Fungal Communities Associated with Peacock and Cercospora Leaf Spots in Olive
Abstract
1. Introduction
2. Results
2.1. Fungal Identification—Structural Diversity
2.2. Multivariate Data Analysis—Fungal Richness
3. Discussion
4. Materials and Methods
4.1. Sampling Collection
4.2. Fungal Isolation and DNA Extraction
4.3. Fungal Molecular Identification
4.4. Data Analysis
Author Contributions
Funding
Conflicts of Interest
References
- IOC. Available online: http://www.internationaloliveoil.org/ (accessed on 4 June 2018).
- Grieco, F.; Alkowni, R.; Saponari, M.; Pantaleo, V.; Savino, V.; Martelli, G.P. Molecular detection of olive-infecting viruses. Acta Hortic. 2002, 586, 737–740. [Google Scholar] [CrossRef]
- Sistani, F.; Ramezanpour, S.; Nasrollanejad, S. Field Evaluation of Different Fungicides Application to Control Olive Leaf Spot. Aust. J. Basic Appl. Sci. 2009, 3, 3341–3345. [Google Scholar]
- Moral, J.; Alsalimiya, M.; Roca, L.; Díez, C.; León, L.; de la Rosa, R.; Barranco, D.; Rallo, L.; Trapero, A. Relative susceptibility of new olive cultivars to Spilocaea oleagina, Colletotrichum acutatum, and Pseudocercospora cladosporioides. Plant Dis. 2015, 99, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Trapero, A.; Blanco, M. Enfermedades. In El cultivo del Olivo; Barranco, D., Fernandez-Escobar, R., Rallo, L., Eds.; Mundi Prensa-Junta de Andalucia: Madrid, Spain, 2004; pp. 510–514. [Google Scholar]
- Vossen, P. Timing sprays for control peacock spot and olive knot disease. Olive News Univ. Calif. Coop. Ext. Glenn Cty. 2004, 7, 11. [Google Scholar]
- Avila, A.; Groenewald, J.; Trapero, A.; Crous, P. Characterisation and epitypification of Pseudocercospora cladosporioides, the causal organism of Cercospora leafspot of olives. Mycol. Res. 2005, 109, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Graniti, A.; Frisullo, S.; Pennisi, A.; Magnano di San Lio, G. Infections of Glomerella cingulata on olive in Italy. EPPO Bull. 1993, 23, 457–465. [Google Scholar] [CrossRef]
- Shabi, E.; Birger, R.; Lavee, S.; Klein, I. Leaf spot (Spilocaea oleaginea) on olive in Israel and its control. Acta Horticult. 1994, 356, 390–394. [Google Scholar] [CrossRef]
- Viruega, J.; Moral, J.; Roca, L.; Navarro, N.; Trapero, A. Spilocaea oleagina in Olive Groves of Southern Spain: Survival, Inoculum Production, and Dispersal. Plant Dis. 2013, 97, 1549–1556. [Google Scholar] [CrossRef]
- Vega, J.; Cabezas, D. El “repilo plomizo” del olivo, causado por Cercospora cladosporioides Sacc., enfermedad presente en Espana. Bol. Sanid. Veg. Plagas 1985, 11, 31–35. [Google Scholar]
- Henricot, B.; Gorton, C.; Denton, J.; Denton, G. Pseudocercospora cladosporioides, the cause of leaf spot on olive, a pathogen new to the United Kingdom. Plant Pathol. 2009, 58, 803. [Google Scholar] [CrossRef]
- Benitez, Y.; Botella, M.; Trapero, A.; Alsalimiya, M.; Caballero, J.; Dorado, G.; Munoz-Blanco, J. Molecular analysis of the interaction between Olea europaea and the biographic fungus Spilocaea oleagina. Mol. Plant Pathol. 2005, 6, 425–438. [Google Scholar] [CrossRef] [PubMed]
- Civantes, M. Olive Pest and Disease Management; IOCC: Towson, MD, USA, 1999. [Google Scholar]
- Figueres, G. Repilos del Olivo: Ataque en Fruto; Phytoma España: Valencia, Spain, 1991; pp. 31–36. [Google Scholar]
- Fragoso, R. Botanica Criptogamica Agrıcola; Espasa Calpe: Madrid, Spain, 1927. [Google Scholar]
- Obanor, F.; Jaspers, M.; Jones, E.; Walter, M. Greenhouse and Field Evaluation of Fungicides for Control of Olive Leaf Spot in New Zealand. Crop Prot. 2008, 27, 1335–1342. [Google Scholar] [CrossRef]
- Sanei, S.; Razav, S. Survey of Spilocaea oleagina, causal agent of olive leaf spot, in North of Iran. J. Yeast Fungal Res. 2011, 2, 33–38. [Google Scholar] [CrossRef]
- Guechi, A.; Girre, L. Sources of Cycloconium Oleaginum (Cast.) conidia for infection of olive leaves and conditions determining leaf spot disease development in the region of Sétif, Algeria. Mycopathologia 1994, 125, 163–171. [Google Scholar] [CrossRef]
- Lops, F.; Frisullo, S.; Rossi, V. Studies on the spread of the olive scab pathogen Spilocaea oleagina. Bull. OEPP/EPPO Bull. 1993, 23, 385–387. [Google Scholar] [CrossRef]
- Romero, J.; Agustí-Brisach, C.; Roca, L.F.; Moral, J.; González-Domínguez, E.; Rossi, V.; Trapero, A. A long-term study on the effect of agroclimatic variables on olive scab in Spain. Crop Prot. 2018, 114, 39–43. [Google Scholar] [CrossRef]
- Roubal, C.; Regis, S.; Nicot, P.C. Field models for the prediction of leaf infection and incubation period of Fusicladium oleagineum on olive based on rain, temperature and relative humidity. Plant Pathol. 2013, 62, 657–666. [Google Scholar] [CrossRef]
- Viruega, J.R.; Roca, L.F.; Moral, J.; Trapero, A. Factors affecting infection and disease development on olive leaves inoculated with Fusicladium oleagina. Plant Dis. 2011, 95, 1139–1146. [Google Scholar] [CrossRef]
- Teviotdale, B.; Sibbett, G.; Harper, D. Several Copper Fungicides Control Olive Leaf Spot. Calif. Agric. 1989, 43, 30–31. [Google Scholar]
- Malavolta, C.; Perdikis, D. IOBC technical guidelines III. Guidelines for integrated production of olives. IOBC/WPRS Bull. 2012, 77, 1–19. [Google Scholar]
- HOA. Hunter Olive Handbook: A Practical Guide for Sustainable Olive Production; Australian eBook Publisher: Acacia Ridge, QLD, Australia, 2016. [Google Scholar]
- Roca, L.; Horchani, H.; Trapero, A. Search for alternatives to copper for the control of olive leaf spot caused by Fusiciadium oleagineum. In Proceedings of the 4th European Meeting of the lOBC/wprs working Group Integrated Protection of Olive Crops, Córdoba, Spain; 2009; p. 54. [Google Scholar]
- Gomes, T.; Pereira, J.A.; Benhadi, J.; Lino-Neto, T.; Baptista, P. Endophytic and Epiphytic Phyllosphere Fungal Communities Are Shaped by Different Environmental Factors in a Mediterranean Ecosystem. Microb. Ecol. 2018, 76, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Martins, F.; Pereira, J.A.; Bota, P.; Bento, A.; Baptista, P. Fungal endophyte communities in above- and belowground olive tree organs and the effect of season and geographic location on their structures. Fungal Ecol. 2016, 20, 193–201. [Google Scholar] [CrossRef]
- Materatski, P.; Varanda, C.; Carvalho, T.; Dias, A.; Campos, M.; Rei, F.; Félix, M. Spatial and temporal variation of fungal endophytic richness and diversity associated to the phyllosphere of olive cultivars. Fungal Biol. 2019, 123, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.; Nicosia, M.; Cacciola, S.; Droby, S.; Schena, L. Metabarcoding analysis of fungal diversity in the phyllosphere and carposphere of olive (Olea europaea). PLoS ONE 2015, 10, e0131069. [Google Scholar] [CrossRef] [PubMed]
- Materatski, P.; Varanda, C.; Carvalho, T.; Dias, A.; Campos, M.; Rei, F.; Félix, M. Diversity of Colletotrichum Species Associated with Olive Anthracnose and New Perspectives on Controlling the Disease in Portugal. Agronomy 2018, 8, 301. [Google Scholar] [CrossRef]
- Varanda, C.M.R.; Oliveira, M.; Materatski, P.; Landum, M.; Clara, M.I.E.; Félix, M.R. Fungal endophytic communities associated to phyllospheres of grapevine cultivars under different types of management. Fungal Biol. 2016, 120, 1525–1536. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Yang, L.; Zhu, X.; Zeng, L.; Li, X. Seasonal and Habitat Dependent Variations in Culturable Endophytes of Camellia sinensis. J. Plant Pathol. Microb. 2013, 4, 169. [Google Scholar] [CrossRef]
- Moral, J.; Xaviér, C.; Roca, L.; Romero, J.; Moreda, W.; Trapero, A. La Antracnosis del olivo y su efecto en la calidad del aceite. Grasas Aceites 2014, 65, e028. [Google Scholar] [CrossRef]
- Ivic, D.; Ivanovic, A.; Milicevic, T.; Cvjetkovic, B. Shoot necrosis of olive caused by Phoma incompta, a new disease of olive in Croatia. Phytopathol. Mediterr. 2010, 49, 414–416. [Google Scholar]
- Koukol, O. New species of Chalara occupying coniferous needles. Fungal Divers. 2011, 49, 75–91. [Google Scholar] [CrossRef]
- Lo Piccolo, S.; Mondello, V.; Giambra, S.; Conigliaro, G.; Torta, L.; Burruano, S. Arthrinium phaeospermum, Phoma cladoniicola and Ulocladium consortiale, New Olive Pathogens in Italy. J. Phytopathol. 2014, 162, 258–263. [Google Scholar] [CrossRef]
- Basim, E.; Basim, H.; Abdulai, M.; Baki, D.; Oztürk, N. Identification and characterization of Alternaria alternata causing leaf spot of olive tree (Olea europaea) in Turkey. Crop Prot. 2017, 92, 79–88. [Google Scholar] [CrossRef]
- Bourbos, V.; Skoundridakis, M.; Metzidakis, I. Alternaria alternata: A new disease of leafy cuttings of olive shoots. Acta Hortic. 1999, 474, 585–587. [Google Scholar] [CrossRef]
- Lagogianni, C.; Tjamos, E.; Antoniou, P.; Tsitsigiannis, D. First Report of Alternaria alternata as the Causal Agent of Alternaria Bud and Blossom Blight of Olives. Plant Dis. 2017, 101, 2151. [Google Scholar] [CrossRef]
- Landum, M.C.; Félix, M.d.R.; Alho, J.; Garcia, R.; Cabrita, M.J.; Rei, F.; Varanda, C.M.R. Antagonistic activity of fungi of Olea europaea L. against Colletotrichum acutatum. Microbiol. Res. 2016, 183, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Dugan, F.; Lupien, S.; Grove, G. Incidence, Aggressiveness and In Planta Interactions of Botrytis cinerea and other Filamentous Fungi Quiescent in Grape Berries and Dormant Buds in Central Washington State. J. Phytopathol. 2002, 150, 375–381. [Google Scholar] [CrossRef]
- Musetti, R.; Polizzotto, R.; Vecchione, A.; Borselli, S.; Zulini, L.; D’Ambrosio, M.; Sanitá di Toppi, L.; Pertot, I. Antifungal activity of diketopiperazines extracted from Alternaria alternata against Plasmopara viticola: An ultrastructural study. Micron 2007, 38, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Larena, I.; Linan, M.; Melgarejo, P. Antibiotic Production of the Biocontrol Agents Epicoccum nigrum and Candida sake. In Proceedings of the 6th Conference EFPP, Prague, Czech Republic, 8–14 September 2002; pp. 205–208. [Google Scholar]
- Martini, M.; Musetti, R.; Grisan, S.; Polizzotto, R.; Borselli, S.; Pavan, F.; Osler, R. DNA-dependent detection of the grapevine fungal endophytes Aureobasidium pullulans and Epicoccum nigrum. Plant Dis. 2009, 93, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.; Harris, R.; Spear, R.; Lau, G.; Nordheim, E. Morphogenesis and adhesion of Aureobasidium pullulans. Can. J. Microbiol. 1994, 40, 6–17. [Google Scholar] [CrossRef]
- Deshpande, M.; Rale, V.; Lynch, J. Aureobasidium pullulans In applied microbiology: A status report. Enzyme Microb. Technol. 1992, 14, 514–527. [Google Scholar] [CrossRef]
- Grabowski, M. The study of new fungus species causing apple sooty blotch. Folia Hort Ann. 2007, 19, 89–97. [Google Scholar]
- Pancher, M.; Ceol, M.; Corneo, P.; Longa, C.; Yousaf, S.; Pertot, I.; Campisano, A. Fungal endophytic communities in grapevines (Vitis vinifera L.) respond to crop management. Appl. Environ. Microbiol. 2012, 78, 4308–4317. [Google Scholar] [CrossRef] [PubMed]
- Hartati, S.; Wiyono, S.; Hidayat, S.; Sinaga, M. Mode of Action of Yeast-Like Fungus Aureobasidium pullulans in Controlling Anthracnose of Postharvest Chili. Int. J. Sci. Basic Appl. Res. 2015, 20, 253–263. [Google Scholar]
- Tashiro, N.; Noguchi, M.; Ide, Y.; Kuchiki, F. Sooty spot caused by Cladosporium cladosporioides in postharvest Satsuma mandarin grown in heated greenhouses. J. Gen. Plant Pathol. 2013, 79, 158–161. [Google Scholar] [CrossRef]
- Wachowska, U.; Głowacka, K. Antagonistic interactions between Aureobasidium pullulans and Fusarium culmorum, a fungal pathogen of winter wheat. BioControl 2014, 59, 635–645. [Google Scholar] [CrossRef]
- Wang, N.; Thomson, M.; Bodles, W.J.A.; Crawford, R.M.M.; Hunt, H.V.; Featherstone, A.W.E.A. Genome sequence of dwarf birch (Betula nana) and cross-species RAD markers. Mol. Ecol. 2013, 22, 3098–3111. [Google Scholar] [CrossRef] [PubMed]
- Munitz, M.; Garrido, C.; Gonzalez, H.; Resnik, S.; Salas, P. Mycoflora and potential mycotoxin production of freshly harvested blueberry in Concordia, Entre Rios province, Argentina. Int. J. Fruit Sci. 2013, 13, 312–325. [Google Scholar] [CrossRef]
- Kido, L.; Uzabakiriho, J.; Chimwamurombe, P. Isolation and identification of pathogenic fungi associated with Aloe zebrina flower malformation—First report. J. Pure Appl. Microbiol. 2012, 6, 125–129. [Google Scholar]
- Frisullo, S.; Elshafie, H.; Mang, S. First report of two phomopsis species on olive trees in Italy. J. Plant Pathol. 2015, 97, 391–403. [Google Scholar]
- Frisullo, S.; Carlucci, A. Minor fungal diseases of olives. In Olive Diseases and Disorders; Schena, L., Agosteo, G., Cacciola, S.O., Eds.; Transworld Research Network: Kerala, India, 2011; pp. 291–304. [Google Scholar]
- Morath, S.; Hung, R.; Bennett, J. Fungal volatile organic compounds: A review with emphasis on their biotechnological potential. Fungal Biol. Rev. 2012, 26, 73–83. [Google Scholar] [CrossRef]
- Siddiquee, S.; Al Azad, S.; Bakar, F.A.; Naher, L.; Kumar, S.V. Separation and identification of hydrocarbons and other volatile compounds from cultures of Aspergillus niger by GC–MS using two different capillary columns and solvents. J. Saudi Chem. Soc. 2015, 19, 243–256. [Google Scholar] [CrossRef]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Clarke, K.R.; Warwick, R.M. Changes in Marine Communities: An Approach to Statistical Analysis and Interpretation, 2nd ed.; Primer-E, P.U.K., Ed.; PRIMER-E Ltd.: Plymouth, UK, 2001. [Google Scholar]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. PERMANOVA A+ for PRIMER: Guide to Software and Statistical Methods; PRIMER-E: Plymouth, UK, 2008. [Google Scholar]
- Clarke, K.; Green, R. Statistical design and analysis for a biological effects study. Marine Ecol. Prog. Ser. 1988, 46, 213–226. [Google Scholar] [CrossRef]

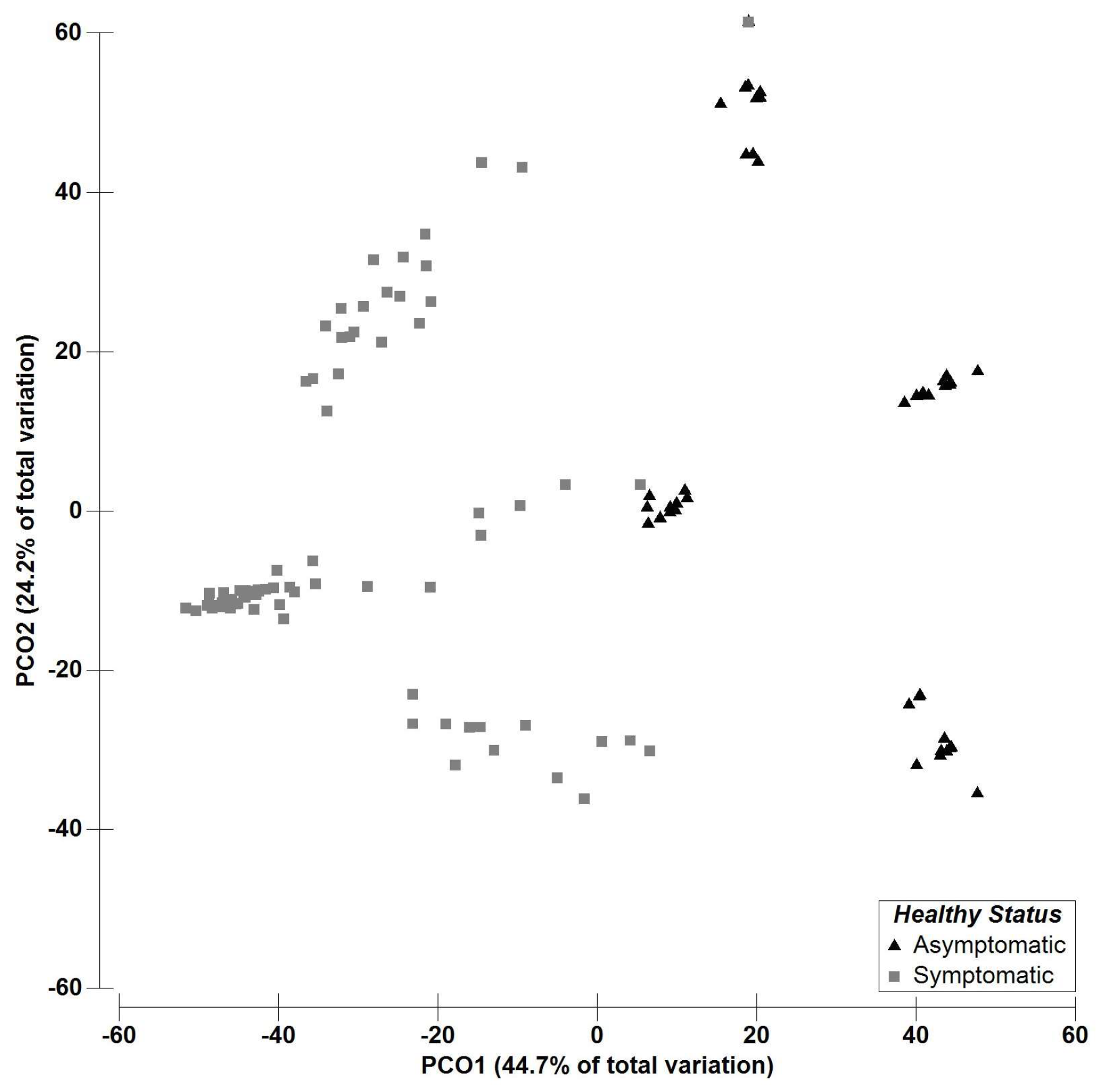
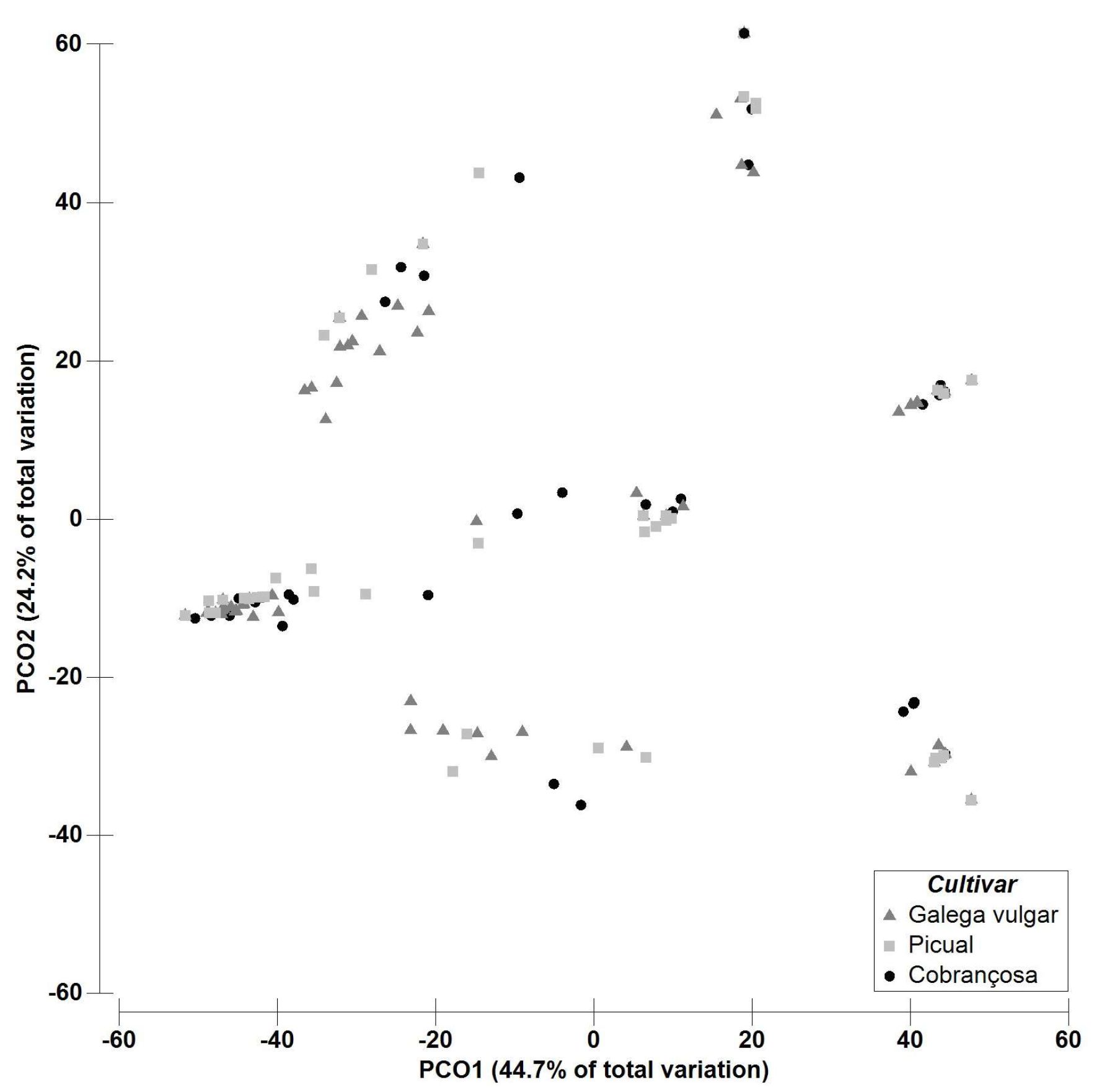
| Fungal OTUs | Asymptomatic | Symptomatic | Galega Vulgar | Cobrançosa | Picual | |||
|---|---|---|---|---|---|---|---|---|
| - | - | Asymptomatic | Symptomatic | Asymptomatic | Symptomatic | Asymptomatic | Symptomatic | |
| Similarity | ||||||||
| 37.40% | 51.20% | 48.09% | 61.54% | 38.18% | 59.37% | 30.78% | 55.24% | |
| Alternaria sp. | 62.81 | 0.75 | 56.18 | 0.92 | 77.48 | 0.74 | 49.28 | 0.22 |
| Epicoccum sp. | 29.11 | 3.37 | 37.42 | 4.2 | 10.1 | 3.22 | 43.82 | 1.41 |
| Arthrinium sp. | 2.06 | 0.00 | 1.81 | 0.00 | 2.35 | 0.00 | 1.33 | 0.00 |
| Chaetomium sp. | 1.83 | 0.00 | 0.97 | 0.00 | 3.35 | 0.00 | 0.93 | 0.00 |
| Diaporthe sp. | 1.68 | 0.00 | 0.86 | 0.00 | 1.28 | 0.00 | 2.71 | 0.00 |
| Aspergillus sp. | 1.10 | 0.00 | 0.19 | 0.00 | 5.28 | 0.00 | 0.00 | 0.00 |
| Fusarium sp. | 0.68 | 0.00 | 0.96 | 0.00 | 0.09 | 0.00 | 0.83 | 0.00 |
| Nigrospora sp. | 0.60 | 0.00 | 0.62 | 0.00 | 0.07 | 0.00 | 1.11 | 0.00 |
| Aureobasidium sp. | 0.14 | 0.21 | 0.99 | 0.02 | 0.00 | 0.12 | 0.00 | 0.42 |
| Botrytis sp. | 0.00 | 1.16 | 0.00 | 6.03 | 0.00 | 0.1 | 0.00 | 0.00 |
| Bullera sp. | 0.00 | <0.05 | 0.00 | 0.23 | 0.00 | 0.00 | 0.00 | 0.00 |
| Chalara sp. | 0.00 | 16.99 | 0.00 | 22.73 | 0.00 | 34.46 | 0.00 | 0.73 |
| Cryptococcus sp. | 0.00 | 0.26 | 0.00 | 1.99 | 0.00 | 0.00 | 0.00 | 0.00 |
| Erythrobasidium sp. | 0.00 | <0.05 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.21 |
| Fusicladium sp. | 0.00 | <0.05 | 0.00 | 0.64 | 0.00 | 0.00 | 0.00 | 0.00 |
| Neofusicoccum sp. | 0.00 | <0.05 | 0.00 | 0.00 | 0.00 | <0.05 | 0.00 | 0.00 |
| Foliophoma sp. | 0.00 | 9.52 | 0.00 | 19.88 | 0.00 | 0.00 | 0.00 | 18.38 |
| P. cladosporioides | 0.00 | 27.14 | 0.00 | 16.49 | 0.00 | 23.97 | 0.00 | 33.36 |
| Saccharata sp. | 0.00 | <0.05 | 0.00 | 0.14 | 0.00 | 0.00 | 0.00 | 0.00 |
| V. oleaginea | 0.00 | 40.42 | 0.00 | 26.52 | 0.00 | 37.35 | 0.00 | 45.27 |
| Sporobolomyces sp. | 0.00 | <0.05 | 0.00 | 0.22 | 0.00 | 0.00 | 0.00 | 0.00 |
| Source of Variation | Degrees of Freedom | Sum of Squares | Mean Squares | Pseudo-F | Perms | P(perm) | |
|---|---|---|---|---|---|---|---|
| Fungal | Health status | 1 | 369990 | 369990 | 23.942 | 38 | 0.0014 |
| richness | Cultivar | 2 | 34596 | 17298 | 9.7174 | 9944 | 0.0001 |
| - | Health status x Cultivar | 2 | 30908 | 15454 | 8.6813 | 9948 | 0.0001 |
| - | Residual | 294 | 523360 | 1780 | - | - | - |
| - | Total | 299 | 958850 | - | - | - | |
| Fungal | Health status | 1 | 914.28 | 914.28 | 0.71023 | 360 | 0.5397 |
| richness* | Cultivar | 2 | 14325 | 7162.5 | 53.698 | 9920 | 0.0001 |
| - | Health status x Cultivar | 2 | 2581 | 1290.5 | 9.6749 | 9941 | 0.0001 |
| - | Residual | 275 | 36681 | 133.39 | - | - | - |
| - | Total | 280 | 54692 | - | - | - | - |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varanda, C.M.R.; Materatski, P.; Landum, M.; Campos, M.D.; Félix, M.d.R. Fungal Communities Associated with Peacock and Cercospora Leaf Spots in Olive. Plants 2019, 8, 169. https://doi.org/10.3390/plants8060169
Varanda CMR, Materatski P, Landum M, Campos MD, Félix MdR. Fungal Communities Associated with Peacock and Cercospora Leaf Spots in Olive. Plants. 2019; 8(6):169. https://doi.org/10.3390/plants8060169
Chicago/Turabian StyleVaranda, Carla M.R., Patrick Materatski, Miguel Landum, Maria Doroteia Campos, and Maria do Rosário Félix. 2019. "Fungal Communities Associated with Peacock and Cercospora Leaf Spots in Olive" Plants 8, no. 6: 169. https://doi.org/10.3390/plants8060169
APA StyleVaranda, C. M. R., Materatski, P., Landum, M., Campos, M. D., & Félix, M. d. R. (2019). Fungal Communities Associated with Peacock and Cercospora Leaf Spots in Olive. Plants, 8(6), 169. https://doi.org/10.3390/plants8060169







