J.; Liesche, J.; Liu, X.; Hu, Y.; Si, W.; Guo, J.; Li, J. Ethylene promotes pollen tube growth by affecting actin filament organization via the cGMP-dependent pathway in Arabidopsis thaliana. Protoplasma 2017, 255, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Teng, X.D.; Zheng, Q.Q.; Zhao, Y.Y.; Lu, J.Y.; Wang, Y.; Guo, H.; Yang, Z.N. Ethylene signaling is critical for synergid cell functional specification and pollen tube attraction. Plant J. 2018, 96, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed]
- Speranza, A.; Crinelli, R.; Scoccianti, V.; Geitmann, A. Reactive oxygen species are involved in pollen tube initiation in kiwifruit. Plant Biol. 2012, 14, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, A.V.; Matveyeva, N.P.; Yermakov, I.P. Reactive oxygen species are involved in regulation of pollen wall cytomechanics. Plant Biol. 2014, 16, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Potocký, M.; Jones, M.A.; Bezvoda, R.; Smirnoff, N.; Zárský, V. Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. New Phytol. 2007, 174, 742–751. [Google Scholar] [CrossRef]
- Kaya, H.; Nakajima, R.; Iwano, M.; Kanaoka, M.M.; Kimura, S.; Takeda, S.; Kawarazaki, T.; Senzaki, E.; Hamamura, Y.; Higashiyama, T.; et al. Ca2+-activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. Plant Cell 2014, 26, 1069–1080. [Google Scholar] [CrossRef]
- Lassig, R.; Gutermuth, T.; Bey, T.D.; Konrad, K.R.; Romeis, T. Pollen tube NAD(P)H oxidases act as a speed control to dampen growth rate oscillations during polarized cell growth. Plant J. 2014, 78, 94–106. [Google Scholar] [CrossRef]
- Duan, Q.; Kita, D.; Johnson, E.A.; Aggarwal, M.; Gates, L.; Wu, H.M.; Cheung, A.Y. Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat. Commun. 2014, 5, 3129. [Google Scholar] [CrossRef]
- Forbes, A.M.; Meier, G.P.; Haendiges, S.; Taylor, L.P. Structure-activity relationship studies of flavonol analogues on pollen germination. J. Agric. Food Chem. 2014, 62, 2175–2181. [Google Scholar] [CrossRef] [PubMed]
- Muhlemann, J.K.; Younts, T.L.B.; Muday, G.K. Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11188–E11197. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.H.; Zou, J.; Feng, J.; Peng, X.B.; Wu, J.Y.; Wu, Y.L.; Palanivelu, R.; Sun, M.X. Exogenous γ-aminobutyric acid (GABA) affects pollen tube growth via modulating putative Ca2+-permeable membrane channels and is coupled to negative regulation on glutamate decarboxylase. J. Exp. Bot. 2014, 65, 3235–3248. [Google Scholar] [CrossRef] [PubMed]
- Palanivelu, R.; Brass, L.; Edlund, A.F.; Preuss, D. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 2003, 114, 47–59. [Google Scholar] [CrossRef]
- Chantha, S.; Tebbji, F.; Matton, D.P. From the Notch signaling pathway to ribosome biogenesis. Plant Signal. Behav. 2007, 2, 168–170. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H. Predicting secretory proteins with SignalP. In Protein Function Prediction; Methods in Molecular Biology; Kihara, D., Ed.; Humana Press: New York, NY, USA, 2017; Volume 1611, pp. 59–73. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. PredGPI: A GPI-anchor predictor. BMC Bioinf. 2008, 9, 392. [Google Scholar] [CrossRef] [PubMed]
- Joly, V.; Matton, D.P. KAPPA, a simple algorithm for discovery and clustering of proteins defined by a key amino acid pattern: A case study of the cysteine-rich proteins. Bioinformatics 2015, 31, 1716–1723. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joly, V.; Tebbji, F.; Nantel, A.; Matton, D.P. Pollination Type Recognition from a Distance by the Ovary Is Revealed Through a Global Transcriptomic Analysis. Plants 2019, 8, 185. https://doi.org/10.3390/plants8060185
Joly V, Tebbji F, Nantel A, Matton DP. Pollination Type Recognition from a Distance by the Ovary Is Revealed Through a Global Transcriptomic Analysis. Plants. 2019; 8(6):185. https://doi.org/10.3390/plants8060185
Chicago/Turabian StyleJoly, Valentin, Faïza Tebbji, André Nantel, and Daniel P. Matton. 2019. "Pollination Type Recognition from a Distance by the Ovary Is Revealed Through a Global Transcriptomic Analysis" Plants 8, no. 6: 185. https://doi.org/10.3390/plants8060185
APA StyleJoly, V., Tebbji, F., Nantel, A., & Matton, D. P. (2019). Pollination Type Recognition from a Distance by the Ovary Is Revealed Through a Global Transcriptomic Analysis. Plants, 8(6), 185. https://doi.org/10.3390/plants8060185




