Non-cultivated Cotton Species (Gossypium spp.) Act as a Reservoir for Cotton Leaf Curl Begomoviruses and Associated Satellites
Abstract
:1. Introduction
2. Results
2.1. Assembly and Analysis of NGS Data Identified Begomovirus and Alpha/Betasatellites in G. mustelinum, G. raimondii and G. thurberi
2.2. Diverse Species and Strains of Begomovirus/Alphasatellite/Betasatellite are Maintained by Non-cultivated Cotton Species
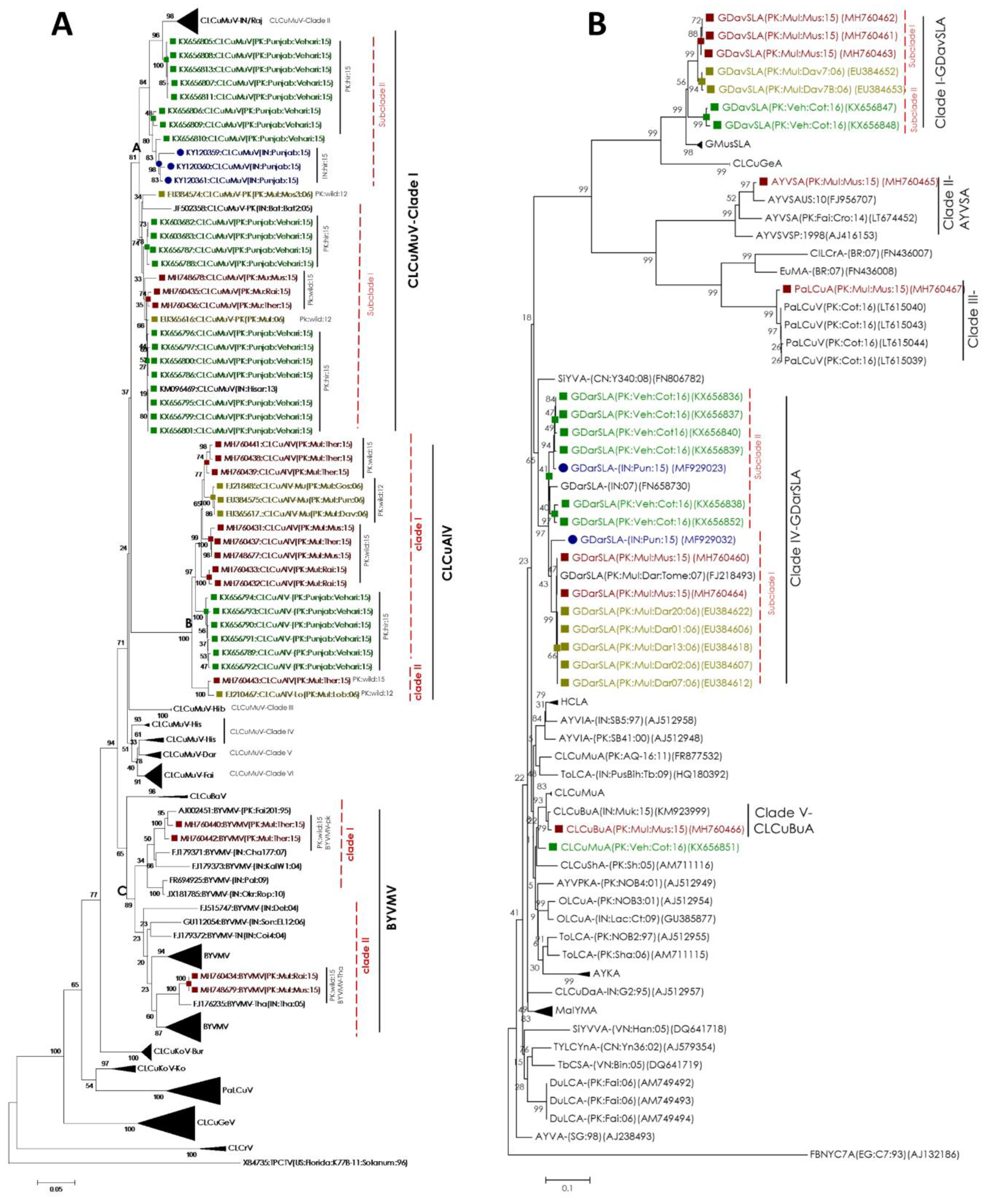
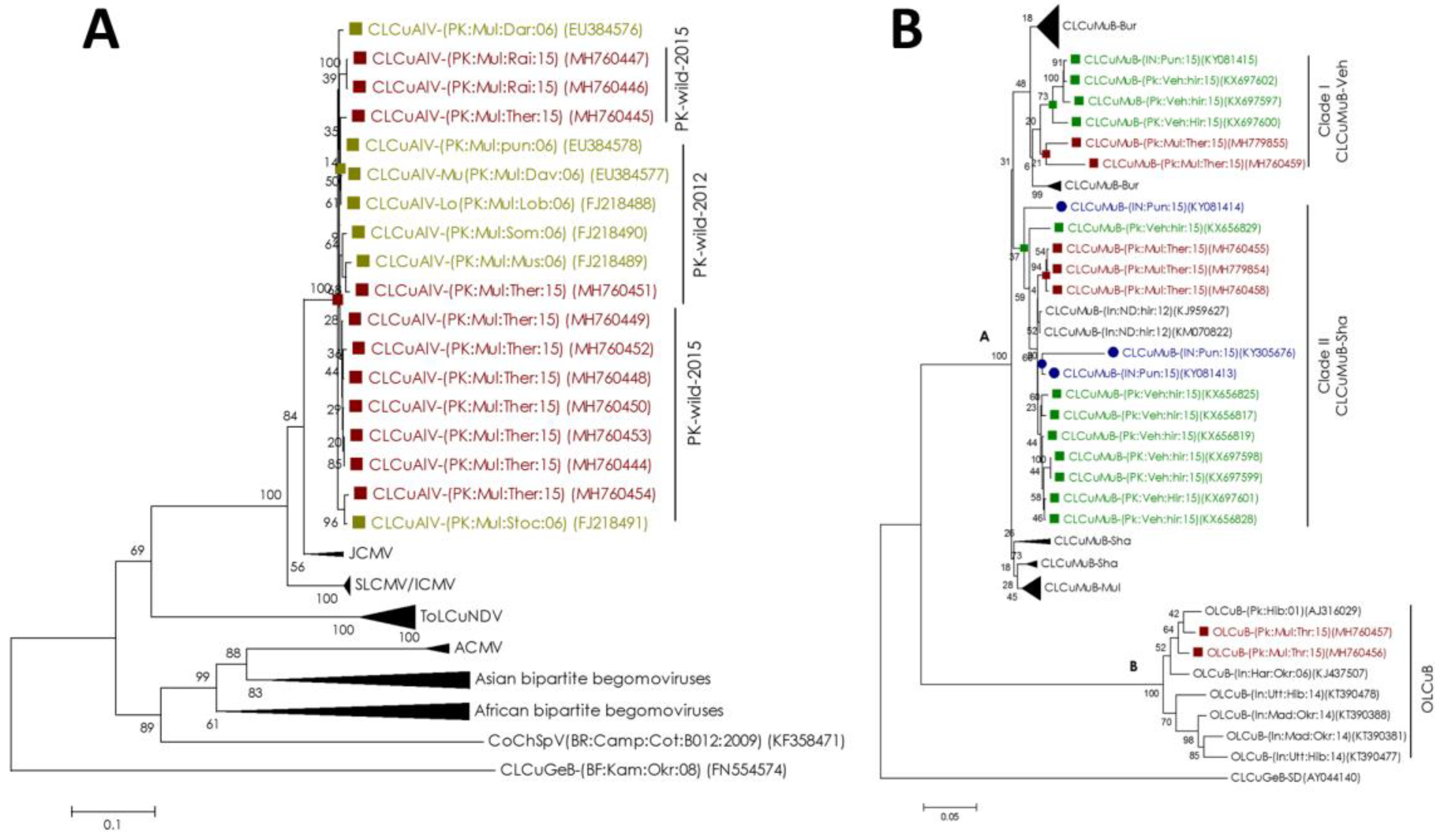
2.3. Begomovirus/Satellites are Phylogenetically Related to the Recent Cotton Leaf Curl Disease Complex
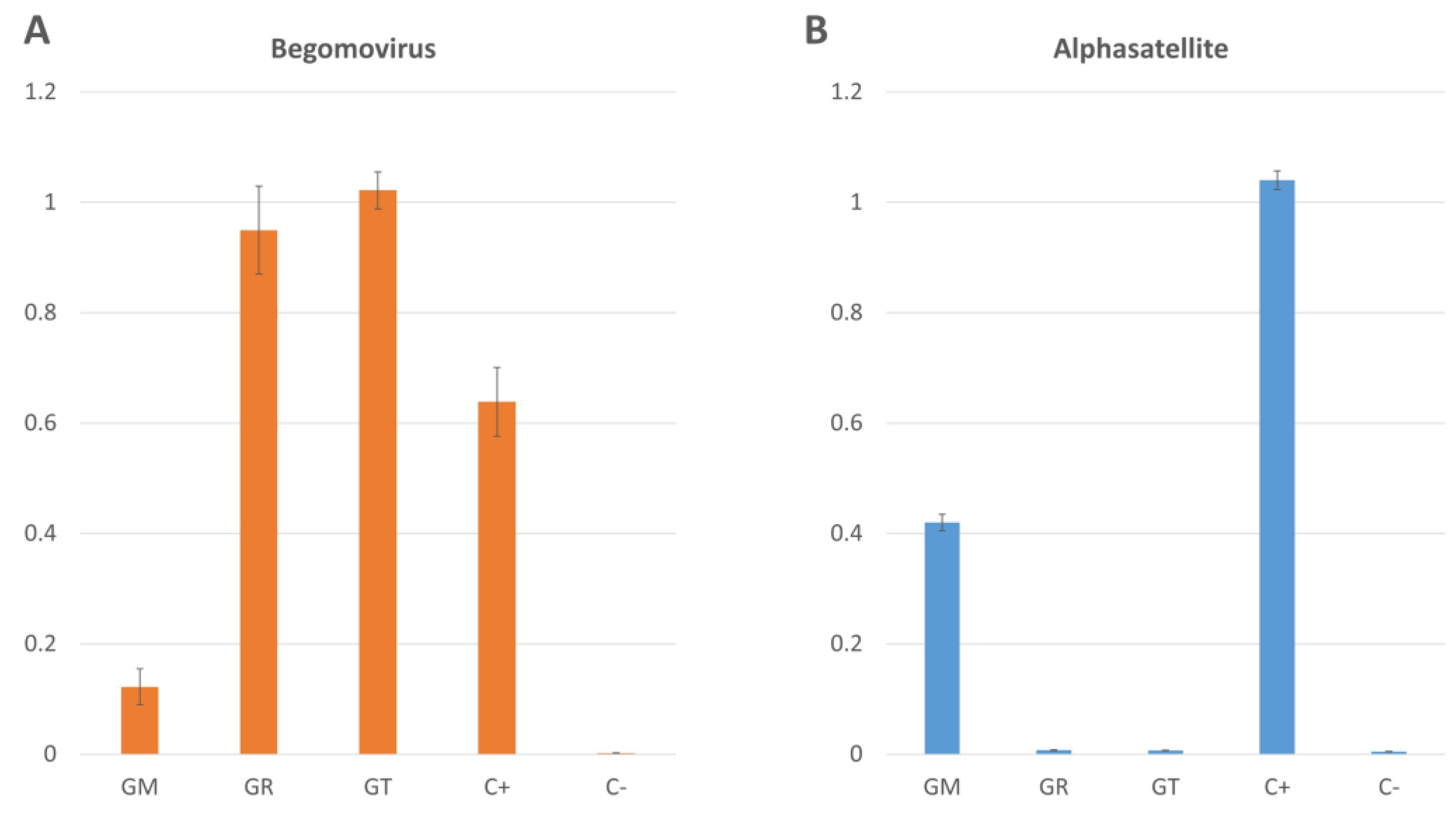
2.4. Begomovirus Levels are Reduced in the Presence of an Alphasatellite
3. Discussion
4. Materials and Methods
4.1. Sample Collection, DNA Extraction and Rolling Circle Amplification
4.2. Library Preparation, Sequencing, Assembly and Analysis
4.3. Alignment, SDT and Phylogeny
4.4. Recombination Analysis
4.5. Quantification with qPCR
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Anthony, V.M.; Ferroni, M. Agricultural biotechnology and smallholder farmers in developing countries. Curr. Opin. Biotechnol. 2012, 23, 278–285. [Google Scholar] [CrossRef]
- Sattar, M.N.; Kvarnheden, A.; Saeed, M.; Briddon, R.W. Cotton leaf curl disease—An emerging threat to cotton production worldwide. J. Gen. Virol. 2013, 94, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Virol. 2015, 2, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Fondong, V.N. Geminivirus protein structure and function. Mol. Plant Pathol. 2013, 14, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Zhou, X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Nawaz-Ul-Rehman, M.S.; Nahid, N.; Mansoor, S.; Briddon, R.W.; Fauquet, C.M. Post-transcriptional gene silencing suppressor activity of two non-pathogenic alphasatellites associated with a begomovirus. Virology 2010, 405, 300–308. [Google Scholar] [CrossRef]
- Rosario, K.; Marr, C.; Varsani, A.; Kraberger, S.; Stainton, D.; Moriones, E.; Polston, J.E.; Breitbart, M. Begomovirus-Associated Satellite DNA Diversity Captured Through Vector-Enabled Metagenomic (VEM) Surveys Using Whiteflies (Aleyrodidae). Viruses-Basel 2016, 8, 36. [Google Scholar] [CrossRef]
- Lozano, G.; Trenado, H.P.; Fiallo-Olive, E.; Chirinos, D.; Geraud-Pouey, F.; Briddon, R.W.; Navas-Castillo, J. Characterization of non-coding DNA satellites associated with sweepoviruses (genus Begomovirus, Geminiviridae)—Definition of a distinct class of begomovirus-associated satellites. Front. Microbiol. 2016, 7, 162. [Google Scholar] [CrossRef] [PubMed]
- Zubair, M.; Zaidi, S.S.; Shakir, S.; Amin, I.; Mansoor, S. An insight into Cotton leaf curl Multan betasatellite, the most important component of cotton leaf curl disease complex. Viruses 2017, 9, 280. [Google Scholar] [CrossRef]
- Wendel, J.F.; Albert, V.A. Phylogenetics of the cotton genus (Gossypium): Character-state weighted parsimony analysis of chloroplast-DNA restriction site data and its systematic and biogeographic implications. Syst. Bot. 1992, 115–143. [Google Scholar] [CrossRef]
- Krapovickas, A.; Seijo, G.; Seijo, J.G. Gossypium ekmanianum (Malvaceae), algodon silvestre de la Republica Dominicana. Bonplandia 2008, 17, 55–63. [Google Scholar]
- Paterson, A.H. Genomics of Cotton; Springer: New York, NY, USA, 2008. [Google Scholar]
- Moulherat, C.; Tengberg, M.; Haquet, J.-F.; Mille, B. First evidence of cotton at neolithic mehrgarh, Pakistan: Analysis of mineralized fbres from a copper bead. J. Archaeol. Sci. 2002, 29, 1393–1401. [Google Scholar] [CrossRef]
- Rahman, M.; Rahmat, Z.; Mahmood, A.; Abdullah, K.; Zafar, Y. Cotton germplasm of Pakistan. In World Cotton Germplasm Resources; IntechOpen: London, UK, 2014; Volume 10, p. 58620. [Google Scholar]
- Azhar, M.T.; Amin, I.; Bashir, A.; Mansoor, S. Characterization of resistance gene analogs from Gossypium arboreum and their evolutionary relationships with homologs from tetraploid cottons. Euphytica 2010, 178, 351–362. [Google Scholar] [CrossRef]
- Hussain, T.; Mahmood, T. A note on leaf curl disease of cotton. Pak. Cottons 1988, 32, 248–251. [Google Scholar]
- Rishi, N.; Chauhan, M.S. Appearance of leaf curl disease of cotton in Northern India. J. Cotton Res. Dev. 1994, 8, 179–180. [Google Scholar]
- Adams, M.J.; Elliot, J.L.; Andrew, M.Q.; King, B.H.; Robert, L.H.; Nick, J.K.; Andrew, M.K.; Mart, K.; Jens, H.K.; Mart, K.; et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2017, 162, 2505–2538. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Briddon, R.W.; Bull, S.E.; Bedford, I.D.; Bashir, A.; Hussain, M.; Saeed, M.; Zafar, Y.; Malik, K.A.; Fauquet, C.; et al. Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA beta. Arch. Virol. 2003, 148, 1969–1986. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, Y.; Robinson, D.J.; Harrison, B.D. Four DNA-A variants among Pakistani isolates of cotton leaf curl virus and their affinities to DNA-A of geminivirus isolates from okra. J. Gen. Virol. 1998, 79, 915–923. [Google Scholar] [CrossRef]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Idris, A.M.; Mansoor, S.; Bedford, I.D.; Dhawan, P.; Rishi, N.; Siwatch, S.S.; Abdel-Salam, A.M.; et al. Diversity of DNA beta, a satellite molecule associated with some monopartite begomoviruses. Virology 2003, 312, 106–121. [Google Scholar] [CrossRef]
- Briddon, R.W.; Mansoor, S.; Bedford, I.D.; Pinner, M.S.; Saunders, K.; Stanley, J.; Zafar, Y.; Malik, K.A.; Markham, P.G. Identification of DNA components required for induction of cotton leaf curl disease. Virology 2001, 285, 234–243. [Google Scholar] [CrossRef]
- Rahman, M.; Hussain, D.; Malik, T.A.; Zafar, Y. Genetics of resistance to cotton leaf curl disease in Gossypium hirsutum. Plant Pathol. 2005, 54, 764–772. [Google Scholar] [CrossRef]
- Mansoor, S.; Amin, I.; Iram, S.; Hussain, M.; Zafar, Y.; Malik, K.A.; Briddon, R.W. Breakdown of resistance in cotton to cotton leaf curl disease in Pakistan. Plant Pathol. 2003, 52, 784. [Google Scholar] [CrossRef]
- Zubair, M.; Zaidi, S.S.; Shakir, S.; Farooq, M.; Amin, I.; Scheffler, J.A.; Scheffler, B.E.; Mansoor, S. Multiple begomoviruses found associated with cotton leaf curl disease in Pakistan in early 1990 are back in cultivated cotton. Sci. Rep. 2017, 7, 680. [Google Scholar] [CrossRef]
- Datta, S.; Budhauliya, R.; Das, B.; Gopalakrishnan, R.; Sharma, S.; Chatterjee, S.; Vanlalhmuaka; Raju, P.S.; Veer, V. Rebound of Cotton leaf curl Multan virus and its exclusive detection in cotton leaf curl disease outbreak, Punjab (India), 2015. Sci. Rep. 2017, 7, 17361. [Google Scholar] [CrossRef] [PubMed]
- Sattar, M.N.; Iqbal, Z.; Tahir, M.N.; Ullah, S. The prediction of a new CLCuD epidemic in the Old World. Front. Microbiol. 2017, 8, 631. [Google Scholar] [CrossRef]
- Akhtar, K.P.; Haidar, S.; Khan, M.K.R.; Ahmad, M.; Sarwar, N.; Murtaza, M.A.; Aslam, M. Evaluation of Gossypium species for resistance to cotton leaf curl Burewala virus. Ann. Appl. Biol. 2010, 157, 135–147. [Google Scholar] [CrossRef]
- Akhtar, K.P.; Ullah, R.; Khan, I.A.; Saeed, M.; Sarwar, N.; Mansoor, S. First symptomatic evidence of infection of Gossypium arboreum with Cotton leaf curl Burewala virus through grafting. Int. J. Agric. Biol. 2013, 15, 157–160. [Google Scholar]
- Nawaz-ul-Rehman, M.S.; Briddon, R.W.; Fauquet, C.M. A melting pot of Old World begomoviruses and their satellites infecting a collection of Gossypium species in Pakistan. PLoS ONE 2012, 7, e40050. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.K.; Zerbini, F.M.; Navas-Castillo, J.; Moriones, E.; Ramos-Sobrinho, R.; Silva, J.C.; Fiallo-Olive, E.; Briddon, R.W.; Hernandez-Zepeda, C.; Idris, A.; et al. Revision of begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015, 160, 1593–1619. [Google Scholar] [CrossRef]
- Saleem, H.; Nahid, N.; Shakir, S.; Ijaz, S.; Murtaza, G.; Khan, A.A.; Mubin, M.; Nawaz-ul-Rehman, M.S. Diversity, Mutation and Recombination Analysis of Cotton Leaf Curl Geminiviruses. PLoS ONE 2016, 11, e0151161. [Google Scholar] [CrossRef]
- Shafiq, M.; Iqbal, Z.; Ali, I.; Abbas, Q.; Mansoor, S.; Briddon, R.W.; Amin, I. Real-time quantitative PCR assay for the quantification of virus and satellites causing leaf curl disease in cotton in Pakistan. J. Vir. Methods 2017, 248, 54–60. [Google Scholar] [CrossRef]
- Ullah, R.; Akhtar, K.P.; Moffett, P.; Akbar, F.; Hassan, I.; Mansoor, S.; Briddon, R.W.; Mumtaz Hassan, H.; Saeed, M. Examination by grafting of the extreme resistance to cotton leaf curl disease in two wild Gossypium hirsutum L. cultivars. J. Plant Pathol. 2017, 99. [Google Scholar] [CrossRef]
- Naqvi, R.Z.; Zaidi, S.S.; Akhtar, K.P.; Strickler, S.; Woldemariam, M.; Mishra, B.; Mukhtar, M.S.; Scheffler, B.E.; Scheffler, J.A.; Jander, G.; et al. Transcriptomics reveals multiple resistance mechanisms against cotton leaf curl disease in a naturally immune cotton species, Gossypium arboreum. Sci. Rep. 2017, 7, 15880. [Google Scholar] [CrossRef]
- Briddon, R.W.; Akbar, F.; Iqbal, Z.; Amrao, L.; Amin, I.; Saeed, M.; Mansoor, S. Effects of genetic changes to the begomovirus/betasatellite complex causing cotton leaf curl disease in South Asia post-resistance breaking. Virus Res. 2014, 186, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.A.; Iqbal, Z.; Amin, I.; Mansoor, S. First report of Tomato leaf curl Gujarat virus, a bipartite begomovirus on cotton showing leaf curl symptoms in Pakistan. Plant Dis. 2015, 99, 1655–1656. [Google Scholar] [CrossRef]
- Zaidi, S.S.; Shafiq, M.; Amin, I.; Scheffler, B.E.; Scheffler, J.A.; Briddon, R.W.; Mansoor, S. Frequent occurrence of Tomato leaf curl New Delhi virus in cotton leaf curl disease affected cotton in Pakistan. PLoS ONE 2016, 11, e0155520. [Google Scholar] [CrossRef]
- Zaidi, S.S.; Martin, D.P.; Amin, I.; Farooq, M.; Mansoor, S. Tomato leaf curl New Delhi virus: A widespread bipartite begomovirus in the territory of monopartite begomoviruses. Mol. Plant. Pathol. 2017, 18, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-J.; Zhou, X.-P. Interaction between a nanovirus-like component and the Tobacco curly shoot virus/satellite complex. Acta Biochim. Biophys. Sin. 2005, 37, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1989, 12, 13–15. [Google Scholar]
- Guenoune-Gelbart, D.; Sufrin-Ringwald, T.; Capobianco, H.; Gaba, V.; Polston, J.E.; Lapidot, M. Inoculation of plants with begomoviruses by particle bombardment without cloning: Using rolling circle amplification of total DNA from infected plants and whiteflies. J. Virol. Methods 2010, 168, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Gnerre, S.; Maccallum, I.; Przybylski, D.; Ribeiro, F.J.; Burton, J.N.; Walker, B.J.; Sharpe, T.; Hall, G.; Shea, T.P.; Sykes, S.; et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 2011, 108, 1513–1518. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1. [Google Scholar] [CrossRef]
| Species | Begomovirus/alphasatellite/betasatellite | Genomic Component | Strain |
|---|---|---|---|
| G. mustelinum (AADD) | Cotton leaf curl Multan virus | Monopartite | Pakistan |
| Bhendi yellow vein mosaic virus | Monopartite | Thanagan | |
| Cotton leaf curl Alabad virus | DNA-A | Multan | |
| Cotton leaf curl Alabad virus | DNA-B | Multan | |
| Cotton leaf curl Burewala alphasatellite | Alphasatellite | N. A | |
| Ageratum yellow vein Singapore alphasatellite | Alphasatellite | N. A | |
| Gossypium darwinii symptomless alphasatellite | Alphasatellite | N. A | |
| Gossypium davidsonii symptomless alphasatellite | Alphasatellite | N. A | |
| Papaya leaf curl alphasatellite | Alphasatellite | N. A | |
| G. raimondii (DD) | Bhendi yellow vein mosaic virus | Monopartite | Thanagan |
| Cotton leaf curl Alabad virus | DNA-A | Multan | |
| Cotton leaf curl Alabad virus | DNA-B | Multan | |
| Cotton leaf curl Multan virus | Monopartite | Pakistan | |
| G. thurberi (DD) | Bhendi yellow vein mosaic virus | Monopartite | Pakistan |
| Cotton leaf curl Alabad virus | DNA-A | Multan | |
| Cotton leaf curl Alabad virus | DNA-B | Multan | |
| Cotton leaf curl Alabad virus | DNA-A | Lobatum | |
| Cotton leaf curl Alabad virus | DNA-B | Lobatum | |
| Cotton leaf curl Multan virus | Monopartite | Pakistan | |
| Cotton leaf curl Multan betasatellite | Betasatellite | Shahdadpur | |
| Cotton leaf curl Multan betasatellite | Betasatellite | Burewala | |
| Okra leaf curl betasatellite | Betasatellite | N. A |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shakir, S.; Zaidi, S.S.-e.-A.; Atiq-ur-Rehman; Farooq, M.; Amin, I.; Scheffler, J.; Scheffler, B.; Shah Nawaz-ul-Rehman, M.; Mansoor, S. Non-cultivated Cotton Species (Gossypium spp.) Act as a Reservoir for Cotton Leaf Curl Begomoviruses and Associated Satellites. Plants 2019, 8, 127. https://doi.org/10.3390/plants8050127
Shakir S, Zaidi SS-e-A, Atiq-ur-Rehman, Farooq M, Amin I, Scheffler J, Scheffler B, Shah Nawaz-ul-Rehman M, Mansoor S. Non-cultivated Cotton Species (Gossypium spp.) Act as a Reservoir for Cotton Leaf Curl Begomoviruses and Associated Satellites. Plants. 2019; 8(5):127. https://doi.org/10.3390/plants8050127
Chicago/Turabian StyleShakir, Sara, Syed Shan-e-Ali Zaidi, Atiq-ur-Rehman, Muhammad Farooq, Imran Amin, Jodi Scheffler, Brian Scheffler, Muhammad Shah Nawaz-ul-Rehman, and Shahid Mansoor. 2019. "Non-cultivated Cotton Species (Gossypium spp.) Act as a Reservoir for Cotton Leaf Curl Begomoviruses and Associated Satellites" Plants 8, no. 5: 127. https://doi.org/10.3390/plants8050127
APA StyleShakir, S., Zaidi, S. S.-e.-A., Atiq-ur-Rehman, Farooq, M., Amin, I., Scheffler, J., Scheffler, B., Shah Nawaz-ul-Rehman, M., & Mansoor, S. (2019). Non-cultivated Cotton Species (Gossypium spp.) Act as a Reservoir for Cotton Leaf Curl Begomoviruses and Associated Satellites. Plants, 8(5), 127. https://doi.org/10.3390/plants8050127





