Expression of Genes Related to Sugar and Amino Acid Transport and Cytokinin Metabolism during Leaf Development and Senescence in Pisum sativum L.
Abstract
:1. Introduction
2. Results
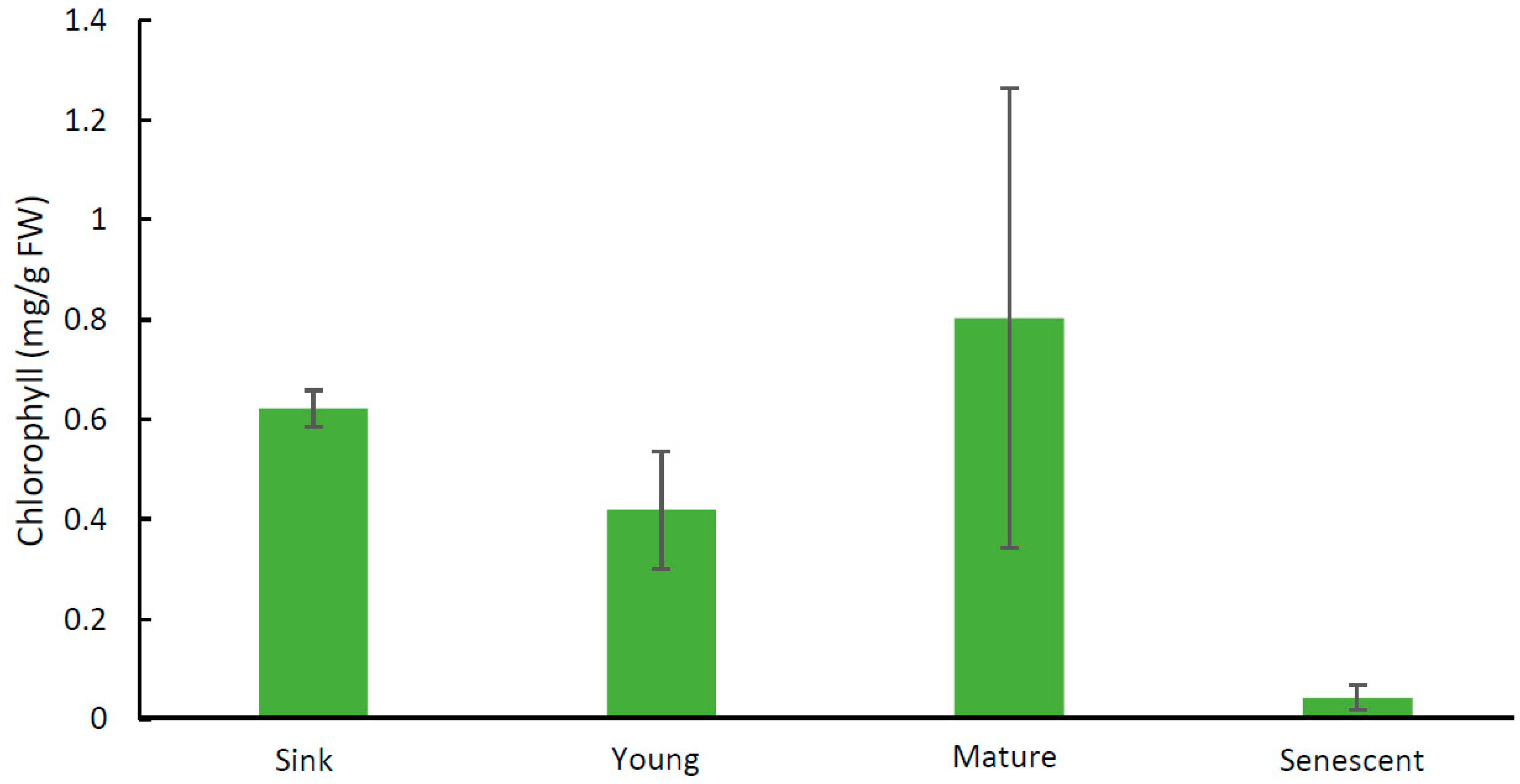
2.1. Chlorophyll Analyses
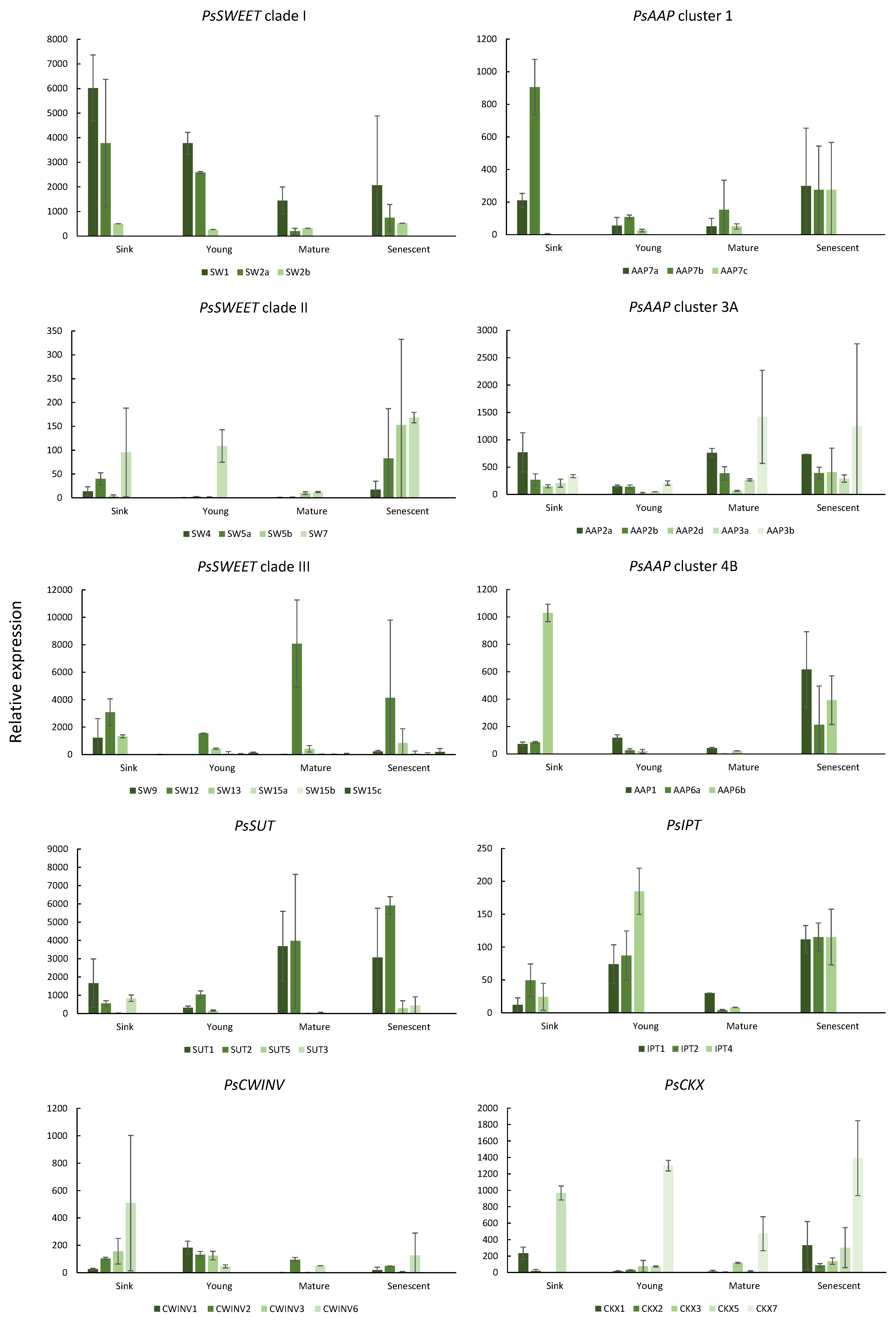
2.2. Gene Expression
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Gene Isolation and Sequence Analysis
4.3. RNA Isolation and cDNA Synthesis
4.4. Quantitative Reverse Transcription Polymerase Chain Reaction
4.5. Chlorophyll Estimation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Woo, H.R.; Masclaux-Daubresse, C.; Lim, P.O. Plant senescence: How plants know when and how to die. J. Exp. Bot. 2018, 69, 715–718. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Earl, S.; Harrison, E.; Mathas, E.; Navabpour, S.; Page, T.; Pink, D. The molecular analysis of leaf senescence—A genomics approach. Plant Biol. J. 2003, 1, 3–22. [Google Scholar]
- Foyer, C.H.; Lam, H.-M.; Nguyen, H.T.; Siddique, K.H.M.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M.; et al. Neglecting legumes has compromised human health and sustainable food production. Nat. Plant 2016, 2, 16112. [Google Scholar]
- Vanacker, H.; Sandalio, L.M.; Jiménez, A.; Palma, J.M.; Corpas, F.J.; Meseguer, V.; Gómez, M.; Sevilla, F.; Leterrier, M.; Foyer, C.H.; et al. Roles for redox regulation in leaf senescence of pea plants grown on different sources of nitrogen nutrition. J. Exp. Bot. 2006, 57, 1735–1745. [Google Scholar] [CrossRef] [Green Version]
- Buchanan-Wollaston, V.; Page, T.; Harrison, E.; Breeze, E.; Lim, P.O.; Nam, H.G.; Lin, J.-F.; Wu, S.-H.; Swidzinski, J.; Ishizaki, K.; et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005, 42, 567–585. [Google Scholar] [CrossRef] [Green Version]
- van der Graaff, E.; Schwacke, R.; Schneider, A.; Desimone, M.; Flügge, U.-I.; Kunze, R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006, 141, 776–792. [Google Scholar] [CrossRef]
- Sade, N.; del Mar Rubio-Wihelmi, M.; Umnajkitikorn, K.; Blumwald, E. Stress-induced senescence and plant tolerance to abiotic stress. J. Exp. Bot. 2018, 69, 845–853. [Google Scholar] [CrossRef]
- Clément, G.; Moison, M.; Soulay, F.; Reisdorf-Cren, M.; Masclaux-Daubresse, C. Metabolomics of laminae and midvein during leaf senescence and source-sink metabolite management in Brassica napus L. leaves. J. Exp. Bot. 2018, 69, 891–903. [Google Scholar] [CrossRef]
- Chrobok, D.; Law, S.R.; Brouwer, B.; Lindén, P.; Ziolkowska, A.; Liebsch, D.; Narsai, R.; Szal, B.; Moritz, T.; Rouchier, N.; et al. Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant Physiol. 2016, 172, 2132–2153. [Google Scholar] [CrossRef]
- Mothes, K.; Engelbrecht, L. On the activity of a kinetin-like root factor. Life Sci. 1963, 11, 852–857. [Google Scholar] [CrossRef]
- Richmond, A.E.; Lang, A. Effect of kinetin on protein content and survival of detached Xanthium leaves. Science 1957, 125, 650–651. [Google Scholar] [CrossRef]
- Balibrea, L.M.E.; Gonzalez, M.C.G.; Fatima, T.; Ehneß, R.; Lee, T.; Proels, R.; Tanner, W.; Roitsch, R. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell 2004, 16, 1276–1287. [Google Scholar] [CrossRef]
- Dhandapani, P.; Song, J.; Novak, O.; Jameson, P.E. Infection by Rhodococcus fascians maintains cotyledons as a sink tissue for the pathogen. Ann. Bot. 2017, 119, 841–852. [Google Scholar]
- Ninan, A.S.; Shah, A.; Song, J.; Jameson, P.E. Differential gene expression in the meristem and during early fruit growth of Pisum sativum L. identifies potential targets for breeding. Int. J. Mol. Sci. 2017, 18, 428. [Google Scholar] [CrossRef]
- Jameson, P.E. Cytokinin metabolism and compartmentation. In Cytokinins Chemistry, Activity and Function; Mok, D.W.S., Mok, M.C., Eds.; CRC Press: Boca Raton, FL, USA, 1994; pp. 113–128. [Google Scholar]
- Šmehilová, M.; Dobrůšková, J.; Novák, P.; Takáč, T.; Galuszka, P. Cytokinin-specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front. Plant Sci. 2016, 7, 1264. [Google Scholar] [CrossRef]
- Brütting, C.; Schäfer, M.; Vanková, R.; Gase, K.; Baldwin, I.T.; Meldau, S. Changes in cytokinins are sufficient to alter developmental patterns of defense metabolites in Nicotiana attenuata. Plant J. 2017, 89, 15–30. [Google Scholar] [CrossRef]
- Edlund, E.; Novak, O.; Karady, M.; Ljung, K.; Jansson, S. Contrasting patterns of cytokinins between years in senescing aspen leaves. Plant Cell Environ. 2017, 40, 622–634. [Google Scholar] [CrossRef] [Green Version]
- Jibran, R.; Hunter, D.A.; Dijkwel, P.P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013, 82, 547–561. [Google Scholar] [CrossRef]
- Yadav, U.P.; Ayre, B.G.; Bush, R. Transgenic approaches to altering carbon and nitrogen partitioning in whole plants: Assessing the potential to improve crop yields and nutritional quality. Front. Plant Sci. 2015, 6, 275. [Google Scholar] [CrossRef]
- Jian, H.; Lu, K.; Yang, B.; Wang, T.; Zhang, L.; Zhang, A.; Wang, J.; Liu, L.; Qu, C.; Li, J. Genome-wide analysis and expression profiling of the SUC and SWEET gene families of sucrose transporters in oilseed rape (Brassica napus L.). Front. Plant Sci. 2016, 7, 1464. [Google Scholar] [CrossRef]
- Chandran, D. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. Int. J. Biochem. Mol. Biol. 2015, 67, 461–471. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–534. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Li, Y.; Feng, S.; Ma, S.; Sui, X.; Zhang, Z. Spatiotemporal expression and substrate specificity analysis of the cucumber SWEET gene family. Front. Plant Sci. 2017, 8, 1855. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Zhang, H.; Zhang, Q.; Zhai, H.; Liu, Q.; He, S. The plasma membrane-localized sucrose transporter IbSWEET10 contributes to the resistance of sweet potato to Fusarium oxysporum. Front. Plant Sci. 2017, 8, 197. [Google Scholar] [CrossRef]
- Seo, P.J.; Park, J.-M.; Ki Kang, S.; Kim, S.-G.; Park, C.-M. An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 2011, 233, 189–200. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, L.; Huang, W.; Yuan, M.; Zhou, F.; Li, X.; Lin, Y. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS ONE 2014, 9, e94210. [Google Scholar] [CrossRef]
- Bezrutczyk, M.; Hartwig, T.; Horschman, M.; Nian Char, S.; Yang, J.; Yang, B.; Frommer, W.B.; Sosso, D. Impaired phloem loading in zmsweet13a,b,c sucrose transporter triple knock-out mutants in Zea mays. New Phytol. 2018, 218, 594–603. [Google Scholar] [CrossRef]
- Jameson, P.E.; Dhandapani, P.; Novak, O.; Song, J. Cytokinins and expression of SWEET, SUT, SWINV and AAP genes increase as pea seeds germinate. Int. J. Mol. Sci. 2016, 17, 2013. [Google Scholar] [CrossRef]
- Guo, Q.; Turnbull, M.H.; Song, J.; Roche, J.; Novak, O.; Späth, J.; Jameson, P.E.; Love, J. Depletion of carbohydrate reserves limits nitrate uptake during early regrowth in Lolium perenne L. J. Exp. Bot. 2017, 68, 1569–1583. [Google Scholar] [CrossRef]
- Gu, J.; Li, Z.; Mao, Y.; Struik, P.C.; Zhang, H.; Liu, L.; Wang, Z.; Yang, J. Roles of nitrogen and cytokinin signals in root and shoot communications in maximizing of plant productivity and their agronomic applications. Plant Sci. 2018, 274, 320–331. [Google Scholar] [CrossRef]
- McKenzie, M.J.; Mett, V.; Reynolds, P.H.S.; Jameson, P.E. Controlled cytokinin production in transgenic tobacco using a copper-inducible promoter. Plant Physiol. 1998, 116, 969–977. [Google Scholar] [CrossRef]
- Gan, S.; Amasino, R.M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science 2014, 270, 1986–1988. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. Translational researches of leaf senescence for enhancing plant productivity and quality. J. Exp. Bot. 2014, 65, 3901–3913. [Google Scholar] [CrossRef]
- Jordi, W.; Schapendonk, A.; Davelaar, E.; Stoopen, G.M.; Pot, C.S.; de Visser, R.; van Rhijn, J.A.; Gan, S.; Amasino, R.M. Increased cytokinin levels in transgenic PSAG12-IPT tobacco plants have large direct and indirect effects on leaf senescence, photosynthesis and N partitioning. Plant Cell Environ. 2000, 23, 279–289. [Google Scholar] [CrossRef]
- Cowan, A.K.; Freeman, M.; Björkman, P.-O.; Nicander, B.; Sitbon, F.; Tillberg, E. Effects of senescence-induced alteration in cytokinin metabolism on source-sink relationships and ontogenic and stress-induced transitions in tobacco. Planta 2005, 221, 801–814. [Google Scholar] [CrossRef]
- Criado, M.V.; Caputo, C.; Roberts, I.N.; Castro, M.A.; Barneix, A.J. Cytokinin-induced changes of nitrogen remobilization and chloroplast ultrastructure in wheat (Triticum aestivum). J. Plant Physiol. 2009, 166, 1775–1785. [Google Scholar] [CrossRef]
- Rosche, E.; Blackmore, D.; Tegeder, M.; Richardson, T.; Schroeder, H.; Higgins, T.J.V.; Frommer, W.B.; Offler, C.E.; Patrick, J.W. Seed-specific overexpression of a potato sucrose transporter increases sucrose uptake and growth rates of developing pea cotyledons. Plant J. 2002, 30, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Rolletschek, H.; Hosein, F.; Miranda, M.; Heim, U.; Götz, K.-P.; Schlereth, A.; Borisjuk, L.; Saalbach, I.; Wobus, U.; Weber, H. Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol. 2005, 137, 1236–1249. [Google Scholar] [CrossRef]
- Tegeder, M. Transporters involved in source to sink partitioning of amino acids and ureides: Opportunities for crop improvement. J. Exp. Bot. 2014, 65, 1865–1878. [Google Scholar] [CrossRef]
- Tegeder, M.; Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2017, 217, 35–53. [Google Scholar] [Green Version]
- Song, J.; Jiang, L.; Jameson, P.E. Expression patterns of Brassica napus genes implicate IPT, CKX, sucrose transporter, cell wall invertase, and amino acid permease gene family members in leaf, flower, silique, and seed development. J. Exp. Bot. 2015, 66, 5067–5082. [Google Scholar]
- Rentsch, D.; Hirner, B.; Schmelzer, E.; Frommer, W.B. Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell 1996, 8, 1437–1446. [Google Scholar] [CrossRef]
- Santiago, J.P.; Tegeder, M. Connecting source with sink: The role of Arabidopsis AAP8 in phloem loading of amino acids. Plant Physiol. 2016, 171, 508–521. [Google Scholar] [CrossRef]
- Tilsner, J.; Kassner, N.; Struck, C.; Lohaus, G. Amino acid contents and transport in oilseed rape (Brassica napus L.) under different nitrogen conditions. Planta 2005, 221, 328–338. [Google Scholar] [CrossRef]
- Liu, Z.; Lv, Y.; Zhang, M.; Liu, Y.; Kong, L.; Zou, M.; Lu, G.C.J.; Yu, X. Identification, expression and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 594. [Google Scholar] [CrossRef]
- Vyroubalová, S.; Václaviková, K.; Turečková, V.; Novák, O.; Šmehilová, M.; Hluska, T.; Ohnoutková, L.; Frébort, I.; Galuszka, P. Characterization of new maize genes putatively involved in cytokinin metabolism and their expression during osmotic stress in relation to cytokinin levels. Plant Physiol. 2009, 151, 433–447. [Google Scholar] [CrossRef]
- Jameson, P.E.; Letham, D.S.; Zhang, R.; Parker, C.W.; Badenoch-Jones, J. Cytokinin translocation and metabolism in lupin species. I. Zeatin riboside introduced into the xylem at the base of Lupinus angustifolius stems. Aust. J. Plant Physiol. 1987, 14, 695–718. [Google Scholar] [CrossRef]
- Hirose, N.; Takei, K.; Huroha, T.; Kamada-Nobusada, T.; Hayashi, H.; Sakakibara, H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 2008, 59, 75–83. [Google Scholar] [CrossRef]
- O’Keefe, D.; Song, J.; Jameson, P.E. Isopentenyl transferase and cytokinin oxidase/dehydrogenase gene family members are differentially expressed during pod and seed development in rapid-cycling brassica. J. Plant Growth Regul. 2011, 30, 92–99. [Google Scholar] [CrossRef]
- Clarke, S.R.; Guy, P.L.; Burritt, D.J.; Jameson, P.E. Changes in the activities of antioxidant enzymes in response to virus infection and hormone treatment. Physiol. Plant 2002, 114, 157–164. [Google Scholar] [CrossRef]
- Zavaleta-Mancera, H.A.; López-Delgado, H.; Loza-Tavera, H.; Mora-Herrera, M.; Trevilla-García, C.; Vargas-Suárez, M.; Ougham, H. Cytokinin promotes catalase and ascorbate peroxidase activities and preserves the chloroplast integrity during dark-senescence. J. Plant Physiol. 2007, 164, 1572–1582. [Google Scholar] [CrossRef]
- Merewitz, E.B.; Du, H.; Yu, W.; Liu, Y.; Gianfagna, T.; Huang, B. Elevated cytokinin content in ipt transgenic creeping bentgrass promotes drought tolerance through regulating metabolite accumulation. J. Exp. Bot. 2012, 63, 1315–1328. [Google Scholar] [CrossRef]
- Huang, J.; Han, B.; Xu, S.; Zhou, M.; Shen, W. Heme oxygenase-1 is involved in the cytokinin-induced alleviation of senescence in detached wheat leaves during dark incubation. J. Plant Physiol. 2011, 168, 768–775. [Google Scholar]
- Xu, Y.; Burgess, P.; Zhang, Z.; Huang, B. Enhancing cytokinin synthesis by overexpressing ipt alleviated drought inhibition of root growth through activating ROS-scavenging systems in Agrostis stolonifera. J. Exp. Bot. 2016, 67, 1979–1992. [Google Scholar] [CrossRef]
- Zimmerman, P.; Zentgraf, U. The correlation between oxidative stress and leaf senescence during plant development. Cell. Mol. Biol. Lett. 2005, 10, 515–534. [Google Scholar]
- Jameson, P.E.; Song, J. Cytokinin: A key driver of seed yield. J. Exp. Bot. 2016, 67, 593–606. [Google Scholar] [CrossRef]
- Ozga, J.A.; Kaur, H.; Savada, R.P.; Reinecke, D.M. Hormonal regulation of reproductive growth under normal and heat-stress conditions in legume and other model crop species. J. Exp. Bot. 2017, 68, 1885–1894. [Google Scholar] [CrossRef]
- Savada, R.P.; Ozga, J.A.; Jayasinghege, C.P.A.; Waduthanthri, K.D.; Reinecke, D.M. Heat stress differentially modifies ethylene biosynthesis and signalling in pea floral and fruit tissues. Plant Mol. Biol. 2017, 95, 313–331. [Google Scholar] [CrossRef]
- Gapper, N.E.; Coupe, S.A.; McKenzie, M.A.; Sinclair, B.K.; Lill, R.E.; Jameson, P.E. Regulation of harvest-induced senescence in broccoli (Brassica oleracea var. italica) by cytokinin, ethylene, and sucrose. J. Plant Growth Regul. 2005, 24, 153–165. [Google Scholar] [CrossRef]
- Song, J.; Jiang, L.; Jameson, P.E. Co-ordinate regulation of gene family members during flag leaf and reproductive development in wheat. BMC Plant Biol. 2012, 12, 78. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucl. Acid Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Evans, T.; Song, J.; Jameson, P.E. Micro-scale chlorophyll analysis and developmental expression of a cytokinin oxidase/dehydrogenase gene during leaf development and senescence. Plant Growth Regul. 2012, 66, 95–99. [Google Scholar] [CrossRef]
- Wellburn, A.R. The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 1994, l44, 307–313. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ninan, A.S.; Grant, J.; Song, J.; Jameson, P.E. Expression of Genes Related to Sugar and Amino Acid Transport and Cytokinin Metabolism during Leaf Development and Senescence in Pisum sativum L. Plants 2019, 8, 76. https://doi.org/10.3390/plants8030076
Ninan AS, Grant J, Song J, Jameson PE. Expression of Genes Related to Sugar and Amino Acid Transport and Cytokinin Metabolism during Leaf Development and Senescence in Pisum sativum L. Plants. 2019; 8(3):76. https://doi.org/10.3390/plants8030076
Chicago/Turabian StyleNinan, Annu S., Jan Grant, Jiancheng Song, and Paula E. Jameson. 2019. "Expression of Genes Related to Sugar and Amino Acid Transport and Cytokinin Metabolism during Leaf Development and Senescence in Pisum sativum L." Plants 8, no. 3: 76. https://doi.org/10.3390/plants8030076
APA StyleNinan, A. S., Grant, J., Song, J., & Jameson, P. E. (2019). Expression of Genes Related to Sugar and Amino Acid Transport and Cytokinin Metabolism during Leaf Development and Senescence in Pisum sativum L. Plants, 8(3), 76. https://doi.org/10.3390/plants8030076




