S-Nitrosoglutathione Reductase—The Master Regulator of Protein S-Nitrosation in Plant NO Signaling
Abstract
1. Introduction
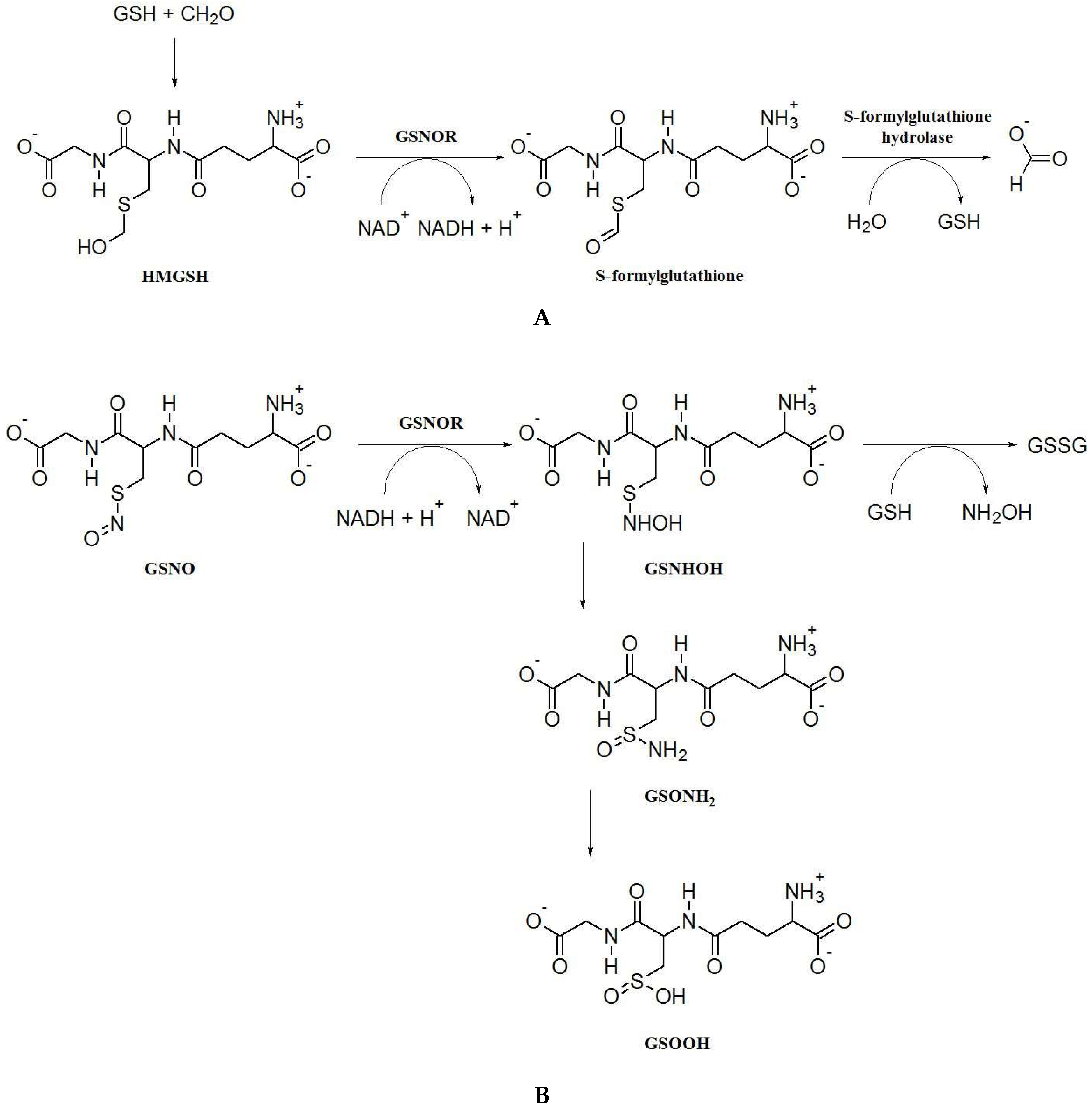
2. S-Nitrosoglutathione Reductase: Key Enzyme of the Regulation of S-Nitrosation and Formaldehyde Detoxification
3. Molecular Properties of S-Nitrosoglutathione Reductase
3.1. GSNOR Structure
3.2. GSNOR Substrate Specificity and Inhibition
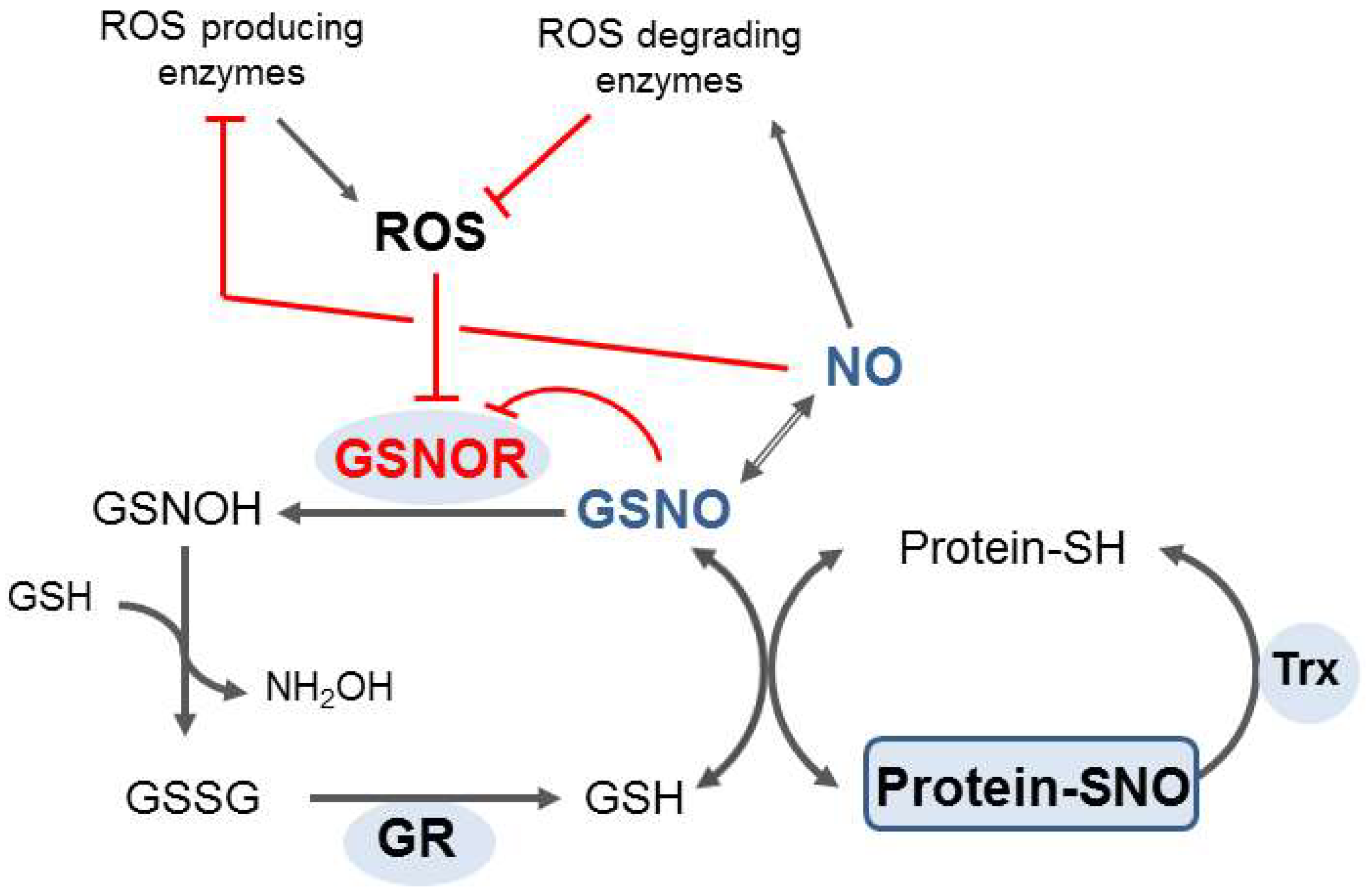
4. The GSNOR Role in Plants
4.1. GSNOR in Plant Growth and Development
4.2. GSNOR in Plant Responses to Abiotic Stress
4.2.1. Mechanical Injury and Wounding
4.2.2. Thermotolerance
4.2.3. Toxic Metals
4.2.4. Soil Salinity and Alkalinity
4.2.5. Other Abiotic Stresses
4.3. GSNOR in Plant Responses to Biotic Stress
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hill, B.G.; Dranka, B.P.; Bailey, S.M.; Lancaster, J.R., Jr.; Darley-Usmar, V.M. What part of NO don’t you under-stand? Some answers to the cardinal questions in nitric oxide biology. J. Biol. Chem. 2010, 285, 19699–19704. [Google Scholar] [CrossRef]
- Groß, F.; Durner, J.; Gaupels, F. Nitric oxide, antioxidants and prooxidants in plant defense responses. Front. Plant Sci. 2013, 4, 419. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Lamattina, L.; Spoel, S.H.; Loake, G.J. Nitric oxide function in plant biology: A redox cue in deconvolution. New Phytol. 2014, 202, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Sakihama, Y.; Nakamura, S.; Yamasaki, H. Nitric oxide production mediated by nitrate reductase in the green alga Chlamydomonas reinhardtii: An alternative NO production pathway in photosynthetic organisms. Plant Cell Physiol. 2002, 43, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.J.; Mandon, J.; Persijn, S.; Cristescu, S.M.; Moshkov, I.E.; Novikova, G.V.; Hall, M.A.; Harren, F.J.M.; Hebelstrup, K.H.; Gupta, K.J. Nitric oxide in plants: An assessment of the current state of knowledge. AoB Plants 2013, 5, pls052. [Google Scholar] [CrossRef]
- Corpas, F.J.; Palma, J.M.; del Río, L.A.; Barroso, J.B. Evidence supporting the existence of L-arginine-dependent nitric oxide synthase activity in plants. New Phytol. 2009, 184, 9–14. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Peroxisomal plant nitric oxide synthase (NOS) protein is imported by peroxisomal targeting signal type 2 (PTS2) in a process that depends on the cytosolic receptor PEX7 and calmodulin. FEBS Lett. 2014, 588, 2049–2054. [Google Scholar] [CrossRef] [PubMed]
- Foresi, N.; Correa-Aragunde, N.; Parisi, G.; Caló, G.; Salerno, G.; Lamattina, L. Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 2010, 22, 3816–3830. [Google Scholar] [CrossRef]
- Jeandroz, S.; Wipf, D.; Stuehr, D.J.; Lamattina, L.; Melkonian, M.; Tian, Z.; Zhu, Y.; Carpenter, E.J.; Wong, G.K.; Wendehenne, D. Occurrence, structure, and evolution of nitric oxide synthase–like proteins in the plant kingdom. Sci. Signal. 2016, 9, re2. [Google Scholar] [CrossRef]
- Gaston, B. Nitric oxide and thiol groups. Biochim. Biophys. Acta 1999, 1411, 323–333. [Google Scholar] [CrossRef]
- Handy, D.E.; Loscalzo, J. Nitric Oxide and Posttranslational Modification of the Vascular Proteome. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- Hess, D.T.; Stamler, J.S. Regulation by S-nitrosylation of Protein Posttranslational Modification. J. Biol. Chem. 2011, 287, 4411–4418. [Google Scholar] [CrossRef]
- Lamotte, O.; Bertoldo, J.B.; Besson-Bard, A.; Rosnoblet, C.; Aimé, S.; Hichami, S.; Terenzi, H.; Wendehenne, D. Protein S-nitrosylation: Specificity and identification strategies in plants. Front. Chem. 2015, 2, 114. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Barroso, J.B. Nitro-oxidative stress vs. oxidative or nitrosative stress in higher plants. New Phytol. 2013, 199, 633–635. [Google Scholar] [CrossRef] [PubMed]
- Frungillo, L.; Skelly, M.J.; Loake, G.J.; Spoel, S.H.; Salgado, I. S-nitrosothiols regulate nitric oxide production and storage in plants through the nitrogen assimilation pathway. Nat. Commun. 2014, 5, 5401. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ruiz, A.; Lamas, S. S-nitrosylation: A potential new paradigm in signal transduction. Cardiovasc. Res. 2004, 62, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Alché, J.D.; Barroso, J.B. Current overview of S-nitrosoglutathione (GSNO) in higher plants. Front. Plant Sci. 2013, 4, 126. [Google Scholar] [CrossRef]
- Liu, L.; Hausladen, A.; Zeng, M.; Que, L.; Heitman, J.; Stamler, J.S. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 2001, 410, 490–494. [Google Scholar] [CrossRef]
- Sakamoto, A.; Ueda, M.; Morikawa, H. Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an S-nitrosoglutathione reductase. FEBS Lett. 2002, 515, 20–24. [Google Scholar] [CrossRef]
- Uotila, L.; Mannervik, B. A steady-state-kinetic model for formaldehyde dehydrogenase from human liver. A mechanism involving NAD+ and the hemimercaptal adduct of glutathione and formaldehyde as substrates and free glutathione as an allosteric activator of the enzyme. Biochem. J. 1979, 177, 869–878. [Google Scholar] [CrossRef]
- Koivusalo, M.; Baumann, M.; Uotila, L. Evidence for the identity of glutathione-dependent formaldehyde dehydrogenase and class III alcohol dehydrogenase. FEBS Lett. 1989, 257, 105–109. [Google Scholar] [CrossRef]
- Jensen, D.; Belka, G.; Du Bois, G. S-nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochem. J. 1998, 331, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Staab, C.A.; Hellgren, M.; Höög, J.O. Medium- and short-chain dehydrogenase/reductase gene and protein families. Cell. Mol. Life Sci. 2008, 65, 3950–3960. [Google Scholar] [CrossRef] [PubMed]
- Feechan, A.; Kwon, E.; Yun, B.W.; Wang, Y.; Pallas, J.A.; Loake, G.J. A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. USA 2005, 102, 8054–8059. [Google Scholar] [CrossRef] [PubMed]
- Staab, C.A.; Ålander, J.; Brandt, M.; Lengqvist, J.; Morgenstern, R.; Grafström, R.C.; Höög, J.O. Reduction of S-nitrosoglutathione by alcohol dehydrogenase 3 is facilitated by substrate alcohols via direct cofactor recycling and leads to GSH-controlled formation of glutathione transferase inhibitors. Biochem. J. 2008, 413, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Staab, C.A.; Ålander, J.; Morgenstern, R.; Grafström, R.C.; Höög, J.O. The Janus face of alcohol dehydrogenase 3. Chem. Biol. Interact. 2009, 178, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Williamson, D.H.; Lund, P.; Krebs, H.A. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J. 1967, 103, 514–527. [Google Scholar] [CrossRef]
- Veech, R.L.; Guynn, R.; Veloso, D. The Time-Course of the Effects of Ethanol on the Redox and Phosphorylation States of Rat Liver. Biochem. J. 1972, 127, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Hanson, A.D.; Gage, D.A.; Shachar-Hill, Y. Plant one-carbon metabolism and its engineering. Trend Plant Sci. 2000, 5, 206–213. [Google Scholar] [CrossRef]
- Espunya, M.C.; Díaz, M.; Moreno-Romero, J.; Martínez, M.C. Modification of intracellular levels of glutathione-dependent formaldehyde dehydrogenase alters glutathione homeostasis and root development. Plant Cell Environ. 2006, 29, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Engeland, K.; Höög, J.O.; Holmquist, B.; Estonius, M.; Jörnvall, H.; Vallee, B.L. Mutation of Arg-115 of human class III alcohol dehydrogenase: A binding site required for formaldehyde dehydrogenase activity and fatty acid activation. Proc. Natl. Acad. Sci. USA 1993, 90, 2491–2494. [Google Scholar] [CrossRef] [PubMed]
- Sanghani, P.C.; Bosron, W.F.; Hurley, T.D. Human Glutathione-Dependent Formaldehyde Dehydrogenase. Structural Changes Associated with Ternary Complex Formation. Biochemistry 2002, 41, 15189–15194. [Google Scholar] [CrossRef] [PubMed]
- Sanghani, P.C.; Robinson, H.; Bosron, W.F.; Hurley, T.D. Human Glutathione-Dependent Formaldehyde Dehydrogenase. Structures of Apo, Binary, and Inhibitory Ternary Complexes. Biochemistry 2002, 41, 10778–10786. [Google Scholar] [CrossRef] [PubMed]
- Kubienová, L.; Kopečný, D.; Tylichová, M.; Briozzo, P.; Skopalová, J.; Šebela, M.; Navrátil, M.; Tâche, R.; Luhová, L.; Barroso, J.B.; et al. Structural and functional characterization of a plant S-nitrosoglutathione reductase from Solanum lycopersicum. Biochimie 2013, 95, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Guerra, D.; Lee, U.; Vierling, E. S-nitrosoglutathione reductases are low-copy number, cysteine-rich proteins in plants that control multiple developmental and defense responses in Arabidopsis. Front. Plant Sci. 2013, 4, 430. [Google Scholar] [CrossRef] [PubMed]
- Sanghani, P.C.; Robinson, H.; Bennett-Lovsey, R.; Hurley, T.D.; Bosron, W.F. Structure–function relationships in human Class III alcohol dehydrogenase (formaldehyde dehydrogenase). Chem. Biol. Interact. 2003, 143–144, 195–200. [Google Scholar] [CrossRef]
- Crotty, J. Crystal Structures and Kinetics of S-Nitrosoglutathione Reductase from Arabidopsis thaliana and Human. Ph.D. Thesis, The University of Arizona, Tuscon, AZ, USA, 2009. [Google Scholar]
- Guerra, D.; Ballard, K.; Truebridge, I.; Vierling, E. S-nitrosation of Conserved Cysteines Modulates Activity and Stability of S-nitrosoglutathione Reductase (GSNOR). Biochemistry 2016, 55, 2452–2464. [Google Scholar] [CrossRef]
- Moulis, J.M.; Holmquist, B.; Vallee, B.L. Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase. Biochemistry 1991, 30, 5743–5749. [Google Scholar] [CrossRef]
- Wagner, F.W.; Burger, A.R.; Vallee, B.L. Kinetic properties of human liver alcohol dehydrogenase: Oxidation of alcohols by class I isoenzymes. Biochemistry 1983, 22, 1857–1863. [Google Scholar] [CrossRef]
- Achkor, H.; Díaz, M.; Fernández, M.R.; Biosca, J.A.; Parés, X.; Martínez, M.C. Enhanced Formaldehyde Detoxification by Overexpression of Glutathione-Dependent Formaldehyde Dehydrogenase from Arabidopsis. Plant Physiol. 2003, 132, 2248–2255. [Google Scholar] [CrossRef]
- Hedberg, J.J.; Griffiths, W.J.; Nilsson, S.J.F.; Höög, J.O. Reduction of S-nitrosoglutathione by human alcohol dehydrogenase 3 is an irreversible reaction as analysed by electrospray mass spectrometry. Eur. J. Biochem. 2003, 270, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Uotila, L.; Koivusalo, M. Purification of formaldehyde and formate dehydrogenases from pea seeds by affinity chromatography and S-formylglutathione as the intermediate of formaldehyde metabolism. Arch. Biochem. Biophys. 1979, 196, 33–45. [Google Scholar] [CrossRef]
- Martínez, M.C.; Achkor, H.; Persson, B.; Fernández, M.R.; Shafqat, J.; Farrés, J.; Jornvall, H.; Parés, X. Arabidopsis Formaldehyde Dehydrogenase. Eur. J. Biochem. 1996, 241, 849–857. [Google Scholar] [PubMed]
- Dolferus, R.; Osterman, J.C.; Peacock, W.J.; Dennis, E.S. Cloning of the Arabidopsis and Rice Formaldehyde Dehydrogenase Genes: Implications for the Origin of Plant ADH Enzymes. Genetics 1997, 146, 1131–1141. [Google Scholar] [PubMed]
- Barroso, J.B.; Corpas, F.J.; Carreras, A.; Rodríguez-Serrano, M.; Esteban, F.J.; Fernández-Ocaña, A.; Chaki, M.; Romero-Puertas, M.C.; Valderrama, R.; Sandalio, L.M.; et al. Localization of S-nitrosoglutathione and expression of S-nitrosoglutathione reductase in pea plants under cadmium stress. J. Exp. Bot. 2006, 57, 1785–1793. [Google Scholar] [CrossRef] [PubMed]
- Chaki, M.; Fernández-Ocaña, A.M.; Valderrama, R.; Carreras, A.; Esteban, F.J.; Luque, F.; Gómez-Rodríguez, M.V.; Begara-Morales, J.C.; Corpas, F.J.; Barroso, J.B. Involvement of reactive nitrogen and oxygen species (RNS and ROS) in sunflower-mildew interaction. Plant Cell Physiol. 2009, 50, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Sun, S.; Wang, C.; Li, Y.; Liang, Y.; An, F.; Li, C.; Dong, H.; Yang, X.; Zhang, J.; et al. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 2009, 19, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- Airaki, M.; Sánchez-Moreno, L.; Leterrier, M.; Barroso, J.B.; Palma, J.M.; Corpas, F.J. Detection and quantification of S-nitrosoglutathione (GSNO) in pepper (Capsicum annuum L.) plant organs by LC-ES/MS. Plant Cell Physiol. 2011, 52, 2006–2015. [Google Scholar] [CrossRef]
- Tichá, T.; Činčalová, L.; Kopečný, D.; Sedlářová, M.; Kopečná, M.; Luhová, L.; Petřivalský, M. Characterization of S-nitrosoglutathione reductase from Brassica and Lactuca spp. and its modulation during plant development. Nitric Oxide 2017, 68, 68–76. [Google Scholar] [CrossRef]
- Lindermayr, C. Crosstalk between reactive oxygen species and nitric oxide in plants: Key role of S-nitrosoglutathione reductase. Free Radic. Biol. Med. 2018, 122, 110–115. [Google Scholar] [CrossRef]
- Barroso, J.B.; Valderrama, R.; Corpas, F.J. Immunolocalization of S-nitrosoglutathione, S-nitrosoglutathione reductase and tyrosine nitration in pea leaf organelles. Acta Physiol. Plant. 2013, 35, 2635–2640. [Google Scholar] [CrossRef]
- Frungillo, L.; de Oliveira, J.F.P.; Saviani, E.E.; Oliveira, H.C.; Martínez, M.C.; Salgado, I. Modulation of mitochondrial activity by S-nitrosoglutathione reductase in Arabidopsis thaliana transgenic cell lines. Biochim. Biophys. Acta 2013, 1827, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Tichá, T.; Lochman, J.; Činčalová, L.; Luhová, L.; Petřivalský, M. Redox regulation of plant S-nitrosoglutathione reductase activity through post-translational modifications of cysteine residues. Biochem. Biophys. Res. Commun. 2017, 494, 27–33. [Google Scholar] [CrossRef]
- Rustérucci, C.; Espunya, M.C.; Díaz, M.; Chabannes, M.; Martínez, M.C. S-nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiol. 2007, 143, 1282–1292. [Google Scholar] [CrossRef] [PubMed]
- Lee, U.; Wie, C.; Fernandez, B.O.; Feelisch, M.; Vierling, E. Modulation of Nitrosative Stress by S-Nitrosoglutathione Reductase Is Critical for Thermotolerance and Plant Growth in Arabidopsis. Plant Cell 2008, 20, 786–802. [Google Scholar] [CrossRef] [PubMed]
- Kwon, E.; Feechan, A.; Yun, B.W.; Hwang, B.H.; Pallas, J.; Kang, J.G.; Loake, G. AtGSNOR1 function is required for multiple developmental programs in Arabidopsis. Planta 2012, 236, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.F.; Wang, D.L.; Wang, C.; Culler, A.H.; Kreiser, M.A.; Suresh, J.; Cohen, J.D.; Pan, J.; Baker, B.; Liu, J.Z. Loss of GSNOR1 Function Leads to Compromised Auxin Signaling and Polar Auxin Transport. Mol. Plant. 2015, 8, 1350–1365. [Google Scholar] [CrossRef]
- Wang, P.; Du, Y.; Hou, Y.J.; Zhao, Y.; Hsu, C.C.; Yuan, F.; Zhu, X.; Tao, W.A.; Song, C.P.; Zhu, J.K. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. USA 2015, 112, 613–618. [Google Scholar] [CrossRef]
- Airaki, M.; Leterrier, M.; Valderrama, R.; Chaki, M.; Begara-Morales, J.C.; Barroso, J.B.; del Río, L.A.; Palma, J.; Corpas, F.J. Spatial and temporal regulation of the metabolism of reactive oxygen and nitrogen species during the early development of pepper (Capsicum annuum) seedlings. Ann. Bot. 2015, 116, 679–693. [Google Scholar] [CrossRef]
- Gong, B.; Wen, D.; Wang, X.; Wei, M.; Yang, F.; Li, Y.; Shi, Q. S-nitrosoglutathione reductase-modulated redox signaling controls sodic alkaline stress responses in Solanum lycopersicum L. Plant Cell Physiol. 2015, 56, 790–802. [Google Scholar] [CrossRef]
- Salgado, I.; Martínez, M.C.; Oliveira, H.; Frungillo, L. Nitric oxide signaling and homeostasis in plants: A focus on nitrate reductase and S-nitrosoglutathione reductase in stress-related responses. Braz. J. Bot. 2013, 36, 89–98. [Google Scholar] [CrossRef]
- Corpas, F.J.; Chaki, M.; Fernández-Ocaña, A.; Valderrama, R.; Palma, J.M.; Carreras, A.; Begara-Morales, J.C.; Airaki, M.; del Río, L.A.; Barroso, J.B. Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol. 2008, 49, 1711–1722. [Google Scholar] [CrossRef] [PubMed]
- Chaki, M.; Valderrama, R.; Fernández-Ocaña, A.M.; Carreras, A.; Gómez-Rodríguez, M.V.; Pedrajas, J.R.; Begara-Morales, J.C.; Sánchez-Calvo, B.; Luque, F.; Leterrier, M.; et al. Mechanical wounding induces a nitrosative stress by downregulation of GSNO reductase and an increase in S-nitrosothiols in sunflower (Helianthus annuus) seedlings. J. Exp. Bot. 2011, 62, 1803–1813. [Google Scholar] [CrossRef] [PubMed]
- Airaki, M.; Leterrier, M.; Mateos, R.M.; Valderrama, R.; Chaki, M.; Barroso, J.B.; del Río, L.A.; Palma, J.M.; Corpas, F.J. Metabolism of reactive oxygen species and reactive nitrogen species in pepper (Capsicum annuum L.) plants under low temperature stress. Plant Cell Environ. 2012, 35, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Ziogas, V.; Tanou, G.; Filippou, P.; Diamantidis, G.; Vasilakakis, M.; Fotopoulos, V.; Molassiotis, A. Nitrosative responses in citrus plants exposed to six abiotic stress conditions. Plant Physiol. Biochem. 2013, 68, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Kubienová, L.; Tichá, T.; Jahnová, J.; Luhová, L.; Mieslerová, B.; Petřivalský, M. Effect of abiotic stress stimuli on S-nitrosoglutathione reductase in plants. Planta 2014, 239, 139–146. [Google Scholar] [CrossRef]
- Espunya, M.C.; De Michele, R.; Gómez-Cadenas, A.; Martínez, M.C. S-nitrosoglutathione is a component of wound- and salicylic acid-induced systemic responses in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 3219–3227. [Google Scholar] [CrossRef]
- Díaz, M.; Achkor, H.; Titarenko, E.; Martínez, M.C. The gene encoding glutathione-dependent formaldehyde dehydrogenase/GSNO reductase is responsive to wounding, jasmonic acid and salicylic acid. FEBS Lett. 2003, 543, 136–139. [Google Scholar] [CrossRef]
- Wünsche, H.; Baldwin, I.T.; Wu, J. S-Nitrosoglutathione reductase (GSNOR) mediates the biosynthesis of jasmonic acid and ethylene induced by feeding of the insect herbivore Manduca sexta and is important for jasmonate-elicited responses in Nicotiana attenuate. J. Exp. Bot. 2011, 62, 4605–4616. [Google Scholar] [CrossRef]
- De Pinto, M.C.; Locato, V.; Sgobba, A.; Romero-Puertas, M.C.; Gadaleta, C.; Delledonne, M.; De Gara, L. S-Nitrosylation of Ascorbate Peroxidase Is Part of Programmed Cell Death Signaling in Tobacco Bright Yellow-2 Cells. Plant Physiol. 2013, 163, 1766–1775. [Google Scholar] [CrossRef]
- Bai, X.G.; Chen, J.H.; Kong, X.X.; Todd, C.D.; Yang, Y.P.; Hu, X.Y.; Li, D.Z. Carbon monoxide enhances the chilling tolerance of recalcitrant Baccaurea ramiflora seeds via nitric oxide-mediated glutathione homeostasis. Free Radic. Biol. Med. 2012, 53, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Sehrawat, A.; Deswal, R. S-nitrosylation analysis in Brassica juncea apoplast highlights the importance of nitric oxide in cold-stress signaling. J. Proteome Res. 2014, 13, 2599–2619. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Chen, J.; Ef, A.A.; Wang, P.; Wang, G.; Hu, X.; Shi, J. Quantitative proteomics analysis reveals that S-nitrosoglutathione reductase (GSNOR) and nitric oxide signaling enhance poplar defense against chilling stress. Planta 2015, 242, 1361–1390. [Google Scholar] [CrossRef] [PubMed]
- Pető, A.; Lehotai, N.; Feigl, G.; Tugyi, N.; Ördög, A.; Gémes, K.; Tari, I.; Erdei, L.; Kolbert, Z. Nitric oxide contributes to copper tolerance by influencing ROS metabolism in Arabidopsis. Plant Cell Rep. 2013, 32, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Leterrier, M.; Airaki, M.; Palma, J.M.; Chaki, M.; Barroso, J.B.; Corpas, F.J. Arsenic triggers the nitric oxide (NO) and S-nitrosoglutathione (GSNO) metabolism in Arabidopsis. Environ. Pollut. 2012, 166, 136–143. [Google Scholar] [CrossRef]
- Lehotai, N.; Kolbert, Z.; Pető, A.; Feigl, G.; Ördög, A.; Kumar, D.; Tari, I.; Erdei, L. Selenite-induced hormonal and signaling mechanisms during root growth of Arabidopsis thaliana L. J. Exp. Bot. 2012, 63, 5677–5687. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tian, D.; Todd, C.D.; Luo, Y.; Hu, X. Comparative Proteome Analyses Reveal that Nitric Oxide Is an Important Signal Molecule in the Response of Rice to Aluminum Toxicity. J. Proteome Res. 2013, 12, 1316–1330. [Google Scholar] [CrossRef]
- Arasimowicz-Jelonek, M.; Floryszak-Wieczorek, J.; Drzewiecka, K.; Chmielowska-Bąk, J.; Abramowski, D.; Izbiańska, K. Aluminum induces cross-resistance of potato to Phytophthora infestans. Planta 2014, 239, 679–694. [Google Scholar] [CrossRef]
- Manai, J.; Gouia, H.; Corpas, F.J. Redox and nitric oxide homeostasis are affected in tomato (Solanum lycopersicum) roots under salinity-induced oxidative stress. J. Plant Physiol. 2014, 171, 1028–1035. [Google Scholar] [CrossRef]
- Tanou, G.; Ziogas, V.; Belghazi, M.; Christou, A.; Filippou, P.; Job, D.; Fotopoulos, V.; Molassiotis, A. Polyamines reprogram oxidative and nitrosative status and the proteome of citrus plants exposed to salinity stress. Plant Cell Environ. 2014, 37, 864–885. [Google Scholar] [CrossRef]
- Zhou, S.; Jia, L.; Chu, H.; Wu, D.; Peng, X.; Liu, X.; Zhang, J.; Zhao, J.; Chen, K.; Zhao, L. Arabidopsis CaM1 and CaM4 Promote Nitric Oxide Production and Salt Resistance by Inhibiting S-nitrosoglutathione Reductase via Direct Binding. PLoS Genet. 2016, 12, e1006255. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tian, D.; Kong, X.; Chen, Q.; Abd-Allah, E.F.; Hu, X.; Jia, A. The role of nitric oxide signaling in response to salt stress in Chlamydomonas reinhardtii. Planta 2016, 244, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, S.; Corpas, F.J.; Borsani, O.; Barroso, J.B.; Monza, J. Water stress induces a differential and spatially distributed nitro-oxidative stress response in roots and leaves of Lotus japonicas. Plant Sci. 2013, 201–202, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yang, L.; Zhao, J.; Wei, J.; Kong, X.; Wang, C.; Zhang, X.; Yang, Y.; Hu, X. Comparative proteomic analysis reveals the role of hydrogen sulfide in the adaptation of the alpine plant Lamiophlomis rotata to altitude gradient in the Northern Tibetan Plateau. Planta 2015, 241, 887–906. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, I.; Holzmeister, C.; Wirtz, M.; Geerlof, A.; Fröhlich, T.; Römling, G.; Kuruthukulangarakoola, G.T.; Linster, E.; Hell, R.; Arnold, G.J.; et al. ROS-Mediated Inhibition of S-nitrosoglutathione Reductase Contributes to the Activation of Anti-oxidative Mechanisms. Front. Plant Sci. 2016, 7, 1669. [Google Scholar] [CrossRef]
- Linh, L.H.; Linh, T.H.; Xuan, T.D.; Ham, L.H.; Ismail, A.M.; Khanh, T.D. Molecular Breeding to Improve Salt Tolerance of Rice (Oryza sativa L.) in the Red River Delta of Vietnam. Int. J. Plant Genom. 2012, 2012, 949038. [Google Scholar] [CrossRef]
- Bai, X.G.; Yang, L.; Tian, M.; Chen, J.; Shi, J.; Yang, Y.; Hu, X. Nitric Oxide Enhances Desiccation Tolerance of Recalcitrant Antiaris toxicaria Seeds via Protein S-Nitrosylation and Carbonylation. PLoS ONE 2011, 6, e20714. [Google Scholar] [CrossRef]
- Malik, S.I.; Hussain, A.; Yun, B.W.; Spoel, S.H.; Loake, G.J. GSNOR-mediated de-nitrosylation in the plant defence response. Plant Sci. 2011, 181, 540–544. [Google Scholar] [CrossRef]
- Janus, Ł.; Milczarek, G.; Arasimowicz-Jelonek, M.; Abramowski, D.; Billert, H.; Floryszak-Wieczorek, J. Normoergic NO-dependent changes, triggered by a SAR inducer in potato, create more potent defense responses to Phytophthora infestans. Plant Sci. 2013, 211, 23–34. [Google Scholar] [CrossRef]
- Thalineau, E.; Truong, H.N.; Berger, A.; Fournier, C.; Boscari, A.; Wendehenne, D.; Jeandroz, S. Cross-Regulation between N Metabolism and Nitric Oxide (NO) Signaling during Plant Immunity. Front. Plant Sci. 2016, 7, 472. [Google Scholar] [CrossRef]
- Kneeshaw, S.; Gelineau, S.; Tada, Y.; Loake, G.J.; Spoel, S.H. Selective protein denitrosylation activity of Thioredoxin-h5 modulates plant Immunity. Mol. Cell 2014, 56, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Ghanta, S.; Bhattacharyya, D.; Sinha, R.; Banerjee, A.; Chattopadhyay, S. Nicotiana tabacum overexpressing γ-ECS exhibits biotic stress tolerance likely through NPR1-dependent salicylic acid-mediated pathway. Planta 2011, 233, 895–910. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, I.; Durner, J.; Lindermayr, C. Crosstalk between nitric oxide and glutathione is required for NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1)-dependent defense signaling in Arabidopsis thaliana. New Phytol. 2015, 208, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Yun, B.W.; Skelly, M.J.; Yin, M.; Yu, M.; Mun, B.G.; Lee, S.U.; Hussain, A.; Spoel, S.H.; Loake, G.J. Nitric oxide and S-nitrosoglutathione function additively during plant immunity. New Phytol. 2016, 211, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Miao, K.; Liu, Y.; Zhao, Y.; Zhang, M.; Pan, S.; Dai, Y. Chemical diversity of biologically active metabolites in the sclerotia of Inonotus obliquus and submerged culture strategies for upregulating their production. Appl. Microbiol. Biotechnol. 2010, 87, 1237–1254. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; He, M.; Ding, J.; Xi, Q.; Loake, G.J.; Zheng, W. Regulation of Anticancer Styrylpyrone Biosynthesis in the Medicinal Mushroom Inonotus obliquus Requires Thioredoxin Mediated Transnitrosylation of S-nitrosoglutathione Reductase. Sci. Rep. 2016, 6, 37601. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jahnová, J.; Luhová, L.; Petřivalský, M. S-Nitrosoglutathione Reductase—The Master Regulator of Protein S-Nitrosation in Plant NO Signaling. Plants 2019, 8, 48. https://doi.org/10.3390/plants8020048
Jahnová J, Luhová L, Petřivalský M. S-Nitrosoglutathione Reductase—The Master Regulator of Protein S-Nitrosation in Plant NO Signaling. Plants. 2019; 8(2):48. https://doi.org/10.3390/plants8020048
Chicago/Turabian StyleJahnová, Jana, Lenka Luhová, and Marek Petřivalský. 2019. "S-Nitrosoglutathione Reductase—The Master Regulator of Protein S-Nitrosation in Plant NO Signaling" Plants 8, no. 2: 48. https://doi.org/10.3390/plants8020048
APA StyleJahnová, J., Luhová, L., & Petřivalský, M. (2019). S-Nitrosoglutathione Reductase—The Master Regulator of Protein S-Nitrosation in Plant NO Signaling. Plants, 8(2), 48. https://doi.org/10.3390/plants8020048





