Functional and Genetic Diversity of Bacteria Associated with the Surfaces of Agronomic Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Culture Media
2.2. Isolation of Bacterial Strains
2.3. 16S rRNA Gene Sequencing
2.4. Phylogenetic Analysis
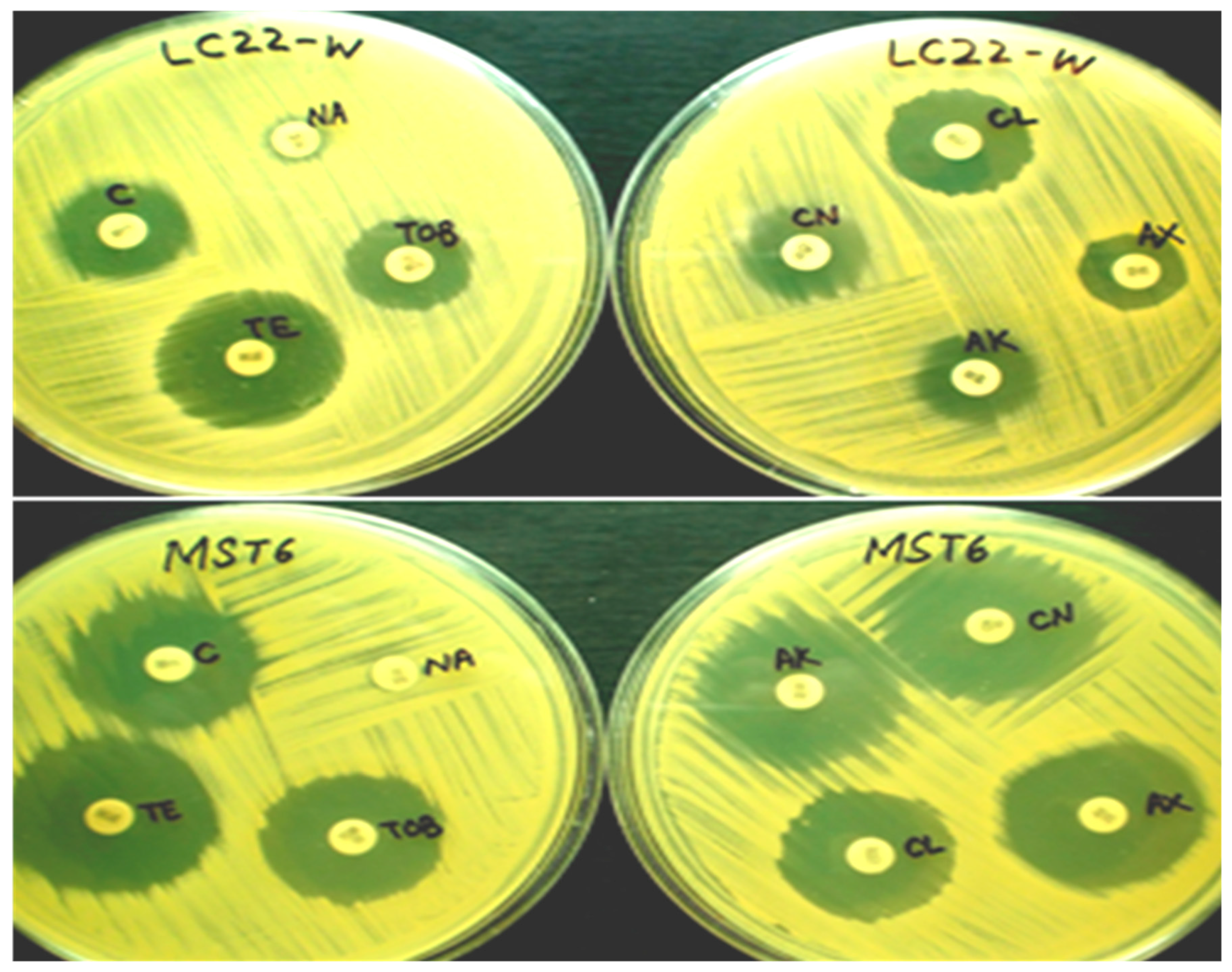
2.5. Antibiotic Susceptibility Pattern of Bacterial Strains
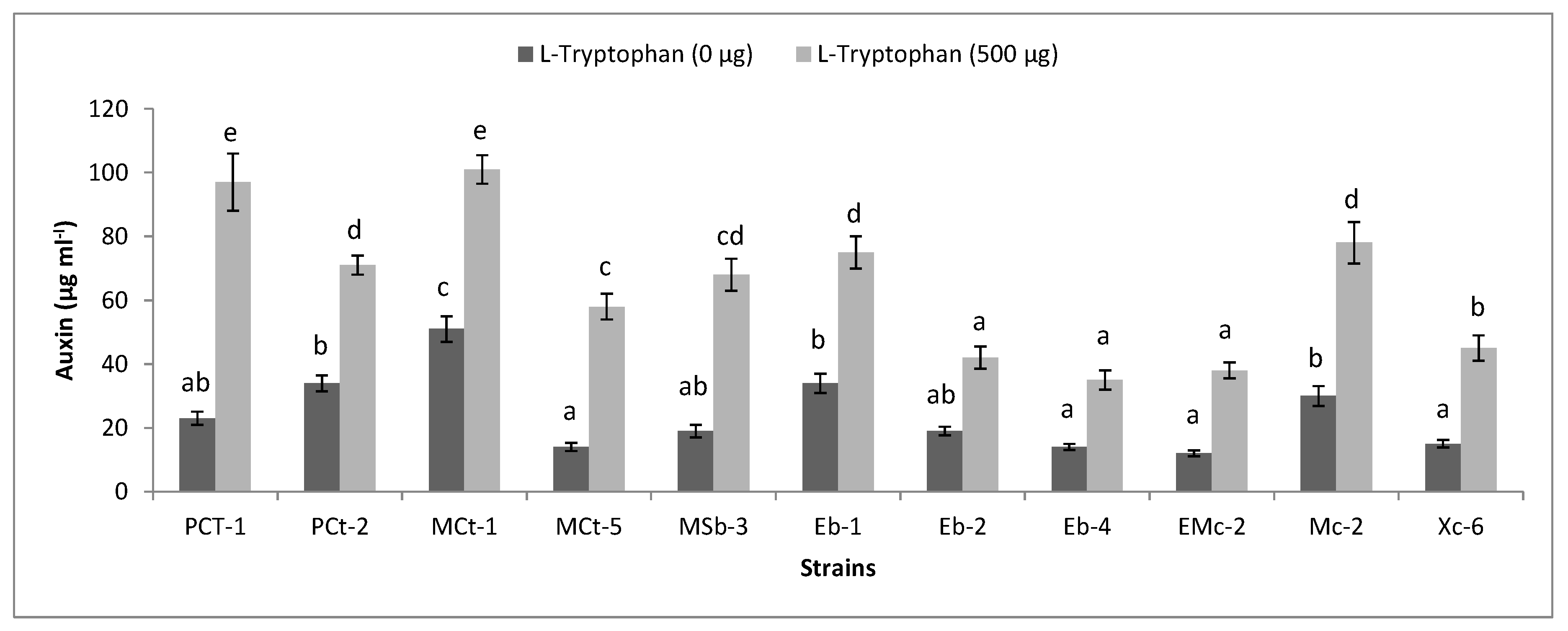
2.6. Functional Diversity of Plant-Associated Bacteria
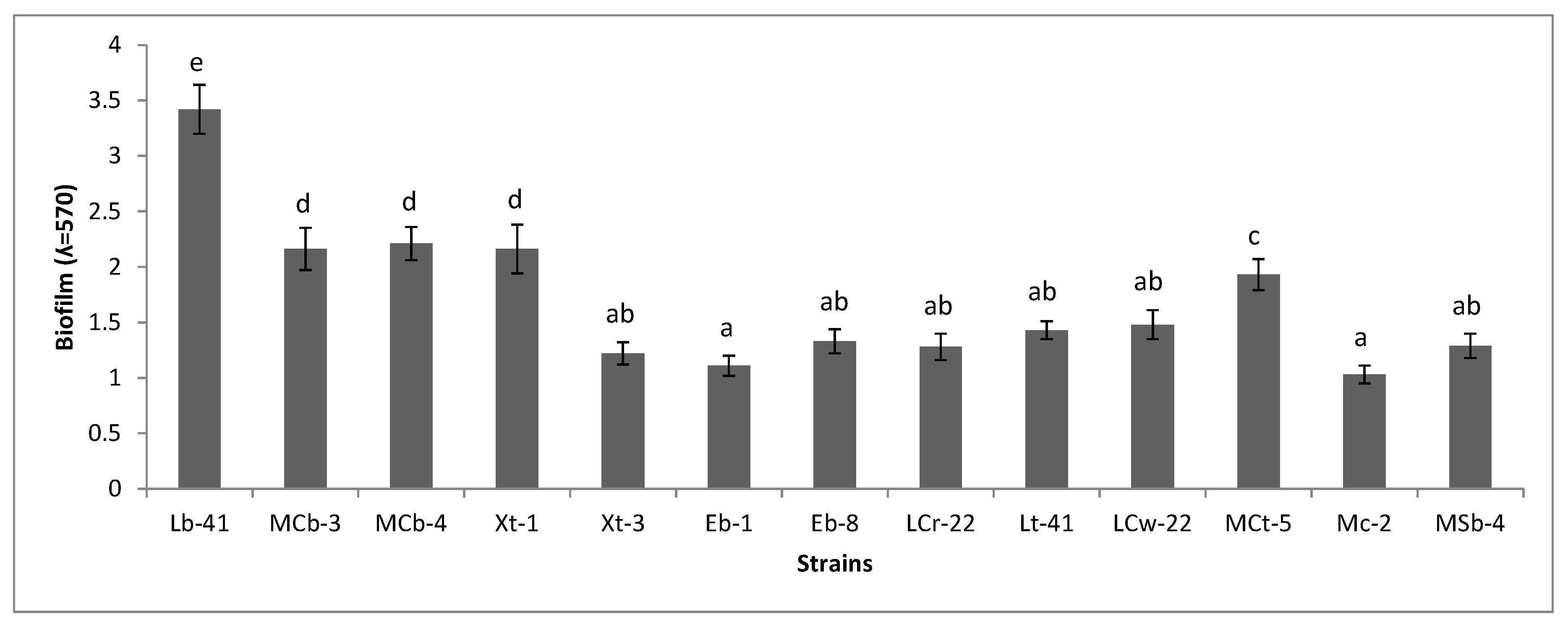
2.7. Biofilm Formation
2.8. Statistical Analysis
3. Results
3.1. 16S rRNA Gene Sequencing
3.2. Phylogenetic Analysis
3.3. Bacterial Diversity of Fresh Vegetables
3.4. Antibiotic Susceptibility of Bacterial Strains
3.5. Functional Diversity of Bacteria
3.6. Biofilm Formation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pem, D.; Jeewon, R. Fruit and vegetable intake: Benefits and progress of nutrition education interventions-Narrative review article. Iran. J. Public Health 2015, 44, 1309–1321. [Google Scholar] [PubMed]
- Sivapalasingam, S.; Friedman, C.R.; Cohen, L.; Tauxe, R.V. Fresh produce: A growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 2004, 67, 2342–2353. [Google Scholar] [CrossRef] [PubMed]
- Heaton, J.C.; Jones, K. Microbial contamination of fruit and vegetables and the behavior of enteropathogens in the phyllosphere: A review. J. Appl. Microbiol. 2008, 102, 613–626. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.A.S.; Wilson, I.G. The occurrence of enteric pathogens and Aeromonas species in organic vegetables. Int. J. Food Microbiol. 2001, 70, 155–162. [Google Scholar] [CrossRef]
- Johannessen, G.S.; Loncarevic, S.; Kruse, H. Bacteriological analysis of fresh produce in Norway. Int. J. Food Microbiol. 2002, 77, 199–204. [Google Scholar] [CrossRef]
- Salleh, N.A.; Rusul, G.; Hassan, Z.; Reezal, A.; Isa, S.H.; Nishibuchi, M.; Radu, S. Incidence of Salmonella spp. in raw vegetables in Selangor, Malaysia. Food Control 2003, 14, 475–479. [Google Scholar] [CrossRef]
- Johnston, L.M.; Jaykus, L.A.; Moll, D.; Martinez, M.C.; Anciso, J.; Mora, B.; Moe, C.L. A field study on the microbiological quality of fresh produce. J. Food Prot. 2005, 68, 1840–1847. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Speh, D.; Dyck, E.; Diez-Gonzalez, F. Preharvest evaluation of coliforms, Escherichia coli, Salmonella and Escherichia coli O157:H7 in organic and conventional produce grown by Minnesota farmers. J. Food Prot. 2004, 67, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Speh, D.; Jones, A.T.; Buesing, K.M.; Diez-Gonzalez, F. Longitudinal microbiological survey of fresh produce grown by farmers in the upper midwest. J. Food Prot. 2006, 69, 1928–1936. [Google Scholar] [CrossRef]
- Holden, N.; Pritchard, L.; Toth, I. Colonization outwith the colon: Plants as an alternative environmental reservoir for human pathogenic enterobacteria. FEMS Microbiol. Rev. 2009, 33, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Suckstorff, I.; Berg, G. Evidence for dose-dependent effects on plant growth by Stenotrophomonas strains from different origins. J. Appl. Microbiol. 2003, 95, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Bernier, S.P.; Silo-Suh, L.; Woods, D.E.; Ohman, D.E.; Sokol, P.A. Comparative analysis of plant and animal models for characterization of Burkholderia cepacia virulence. Infect. Immun. 2003, 71, 5306–5313. [Google Scholar] [CrossRef] [PubMed]
- Teplitski, M.; Warriner, K.; Bartz, J.; Schneider, K.R. Untangling metabolic and communication networks: Interactions of enterics with phytobacteria and their implications in produce safety. Trends Microbiol. 2011, 19, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Garbeva, P.; Raajimakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil. 2009, 321, 341–361. [Google Scholar] [CrossRef]
- Ahmed, A.; Hasnain, S. Auxin-producing Bacillus sp.: Auxin quantification and effect on the growth of Solanum tuberosum. Pure Appl. Chem. 2010, 82, 313–319. [Google Scholar] [CrossRef]
- Lwin, K.M.; Myint, M.M.; Tar, T.; Aung, W.Z.M. Isolation of plant hormone (indole-3-acetic acid—IAA) producing rhizobacteria and study on their effects on maize seedling. Eng. J. 2012, 16, 137–144. [Google Scholar] [CrossRef]
- Talboys, P.J.; Owen, D.W.; Healey, J.R.; Withers, P.J.; Jones, D.L. Auxin secretion by Bacillus amyloliquefaciens FZB42 both stimulates root exudation and limits phosphorus uptake in Triticum aestivum. BMC Plant Biol. 2014, 14, 51. [Google Scholar] [CrossRef]
- Akhtar, S.; Ali, B. Evaluation of rhizobacteria as non-rhizobial inoculants for mung beans. Aust. J. Crop Sci. 2011, 5, 1723–1729. [Google Scholar]
- Cappuccino, J.G.; Sherman, N. Microbiology: A Laboratory Manual; Pearson Education: Singapore, 2002. [Google Scholar]
- Park, S.-H.; Ryu, S.; Kang, D.-H. Development of an improved selective and differential medium for isolatin of Salmonella spp. J. Clin. Microbiol. 2012, 50, 3222–3226. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.L. Similarity analysis of rRNA. In Methods for General and Molecular Bacteriology; Gerhanrdt, P., Murray, R.G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 625–700. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.Y.; Borner, J. Enzymes involved in synthesis and breakdown of indoleacetic acid. In Modern Methods of Plant Analysis; Paech, K., Tracey, M.V., Eds.; Springer: Heidelberg, Germany, 1979; Volume 7, pp. 238–241. [Google Scholar]
- Pikovskaya, R.I. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Microbiologiya 1948, 17, 362–370. [Google Scholar]
- Ahmad, F.; Ahmad, I.; Khan, M.S. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase negative staphylococci to plastic tissue culture plates: A quantitive model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar]
- Berger, C.N.; Sodha, S.V.; Shaw, R.K.; Pink, D.; Hand, P.; Frankel, G. Fresh fruits and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 2010, 12, 2385–2397. [Google Scholar] [CrossRef]
- Critzer, F.J.; Doyle, M.P. Microbial ecology of foodborne pathogens associated with produce. Curr. Opin. Biotechnol. 2010, 21, 125–130. [Google Scholar] [CrossRef]
- Schikora, A.; Gracia, A.V.; Hirt, H. Plants as alternative hosts for Salmonella. Trends Plant Sci. 2012, 17, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Arnesen, S.L.P.; Fagerlund, A.; Granum, P.E. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 2008, 32, 579–606. [Google Scholar] [CrossRef] [PubMed]
- Schoeni, J.L.; Wong, A.C. Bacillus cereus food poisoning and its toxins. J. Food Prot. 2005, 68, 636–648. [Google Scholar] [CrossRef] [PubMed]
- Falomir, M.P.; Rico, H.; Gozalbo, D. Enterobacter and Klebsiella species isolated from fresh vegetables marketed in Valencia (Spain) and their clinically relevant resistances to chemotherapeutic agents. Foodborne Pathog. Dis. 2013, 10, 1002–1007. [Google Scholar] [CrossRef] [PubMed]
- Argudin, M.A.; Mendoza, M.C.; Rodicio, M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef] [PubMed]
- Olsen, S.J.; MacKinon, L.C.; Goulding, J.S.; Slutsker, L. Surveillance for foodborne disease outbreaks–United States, 1993–1997. MMWR 2000, 49, 1–62. [Google Scholar] [PubMed]
- Greene, S.K.; Daly, E.R.; Talbot, E.A.; Demma, L.J.; Holzbauer, S.; Patel, N.J.; Hill, T.A.; Walderhaug, M.O.; Hoekstra, R.M.; Lynch, M.F.; et al. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol. Infect. 2008, 136, 157–165. [Google Scholar] [CrossRef]
- Soon, J.M.; Manning, L.; Davies, W.P.; Baines, R. Fresh produce-associated outbreaks: A call for HACCP on farms? Br. Food J. 2010, 114, 553–597. [Google Scholar] [CrossRef]
- Gumiere, T.; Ribeiro, C.M.; Vasconcellos, R.L.; Cardoso, E.J. Indole-3-acetic acid producing root-associated bacteria on growth of Brazil pine (Araucaria angustifolia) and slash pine (Pinus elliottii). Antonie Van Leeuwenhoek 2014, 105, 663–669. [Google Scholar] [CrossRef]
- Fierro-Coronado, R.F.; Quiroz-Figueroa, F.R.; García-Pérez, L.M.; Ramírez-Chávez, E.; Molina-Torres, J.; Maldonado-Mendoza, I.E. IAA-producing rhizobacteria from chickpea (Cicer arientinum L.) induce changes in root architecture and increase root biomass. Can. J. Microbiol. 2014, 60, 639–648. [Google Scholar] [CrossRef]
- Raheem, A.; Ali, B. Halotolerant rhizobacteria: Beneficial plant metabolites and growth enhancement of Triticum aestivum L. in salt amended soils. Arch. Agron. Soil Sci. 2015, 61, 1691–1705. [Google Scholar] [CrossRef]
- Zaidi, A.; Ahmad, E.; Khan, M.S.; Saif, S.; Rizvi, A. Role of plant growth promoting rhizobacteria in sustainable production of vegetables: Current perspective. Sci. Hortic. 2015, 193, 231–239. [Google Scholar] [CrossRef]
- Ali, B.; Sabri, A.N.; Hasnain, S. Rhizobacterial potential to alter auxin content and growth of Vigna radiata (L.). World J. Microniol. Biotechnol. 2010, 26, 1379–1384. [Google Scholar] [CrossRef]
- Ramey, B.E.; Koutsoudis, M.; Bodman, S.B.; Fuqua, C. Biofilm formation in plant-microbe associations. Curr. Opin. Microbiol. 2004, 7, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Bartz, F.E.; Lickness, J.S.; Heredia, N.; De Aceituno, A.F.; Newman, K.L.; Hodge, D.W.; Jaykus, L.-A.; García, S.; Leon, J.S. Contamination of fresh produce by microbial indicators on farm and in packing facilities: Elucidation of environmental routes. Appl. Environ. Microbiol. 2017, 83, e02984-16. [Google Scholar] [CrossRef] [PubMed]
- Aslam, F.; Ali, B. Halotolerant bacterial diversity associated with Suaeda fruticosa (L.) Forssk. improved growth of Maize under salinity stress. Agronomy 2018, 8, 131. [Google Scholar] [CrossRef]
- Anwar, S.; Ali, B.; Sajid, I. Screening of rhizospheric actinomycetes for various in-vitro and in-vivo plant growth promoting (PGP) traits and for agroactive compounds. Front. Microbiol. 2016, 7, 1334. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Kamilova, F.; Validov, S.; Gafurova, L.; Kucharova, Z.; Lugtenberg, B. High incidence of plant growth-stimulating bacteria associated with the rhizosphere of wheat grown on salinated soil in Uzbekistan. Environ. Microbiol. 2008, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Noreen, S.; Ali, B.; Hasnain, S. Growth promotion of Vigna mungo (L.) by Pseudomonas spp. exhibiting auxin production and ACC-deaminase activity. Ann. Microbiol. 2012, 62, 411–417. [Google Scholar] [CrossRef]
- Sadiq, A.; Ali, B. Growth and yield enhancement of Triticum aestivum L. by rhizobacteria isolated from agronomic plants. Aust. J. Crop Sci. 2013, 7, 1544–1550. [Google Scholar]

| Serial No. | Isolates | Temperature for Isolation | Culture Media | Identified as | Accessions |
|---|---|---|---|---|---|
| 1 | BPc-4 | 30 °C | L-agar | Bacillus cereus BPc-4 | KJ865556 |
| 2 | EMc-3 | 37 °C | L-agar | B. anthracis EMc-3 | KJ865553 |
| 3 | LCw-22 | 30 °C | L-agar | B. cereus LCw-22 | KJ865598 |
| 4 | MSc-5 | 37 °C | MSA | Staphylococcus warneri MSc-5 | KJ865590 |
| 5 | Xc-7 | 30 °C | L-agar | Lysinibacillus fusiformis Xc-7 | KJ865599 |
| 6 | BPc-1 | 37 °C | MAC | Serratia rubidaea BPc-1 | KJ865576 |
| 7 | BPc-3 | 37 °C | MAC | Pantoea dispersa BPc-3 | KJ865552 |
| 8 | EMc-2 | 37 °C | MAC | Se. rubidaea EMc-2 | KJ865581 |
| 9 | EMc-4 | 37 °C | L-agar | Acinetobacter calcoaceticus EMc-4 | KJ865567 |
| 10 | Lc-52 | 37 °C | L-agar | Ac. calcoaceticus Lc-52 | KJ865566 |
| 11 | Lcr-22 | 37 °C | MAC | Se. rubidaea Lcr-22 | KJ865575 |
| 12 | Mc-2 | 37 °C | MAC | Se. rubidaea Mc-2 | KJ865602 |
| 13 | Mc-3 | 37 °C | L-agar | Stenotrophomonas maltophilia Mc-3 | KJ865587 |
| 14 | Mc-4 | 37 °C | L-agar | Ac. calcoaceticus Mc-4 | KJ865588 |
| 15 | MSc-1 | 37 °C | MAC | Pantoea sp. MSc-1 | KJ865571 |
| 16 | Xc-3 | 37 °C | EMB | Enterobacter cloacae Xc-3 | KJ865572 |
| 17 | Xc-5 | 30 °C | L-agar | Pseudomonas putida Xc-5 | KJ865551 |
| 18 | Xc-6 | 37 °C | EMB | Citrobacter freundii Xc-6 | KJ865549 |
| Serial No. | Isolates | Temperature for Isolation | Culture Media | Identified as | Accessions |
|---|---|---|---|---|---|
| 1 | BPb-5 | 37 °C | MSA | Staph. aureus BPb-5 | KJ865591 |
| 2 | Eb-9 | 30 °C | L-agar | B. cereus Eb-9 | KJ865594 |
| 3 | Eb-10 | 30 °C | L-agar | B. thuringiensis Eb-10 | KJ865560 |
| 4 | Lb-41 | 30 °C | L-agar | Arthrobacter nicotianae Lb-41 | KJ865583 |
| 5 | Lb-61 | 30 °C | L-agar | B. subtilis Lb-61 | KJ865595 |
| 6 | MCb-3 | 37 °C | L-agar | Staph. arlettae MCb-3 | KJ865592 |
| 7 | MCb-4 | 37 °C | L-agar | Exiguobacterium mexicanum MCb-4 | KJ865577 |
| 8 | MCb-6 | 30 °C | L-agar | B. cereus MCb-6 | KJ865559 |
| 9 | MCb-8 | 30 °C | L-agar | B. subtilis MCb-8 | KJ865584 |
| 10 | MSb-3 | 30 °C | L-agar | B. cereus MSb-3 | KJ865596 |
| 11 | MSb-4 | 37 °C | L-agar | B. anthracis MSb-4 | KJ865558 |
| 12 | Xb-6 | 30 °C | L-agar | B. anthracis Xb-6 | KJ865582 |
| 13 | BPb-3 | 37 °C | L-agar | Ac. calcoaceticus BPb-3 | KJ865562 |
| 14 | Eb-1 | 37 °C | MAC | Klebsiella pneumoniae Eb-1 | KJ865601 |
| 15 | Eb-2 | 37 °C | MAC | Pa. vagans Eb-2 | KJ865561 |
| 16 | Eb-4 | 30 °C | L-agar | Ac. calcoaceticus Eb-4 | KJ865586 |
| 17 | Eb-6 | 30 °C | L-agar | Burkholderia cepacia Eb-6 | KJ865578 |
| 18 | Eb-8 | 37 °C | L-agar | Ac. calcoaceticus Eb-8 | KJ865568 |
| 19 | Xb-3 | 37 °C | MAC | Se. rubidaea Xb-3 | KJ865564 |
| Serial No. | Isolates | Temperature for Isolation | Culture Media | Identified as | Accessions |
|---|---|---|---|---|---|
| 1 | BPt-5 | 37 °C | MSA | Staph. equorum BPt-5 | KJ865579 |
| 2 | Lt-41 | 37 °C | MSA | Staph. xylosus Lt-41 | KJ865585 |
| 3 | Lt-73 | 37 °C | MSA | Staph. warneri Lt-73 | KJ865565 |
| 4 | MSt-1 | 37 °C | MSA | Staph. xylosus MSt-1 | KJ865600 |
| 5 | MSt-3 | 37 °C | MSA | Staph. gallinarum MSt-3 | KJ865580 |
| 6 | MSt-7 | 30 °C | L-agar | B. cereus MSt-7 | KJ865557 |
| 7 | MSt-8 | 30 °C | L-agar | B. cereus MSt-8 | KJ865589 |
| 8 | PCt-1 | 30 °C | L-agar | B. cereus PCt-1 | KJ865573 |
| 9 | Xt-1 | 37 °C | MSA | Staph. xylosus Xt-1 | KJ865597 |
| 10 | Xt-6 | 30 °C | L-agar | L. fusiformis Xt-6 | KJ865555 |
| 11 | EMt-1 | 37 °C | L-agar | Ac. bouvetii EMt-1 | KJ865593 |
| 12 | EMt-5 | 37 °C | EMB | E. amnigenus EMt-5 | KJ865563 |
| 13 | MCt-1 | 37 °C | L-agar | St. maltophilia MCt-1 | KJ865603 |
| 14 | MCt-5 | 37 °C | MAC | Kluyvera cryocrescens MCt-5 | KJ865554 |
| 15 | MCt-6 | 37 °C | MAC | Se. ureilytica MCt-6 | KJ865570 |
| 16 | MSt-6 | 37 °C | L-agar | Ac. calcoaceticus MSt-6 | KJ865569 |
| 17 | PCt-2 | 37 °C | EMB | E. cloacae PCt-2 | KJ865574 |
| 18 | Xt-3 | 37 °C | EMB | C. werkmannii Xt-3 | KJ865550 |
| Strains | Antibiotics | |||||||
|---|---|---|---|---|---|---|---|---|
| AK | AX | CL | CN | NA | TOB | TE | C | |
| Zones of Inhibition (mm) * | ||||||||
| Bacillus cereus BPc-4 | 18 (S) | 10 (R) | 8 (R) | 14 (I) | 14 (I) | 15 (S) | 22 (S) | 10 (R) |
| Staphylococcus equorum BPt-5 | 26 (S) | 24 (R) | 11 (R) | 20 (S) | 0 (R) | 16 (S) | 16 (I) | 26 (S) |
| S. aureus BPb-5 | 18 (S) | 8 (R) | 18 (S) | 18 (S) | 14 (I) | 16 (S) | 26 (S) | 24 (S) |
| B. anthracis EMc-3 | 20 (S) | 16 (I) | 14 (I) | 24 (S) | 16 (I) | 24 (S) | 24 (S) | 24 (S) |
| B. cereus Eb-9 | 16 (S) | 20 (S) | 0 (R) | 22 (S) | 20 (S) | 12 (R) | 18 (I) | 24 (S) |
| B. thuringiensis Eb-10 | 18 (S) | 20 (S) | 0 (R) | 14 (I) | 0 (R) | 12 (R) | 20 (S) | 12 (R) |
| S. xylosus Lt-41 | 20 (S) | 24 (R) | 22 (S) | 20 (S) | 0 (R) | 18 (S) | 22 (S) | 20 (S) |
| S. warneri Lt-73 | 20 (S) | 0 (R) | 0 (R) | 12 (R) | 0 (R) | 10 (R) | 16 (I) | 24 (S) |
| Arthrobacter nicotianae Lb-41 | 22 (S) | 26 (S) | 16 (S) | 16 (S) | 0 (R) | 14 (I) | 20 (S) | 30 (S) |
| B. subtilis Lb-61 | 28 (S) | 20 (S) | 14 (I) | 30 (S) | 12 (R) | 22 (S) | 26 (S) | 14 (I) |
| B. cereus LCw-22 | 16 (I) | 14 (I) | 20 (S) | 16 (S) | 10 (R) | 16 (S) | 24 (S) | 18 (S) |
| S. arlettae MCb-3 | 32 (S) | 14 (I) | 22 (S) | 28 (S) | 0 (R) | 22 (S) | 8 (R) | 24 (S) |
| Exiguobacterium mexicanum MCb-4 | 22 (S) | 38 (S) | 28 (S) | 20 (S) | 18 (I) | 16 (S) | 26 (S) | 24 (S) |
| B. cereus MCb-6 | 20 (S) | 14 (I) | 18 (S) | 16 (S) | 0 (R) | 14 (I) | 24 (S) | 16 (I) |
| B. subtilis MCb-8 | 16 (I) | 0 (R) | 0 (R) | 10 (R) | 12 (R) | 10 (R) | 16 (I) | 10 (R) |
| S. xylosus MSt-1 | 24 (S) | 22 (R) | 10 (R) | 20 (S) | 0 (R) | 20 (S) | 20 (S) | 24 (S) |
| S. gallinarum MSt-3 | 14 (I) | 12 (R) | 16 (S) | 14 (I) | 0 (R) | 10 (R) | 20 (S) | 18 (S) |
| B. cereus MSt-7 | 26 (S) | 14 (I) | 32 (S) | 24 (S) | 18 (I) | 18 (S) | 14 (R) | 20 (S) |
| B. cereus MSt-8 | 18 (S) | 8 (R) | 14 (I) | 12 (R) | 16 (I) | 18 (S) | 24 (S) | 28 (S) |
| B. cereus MSb-3 | 34 (S) | 42 (S) | 24 (S) | 30 (S) | 12 (R) | 18 (S) | 24 (S) | 30 (S) |
| B. anthracis MSb-4 | 14 (I) | 16 (I) | 12 (I) | 14 (I) | 14 (I) | 20 (S) | 18 (I) | 20 (S) |
| S. warneri MSc-5 | 24 (S) | 40 (S) | 38 (S) | 30 (S) | 14 (I) | 24 (S) | 28 (S) | 24 (S) |
| B. cereus PCt-1 | 18 (S) | 0 (R) | 0 (R) | 14 (I) | 16 (I) | 12 (R) | 16 (I) | 16 (I) |
| S. xylosus Xt-1 | 20 (S) | 20 (R) | 20 (S) | 18 (S) | 0 (R) | 18 (S) | 22 (S) | 20 (S) |
| Lysinibacillus fusiformis Xt-6 | 16 (I) | 0 (R) | 0 (R) | 14 (I) | 14 (I) | 12 (R) | 18 (I) | 20 (S) |
| B. anthracis Xb-6 | 22 (S) | 12 (R) | 14 (I) | 16 (S) | 12 (R) | 16 (S) | 20 (S) | 20 (S) |
| L. fusiformis Xc-7 | 34 (S) | 0 (R) | 0 (R) | 25 (S) | 0 (R) | 8 (R) | 32 (S) | 40 (S) |
| Strains | Antibiotics | |||||||
|---|---|---|---|---|---|---|---|---|
| AK | AX | CL | CN | NA | TOB | TE | C | |
| Zone of Inhibition (mm) * | ||||||||
| Serratia rubidaea BPc-1 | 18 (S) | 12 (R) | 0 (R) | 14 (I) | 14 (I) | 12 (R) | 14 (R) | 18 (S) |
| Pantoea dispersa BPc-3 | 22 (S) | 14 (I) | 14 (I) | 18 (S) | 12 (R) | 14 (I) | 20 (S) | 12 (R) |
| Acinetobacter calcoaceticus BPb-3 | 16 (I) | 14 (I) | 12 (I) | 18 (S) | 14 (I) | 20 (S) | 26 (S) | 22 (S) |
| Klebsiella penumoniae Eb-1 | 18 (S) | 0 (R) | 14 (I) | 18 (S) | 18 (I) | 12 (R) | 20 (S) | 22 (S) |
| P. vagans Eb-2 | 20 (S) | 22 (S) | 20 (S) | 20 (S) | 18 (I) | 22 (S) | 22 (S) | 10 (R) |
| A. calcoaceticus Eb-4 | 20 (S) | 0 (R) | 10 (R) | 12 (R) | 14 (I) | 16 (S) | 18 (I) | 20 (S) |
| Burkholderia cepacia Eb-6 | 24 (S) | 16 (I) | 20 (S) | 18 (S) | 0 (R) | 20 (S) | 20 (S) | 14 (I) |
| A. calcoaceticus Eb-8 | 18 (S) | 8 (R) | 0 (R) | 16 (S) | 16 (I) | 14 (I) | 20 (S) | 12 (R) |
| A. bouvetii EMt-1 | 18 (S) | 22 (S) | 26 (S) | 14 (I) | 18 (I) | 24 (S) | 26 (S) | 22 (S) |
| Enterobacter amnigenus EMt-5 | 18 (S) | 0 (R) | 0 (R) | 12 (R) | 12 (R) | 10 (R) | 16 (I) | 22 (S) |
| S. rubidaea EMc-2 | 26 (S) | 12 (R) | 16 (S) | 16 (S) | 0 (R) | 14 (I) | 22 (S) | 16 (I) |
| A. calcoaceticus EMc-4 | 28 (S) | 14 (I) | 22 (S) | 16 (S) | 16 (I) | 18 (S) | 18 (I) | 20 (S) |
| A. calcoaceticus Lc-52 | 16 (I) | 12 (R) | 12 (I) | 14 (I) | 12 (R) | 14 (I) | 18 (I) | 18 (S) |
| S. rubidaea Lcr-22 | 22 (S) | 14 (I) | 0 (R) | 16 (S) | 18 (I) | 12 (R) | 18 (I) | 20 (S) |
| Stenotrophomonas maltophilia MCt-1 | 16 (I) | 0 (R) | 0 (R) | 14 (I) | 22 (S) | 10 (R) | 20 (S) | 22 (S) |
| Kluyvera cryocrescens MCt-5 | 12 (R) | 10 (R) | 10 (R) | 14 (I) | 12 (R) | 16 (S) | 18 (I) | 18 (S) |
| S. ureilytica MCt-6 | 14 (I) | 10 (R) | 14 (I) | 12 (R) | 14 (I) | 16 (S) | 22 (S) | 20 (S) |
| S. rubidaea Mc-2 | 24 (S) | 18 (S) | 14 (I) | 16 (S) | 12 (R) | 18 (S) | 16 (I) | 22 (S) |
| S. maltophilia Mc-3 | 16 (I) | 0 (R) | 0 (R) | 10 (R) | 0 (R) | 0 (R) | 10 (R) | 12 (R) |
| A. calcoaceticus Mc-4 | 26 (S) | 20 (S) | 28 (S) | 28 (S) | 0 (R) | 26 (S) | 20 (S) | 28 (S) |
| A. calcoaceticus MSt-6 | 28 (S) | 26 (S) | 24 (S) | 28 (S) | 0 (R) | 24 (S) | 28 (S) | 24 (S) |
| Pantoea sp. MSc-1 | 28 (S) | 20 (S) | 18 (S) | 22 (S) | 20 (S) | 25 (S) | 26 (S) | 18 (S) |
| E. cloacae PCt-2 | 12 (R) | 14 (I) | 10 (R) | 10 (R) | 0 (R) | 10 (R) | 18 (I) | 18 (S) |
| E. cloacae Xc-3 | 18 (S) | 0 (R) | 0 (R) | 16 (S) | 20 (S) | 10 (R) | 20 (S) | 22 (S) |
| Pseudomonas putida Xc-5 | 34 (S) | 12 (R) | 10 (R) | 28 (S) | 16 (I) | 30 (S) | 36 (S) | 16 (I) |
| Citrobacter freundii Xc-6 | 18 (S) | 10 (R) | 0 (R) | 16 (S) | 14 (I) | 10 (R) | 14 (R) | 20 (S) |
| C. werkmannii Xt-3 | 16 (I) | 0 (R) | 0 (R) | 18 (S) | 16 (I) | 12 (R) | 18 (I) | 20 (S) |
| S. rubidaea Xb-3 | 22 (S) | 12 (R) | 0 (R) | 18 (S) | 22 (S) | 14 (I) | 16 (I) | 22 (S) |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, B. Functional and Genetic Diversity of Bacteria Associated with the Surfaces of Agronomic Plants. Plants 2019, 8, 91. https://doi.org/10.3390/plants8040091
Ali B. Functional and Genetic Diversity of Bacteria Associated with the Surfaces of Agronomic Plants. Plants. 2019; 8(4):91. https://doi.org/10.3390/plants8040091
Chicago/Turabian StyleAli, Basharat. 2019. "Functional and Genetic Diversity of Bacteria Associated with the Surfaces of Agronomic Plants" Plants 8, no. 4: 91. https://doi.org/10.3390/plants8040091
APA StyleAli, B. (2019). Functional and Genetic Diversity of Bacteria Associated with the Surfaces of Agronomic Plants. Plants, 8(4), 91. https://doi.org/10.3390/plants8040091




