Abstract
Plant viruses are globally responsible for the significant crop losses of economically important plants. All common approaches are not able to eradicate viral infection. Many non-conventional strategies are currently used to control viral infection, but unfortunately, they are not always effective. Therefore, it is necessary to search for efficient and eco-friendly measures to prevent viral diseases. Since the genomic material of 90% higher plant viruses consists of single-stranded RNA, the best way to target the viral genome is to use ribonucleases (RNase), which can be effective against any viral disease of plants. Here, we show the importance of the search for endophytes with protease and RNase activity combined with the capacity to prime antiviral plant defense responses for their protection against viruses. This review discusses the possible mechanisms used to suppress a viral attack as well as the use of local endophytic bacteria for antiviral control in crops.
1. Introduction
Viruses are obligate intracellular parasites that infect almost every living creature [1], including all cultivated crops. The majority of viruses that infect agricultural plants (at least 450 different species) are RNA viruses [2]. More than 40 of them infect potatoes causing cultivar damage which results in a significant loss in their productivity and deterioration of the tuber quality [3]. It is estimated that about 40% of total crop losses is caused by viral infection. DNA viruses are relatively rare in plants, compared to RNA viruses [4]. The effective viral disease management needs integration of all available strategies, which include avoidance of source of infection, vector control by various means, modification in cultivation practices, and the use of host resistance for viruses and vectors [5].
In the last two decades, RNA interference, a post-transcriptional gene-silencing approach, has been used to induce antiviral responses in plants with the help of genome-editing technologies, such as genetic transformation and the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/Cas9 nuclease) system [6,7,8]. However, antisense mechanisms usually work for the RNA replicating inside the nucleus, like dsDNA-RT (Double-stranded DNA reverse transcriptase)viruses (caulimo- and badna-viruses) [8,9]. For plant protection against RNA viruses, ribozymes that are able to cleave viral RNA can be used [1,2].
Most of the viral diseases are spread by the vectors (insects [3,10], nematodes [3], pathogenic fungi [11,12], soil-born fungi [13], oomycetes [14], etc.), and therefore, they are a good target for preventing viral disease. One of the effective means to control virus vectors is the application of bactericides, insecticides, and fungicides.
Another approach is to develop virus-resistant (transgenic) plants capable of eliminating vectors by producing insect toxins, Cry and Vip proteins from plant-growth promoting microorganism (PGPM) B. thuringiensis, proteinase inhibitors, and antibiotics [9]. In contrast, currently there are no effective chemical means to eliminate viral particles in plants. Chemical antiviral drugs are being actively developed in India [15] and in the Netherlands [16]. The nucleosides’ analogs are potent antiviral agents, which effectively inhibit viral replication [17]. Besides the notable effects of these chemicals, they have some major drawbacks: expensiveness, extreme toxicity, and teratogenic effect on animals and humans [18].
Recently, signaling molecules which trigger the plant defense reactions against viruses have attracted the attention of researchers, since these molecules are less toxic, can be easily utilized by plants, and are destructed in the environment without accumulation of dangerous residues [19]. The application of chitin oligomers and PGPM contributed to a significant reduction of viral infection in plants [20,21,22]. The synthetic analogue of the salicylic acid (SA), benzo-(1,2,3)-thiadiazole-7-carbothioic S-methyl ester (benzothiadiazole), effectively reduced the spread of potato virus Y (PVY) in tomato plants [19,23]. It should be noted that the concentration of antiviral compounds in plants may decrease rapidly. Since viral particles are constantly present on the surface of plant cells, viruses may re-infect plants as soon as the inhibitory factor decreases [24].
Thus, the ability of PGPM to protect plants against pathogens and pests [25,26,27,28] makes them a significant resource for the development of biocontrol agents against plant viruses.
2. Microorganisms as the Means of Biocontrol of Plant Viral Infections
PGPM are beneficial microorganisms present in the rhizosphere that colonize plant roots. PGPM provide the host plant resistance to various biotic and abiotic stresses, in return the host plants provide them shelter and nutrients. Bacteria and fungi can live inside the plant tissues and these endophytic microorganisms form the closest relations with the host plant. In 1988, Clay [25] proposed that the plant pathogens may be inhibited by the endophytes, and the experimental confirmations were obtained by Siegel and Latch in 1991 [26].
Currently, several studies have confirmed that endophytic bacteria are highly effective against various pathogens and pests [27,28]. A systematic representation of different types of insecticidal metabolic products produced by endophytic strains of PGPM against different vectors is shown in Figure 1.
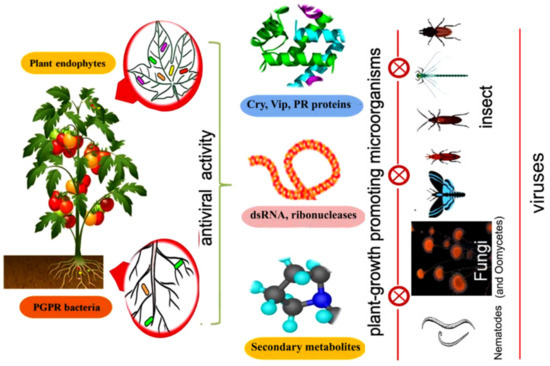
Figure 1.
Effectiveness of different biocidal metabolites produced by plant-growth promoting microorganism (PGPM) for the control of insect vectors and their associated phytopathogens.
Interestingly, Bouizgarne [28] suggested that in some cases, the use of endophytes can be more beneficial than the generation of transgenic plants in terms of crop yield and virus protection. Modeling an artificial plant microbiome with PGPM that produce antiviral compounds and promote the defense potential of plants upon contact with viral particles can also become a promising alternative in the selection for virus tolerance [29].
The biocidal activity of rhizospheric and endophytic PGPM, which produce antibiotics (bacteriocins and/or lipopeptides), suggests that they display the indirect antiviral activity since phytopathogenic bacteria, protozoa, fungi, nematodes, and pests are vectors of a large number of viruses in plants (Figure 2).
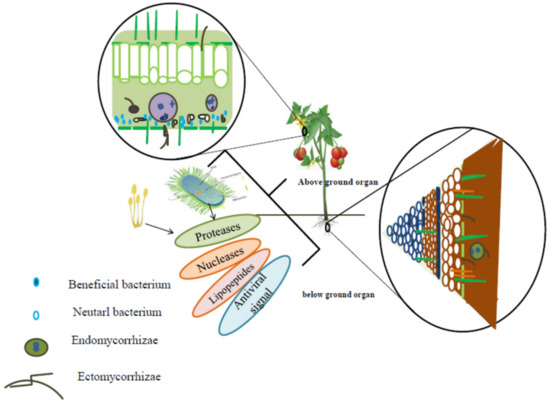
Figure 2.
Location of PGPM in plant tissues and their secondary metabolites, which are of benefit for plants protection against biotic stressors.
Monitoring of world antiviral biocontrol agents market revealed no antiviral bio preparations in the classification of biopesticides or reports on direct acting of antiviral agents of biological origin [30]. The use of insecticidal and other biocidal microorganisms is a promising approach to control viral pest vectors. The agriculture industries are using bio preparations. For example, at the end of 2013, only in China, 132 bio preparations based on Bacillus thuringiensis were registered by 85 companies [31]. The biological preparation “Bitoxibacillin” (Sibbiopharm, Novosibirsk, Russian Federation) based on B. thuringiensis subsp. thuringiensis 98 (BtH1 98) is well known in Russian Federation and the Commonwealth of Independent States [32]. But no data are available on the interactions of these biological agents with plants, and, as we suggest, a low efficiency of these agents observed in some cases arises from the lack of plant/PGPM relations resulting in a negative environmental influence on microorganisms.
The application of bio preparations based on bacteria with fungicidal and insecticidal activities against phytopathogens and insect pests should also help to reduce virus reproduction. Indeed, the treatment of sugar beet with the bacterium Bacillus amylolequifaciens drastically reduced the infection of sugar beet by Phoma betae, which is the vector of the Beet necrotic yellow vein virus (BNYVV) [12]. The application of B. subtilis BS3A25 strain has been found to reduce the CMV infection by inhibiting the development of its vector Aphis gossipi [33]. Colonization of the internal tissues of onion plants with the endophytic fungus Hypocrea lixii (F3ST1) reliably reduced the replication of IYSV as well as the feeding activity of its main vector, Thrips tabaci Lindeman [34].
A large number of studies documented the activity of PGPM against viral infection, virus spread, and reproduction in plants (Table 1). Unfortunately, no reports are available on the interactions of these bacteria with host plants, including their possible penetration into the internal tissues of plants.

Table 1.
Protection of plants by plant growth-promoting microorganisms (PGPM) against viral diseases.
Thus, the soil drench, seed or root treatment, and foliar spraying with PGPM (probably, endophytic, no data) microorganisms contributed to a lesser degree of viral disease symptoms in plants as well as to a reduction of virus concentration (Table 1). This information is very important for PGPM’s practical application. Thus, soil treatment with P. putida A3 prior to sowing reduced TMV infection in tobacco plants more effectively than soil treatment with this PGPM after sowing [51]. The effectiveness of seed treatment is demonstrated in many cases (Table 1).
It should be noted that some strains are able to protect a broad spectrum of plant species, enabling the development of versatile biocontrol agents on the base of these strains. Thus, treatment of plants with P. fluorescens strain CHA0 induced resistance against TNV in tobacco [44] as well as against ULCV in black gram (Vigna mungo) [45] and Musa sp. [20]. Cucumber and tomato seed treatment by Serattia marcescens 90-166 strain in combination with P. putida 89B-61 strain [58] or B. pumilus SE34 strain [59] respectively, induced resistance against CMV, markedly reducing the disease symptoms.
Importantly, combinations of several strains can be more effective against viruses than individual strains. Thus, the dual PGPM combinations, each including strain Bacillus subtilis GB03 and one of the following strains: SE34 (B. pumilus), IN937a (B. amyloliquefaciens), IN937b (B. subtilis), INR7 (B. pumilus), or T4 (B. pumilus), effectively protected tomato plants against CMV [57]. Plant growth-promoting microbial consortium, including Bacillus licheniformis MML2501 + Bacillus sp. MML2551 + Pseudomonas aeruginosa MML2212 + Streptomyces fradiae MML1042, reduced the damage to sunflower plants caused by sunflower necrosis virus disease (SNVD), much stronger than did individually tested strains [60]. Moreover, the addition of Streptomyces sp. PM5 and Trichothecium roseum MML005 to this microbial consortium enhanced its defense effect [60]. Treatment of papaya and tomato seeds with PGPM mixture, consisting of B. amyloliquefaciens IN937a, B. pumilus SE34, and B. pumilus T4, contributed to the subsequent protection of papaya and tomato plants from Papaya ringspot virus (PRSV-W) and tomato chlorotic spot virus (TCSV), respectively [61].
Individual strains can be used in combination with ecologically friendly compounds, which are effective against plant diseases, such as well-studied chitin and chitosan. The combination of chitin oligomers and Pseudomonas fluorescens CHA0 allowed for inducing systemic resistance against BBTV in banana plants [20]. Similarly, seed treatment with PGPM solution containing Bacillus polymixa and Pseudomonas fluorescens mixed with chitosan reduced SqMV infection in cucumber plants [22], while the application of Pseudomonas sp. (206 (4) + B-15 + JK-16) in combination with chitosan enhanced the protection of tomato plants against ToLCV [21].
We suggest that the ability of PGPM to act jointly in consortiums is an opportunity for the development of effective and diversified microorganisms containing antiviral products for plant protection.
4. Signal Pathways and Mechanisms of Plant Resistance to Viruses Induced by Microorganisms
4.1. Virus Recognition and Systemic Resistance in Plants
The plants themselves have quite effective defense mechanisms that prevent viral infection and virus spread. For instance, there are two main types of virus resistance in Solanaceae: extreme resistance and localized hypersensitivity [3]. Extreme resistance provides high resistance to all strains of the virus while localized hypersensitivity is strain-specific. It is necessary to note that Solanum tuberosum, the most important cultivated Solanaceae, have no defense genes against the most harmful PVX and PVY.
Hypersensitive response (HR) is characterized by necrosis and disruption of the virus systemic spread in plants. In potato plants, HR in response to strains PVYC and PVYO of PVY is controlled by potato Nytbr and Nctbr genes, respectively [3,78].
It was shown that the avirulence factor of the PVY virus is the helper component proteinase (HC-Pro) cistron of PVY, while Nx-mediated hypersensitivity and Rx-mediated extreme resistance were elicited by different subunits of coat protein (CP) of PVX [79]. CP of a virus as well as viral RNA, the so-called pathogen-associated molecular patterns (PAMPs), are recognized by plant cell receptors, leading to the development of defense responses in plants [80], including rapid generation of reactive oxygen species (ROS), changes in the content of phytohormones, synthesis of other metabolites, as well as the induction of local and systemic expression of defense genes [81]. This mechanism does not appear to be associated with the RNA interference, which is also an important strategy to protect plants against RNA-containing viruses [82].
When analyzing the protective effect against viral infection, it should be considered that the substances produced by PGPM may interact with the host’s immune system, thereby inducing specific responses in plants. For instance, endophytic bacteria themselves induce the systemic resistance in plants against pathogens [83,84], i.e., they trigger plant defense responses as weak pathogens [85]. This is likely due to the fact that various PAMPs, including flagellin and lipopeptides of endophytic bacteria [39] or CP of viruses [86,87], are recognized by receptors containing leucine-rich repeats (LRR) [88]. Genes encoding pathogenesis related (PR) proteins PR-4 and PR-10 with antiviral activity, including RNase activity, are known to be expressed in plants under the influence of rhizobacteria and their metabolites [56,89], as well as in response to viral [89] and fungal [90] infections. Thus, PGPM can prime plant reactions to viral infection.
4.2. Plant-Growth Promoting Microorganism (PGPM) and Regulation of Plant Defense Mechanisms Against Viruses
Pro-/antioxidant enzymes and phenolpropanoid metabolism enzymes were shown to be involved in plant defence reactions induced by PGRB and their metabolites. Thus, treatment of banana plants by the mixture of rhizospheric P. fluorescens Pf1 and endophytic Bacillus spp. EPB22 resulted in the activation of peroxidase, polyphenol oxidase, and phenylalanine ammonia lyase (PAL), as well as the accumulation of phenolic compounds, which contributes to multiple decreases in BBTV incidence with a final efficiency of up to 80% [62]. Similar changes were observed in BBTV-infected banana [20], ULCV-infected black gram [45], and TSWV-infected tomato plants [91].
According to modern concepts, the plant defense response against pathogens and pests with different lifestyles is regulated by the balance of jasmonic acid (JA)- and salicylic acid (SA)-mediated signaling pathways. In most of the studies, PGPM are characterized as microorganisms that activate resistance to a wide variety of herbivores and necrotrophic pathogens by the JA-dependent signaling pathway, leading to the development of induced systemic resistance (ISR). At the same time, SA induces biochemical processes in plants, leading to resistance against biotrophic pathogens, viruses [1,3], and pests (hemiptera and aphids) [92]. The ability of the B. subtilis BS3A25 isolate to reduce the melon aphid Aphis gossipi (CMV vector) population [33] may be due to aphicidal activity of bacteria-produced surfactants [92,93]. Surfactin of B. subtilis BMG02 increased the resistance of tomato plants to tomato mosaic virus (ToMV) by triggering rapid H2O2 generation and the expression of salicylate-sensitive genes encoding PR-2 protein and PAL, the latter participating in SA biosynthesis [94].
Maurhofer et al. [46] showed that the ability of P. fluorescens CHA0 to protect tobacco plants from TMV is associated with the systemic accumulation of SA in plants as well as with the accumulation of PR-1a, PR-1b, and PR-1c proteins. It can be assumed that the ability of Pseudomonas spp. to protect plants against viral infection is due to SA-induced systemic resistance and is associated with the local generation of ROS in the infection zone. However, the use of bacterial mutants with disrupted production of SA and pseudobactin allowed to show that production of these metabolites by P. fluorescens WCS374r are not required for eliciting Induced systemic resistance (ISR) in Arabidopsis against P. siringae [95], suggesting a fundamental role of JA in this process.
Bacillus spp. associated with tobacco plants induced the development of systemic resistance against TMV by inhibiting the synthesis of CP and enhancing the expression of genes encoding JA- and SA-signaling pathways proteins, Coil and NPR1, defense proteins PR-1a and PR-1b, and cell-wall expansins NtEXP2 and NtEXP6 [39]. The Rhodopseudomonas palustris GJ-22 strain, capable of producing Indolil-acetic acid and 5-aminolevulinic acid, reduced TMV incidence in tobacco plants in the field conditions. Genes of both salicylate-(NbPR1a and NbPR5) and jasmonate-mediated (NbPR3 and NbPDF1.2) signaling pathways were activated after the treatment with this strain [54]. This is somewhat contrary to the data showing that the treatment of tomato with the B. amyloliquefaciens MBI600 strain induced plant resistance to TSWV and PVY accompanied by gene expression of only the SA-induced signaling pathway [24].
Meanwhile, Ryu et al. [55] have revealed that the application of the S. marcescens 90-166 strain to Arabidopsis plants induced resistance against CMV independently of SA, but dependent on JA. It was revealed that the regulatory activity of B. amyloliquefaciens FZB42 [96] and B. cereus AR156 [97] is associated with their ability to inhibit the mechanism of RNA interference of the suppressor genes of the JA defence pathway involving micro RNAs, miR846 and miR825/miR825*, respectively. Nazari et al. [98] revealed the upregulation of miRNAs, nta-miR167 and nta-miR393, and the accumulation of flavanoid compounds in tobacco plants inoculated with B. subtilis ATCC21332 and subsequently infected by Agrobacterium tumefaciens IBRCM10701. The authors suggested that the expression of these miRNAs as well as the accumulation of flavonoid derivatives may be used as markers to assess the efficiency of the PGPM defense effect [98].
The treatment of pepper plants with B. amyloliquefaciens 5B6 reduced the CMV incidence in the field conditions [56] associated with the induction of transcription of genes encoding PR-4, PR-5, and PR-10 proteins. For instance, in hot pepper Capsicum annuum, bacterial derived 2,3-butanediol has been shown to develop a defense response to CMV and TMV, evidenced by the accumulation of transcripts of various defense marker genes, such as Capsicum annuum pathogenesis-related 4 (CaPR4), Ca chitinase 2 (CaChi2), Ca phenylalanine-I ammonia-lyase (CaPAL), CaSAR8.2, Ca 1-aminocyclopropane-1-carboxylic acid oxidase (CaACC), and Ca proteinase inhibitor 2 (CaPIN2), which was similar to the increase in expression of those genes in the benzothiadiazole-treated plants [99].
Beris et al. [24] explained the development of PGRB-induced tomato resistance to the spotted wilt virus and PVY by simultaneous expression of defense genes predominantly of SA-, and to a lesser degree, of JA-signaling cascades. Thus, it should be noted that the treatment of plants with benzothiadiazole, which is used as a reference in many studies of plant resistance to viruses, despite a decrease in the incidence of viral particles in plants, in some cases, suppressed growth and reduced the weight of plants during viral infection compared with bacterial cultures [54,55]. As it was shown by Kumar and co-workers [49], soil application of Paenibacillus lentimorbus B-30488 enhanced the resistance of tobacco plants to CMV while it maintained photosynthetic activity and plant growth. At the same time, the activity of antioxidant enzymes decreased, expression of genes encoding pathogen-induced proteins increased, and polyphenols accumulated, which subsequently prevented the virus spread through plant tissues [49]. Consequently, continued research is needed to develop new approaches to enhance the efficiency of PGPM for improving plant immune potential.
5. Endophytic PGPM as Vectors of RNA Insecticides: Future Approaches
RNA interference (RNAi), which acts at the transcriptional level through RNA-directed DNA methylation and at the post-transcriptional level is mediated by Dicer-like RNase III and small interfering RNA (siRNA), which recognize and inactivate viral RNA, may be used to obtain viral-resistant plants [100]. Zhan and colleagues [101] showed that potato lines, which express CRISPR/Cas13a constructs containing small guide RNA (sgRNA) against coding regions of PVY, were distinguished by lower amounts of virus particles in the tissues and, respectively, by lowered disease symptoms.
Besides the potent effectiveness against virus infection, for example, of tomatoes against the DNA-containing tomato yellow leaf curl virus (TYLCV), expression of the RNAi transgenes affected the host plant transcriptome, resulting in slight phenotypic and developmental abnormalities of the transgenic plants [102]. The bio-insecticides which work on RNA interference are the best way to control the plant virus spreading pest [103,104]. In 2016, Whitten et al. [105] considered that it is necessary to use RNA interference for plant protection against insects, viruses, and fungal phytopathogens. However, it is too tedious and impractical to design such an oversize construct (comprising the defense genes against insects, viruses, and fungi), to integrate it into the plant, preserve it in the plant cells, achieve its biosafety, and subsequently develop the “super resistance” in the target plants. Interestingly, there are new means to deliver “RNA insecticides” in pests’ organisms for their elimination with the help of highly specific microsymbionts exclusively from certain insect species [106]. Monsanto announced the launch of a first insecticide based on RNA silencing technology in the 2020s [107]. This scheme is fundamentally different from plant protection with the use of genetic modifications; however, it requires methods to transfer “RNA insecticide”, “RNA fungicide”, or “RNA viricide” into the plant as well as to protect RNA molecules from sunlight and rain washout [106]. It is likely that endophytic bacteria may also be used as delivery vectors known to successfully colonize plant tissues and form protective biofilms in the apoplast.
6. Conclusions
Currently, a large body of data has accumulated on the positive effect of bacteria and their metabolites on the enhancement of plant defense against viral infection. At the same time, it should be noted that there are different mechanisms of plant defense against viral infection induced by rhizospheric, endophytic, and symbiotic bacteria, as well as their metabolites. PGPM-induced plant defense against common pests, fungal, and bacterial pathogens [85] play an important role in preventing the transmission of viral infections. Several studies have suggested the possibility of using bacterial as well as plant RNase to protect plants from pathogenic viruses. Thus, the identification of the biological properties and the role of PGPM (in particular, endophytic strains) in plant microbiome with the aim of developing biological products with comprehensive activities (antiviral, insecticidal, fungicidal, bactericidal, immune, and growth-promoting), which will be environmentally safe products for plant protection against diseases and pests, is a promising approach of plant defense against viruses (Figure 3). In order to develop a complex multifunctional biological product of a triple action (insecticide + fungicide + viricide), it is important to investigate the plant signaling pathways induced after the influence of these preparations. Thus, it is necessary to study the crosstalk between signaling pathways involved in the development of resistance induced by endophytic bacteria with different biological activities that will eventually contribute to the development of preparations against a wide range of pathogens and herbivores.
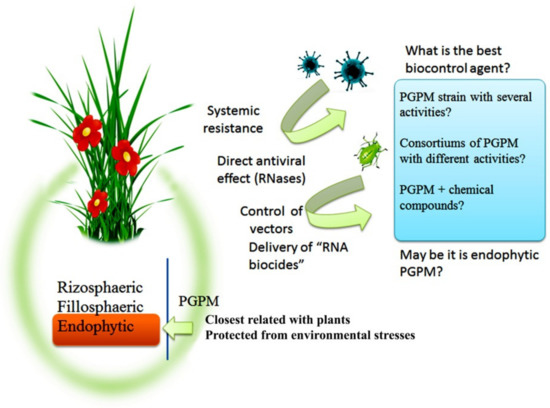
Figure 3.
Effects of PGPM that may promote the development of plant protection to viral diseases and prospects of PGPM use.
PGPM can influence the virus spread by direct antiviral effects of RNase-producing microorganisms or systemic resistance-inducing microorganisms, which live on the surfaces and/or in the internal tissues of plants. These microorganisms can indirectly decrease viral load in agroecosystems by the control of vectors, in particular, by “RNA biocides” specific for pests. PGPM can be “useful” in various combinations for the development of biocontrol agents that will combine direct and indirect activities with close relations with host plants.
Author Contributions
I.V.M. wrote the original draft of the manuscript. A.V.S., G.F.B., S.V.V., V.Y.A., and M.Y.S. made substantial contributions to draft the manuscript and critically revised the final version, P.D.D., G.T.M., and R.M.K. made intellectual collaboration in drafting the manuscript. A.M.A. and B.P.S. made a contribution in editing the text and references. All authors have read and agree to the published version of the manuscript.
Funding
The work was carried out by a joint international grant from the Russian Science Foundation, project number 19-46-02004 and the Department of Science and Technology (DST) of the Government of India, project number C/1756/IFD/2019-20.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design, collection of data, and its interpretation; in the writing of the manuscript; or in the decision to publish the manuscript.
References
- Kunh, J.H. Classify viruses—the gaint is worth the pain. Nature 2019, 566, 318–320. [Google Scholar]
- Soosaar, J.L.; Burch-Smith, T.M.; Dinesh-Kumar, S.P. Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 2005, 3, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Makarova, S.S.; Makarov, V.V.; Taliansky, M.E.; Kalinina, N.O. Resistance to viruses of potato: Current status and prospects. Vavilov J. Genet. Breed. 2017, 21, 62–73. [Google Scholar] [CrossRef][Green Version]
- Borah, B.K.; Sharma, S.; Kant, R.; Johnson, A.M.A.; Venkata, D.; Saigopal, R.; Dasgupta, I. Bacilliform DNA-containing plant viruses in the tropics: Commonalities within a genetically diverse group. Mol. Plant Pathol. 2013, 14, 759–771. [Google Scholar] [CrossRef]
- Sanford, J.C.; Johnston, S.A. The concept of parasite-derived resistance–deriving reistance genes from the parasite’s own genome. J. Theor. Biol. 1985, 113, 395–405. [Google Scholar] [CrossRef]
- Prins, M.; Laimer, M.; Noris, E.; Schubert, J.; Wassenegger, M.; Tepfer, M. Strategies for anti-viral resistance in transgenic plants. Mol. Plant Pathol. 2008, 9, 73–83. [Google Scholar]
- Romay, G.; Bragard, C. Antiviral Defenses in Plants through Genome Editing. Front. Microbiol. 2017, 8, 47. [Google Scholar] [CrossRef]
- Arif, M.; Azhar, U.; Arshad, M.; Zafar, Y.; Mansoor, S.; Asad, S. Engineering broad-spectrum resistance against RNA viruses in potato. Transgenic Res. 2012, 21, 303–311. [Google Scholar] [CrossRef]
- Chung, B.N.; Yoon, J.Y.; Palukaitis, P. Engineered resistance in potato against potato leafroll virus, potato virus A and potato virus Y. Virus Genes. 2013, 47, 86–92. [Google Scholar] [CrossRef]
- Matthews, R.E.F. Plant Virology, 2nd ed.; Academic Press: Cambridge, MA, USA, 1981. [Google Scholar]
- Ali, X.X.; Keldish, M.A.; Pomazkov, U.I. A new carrier of potato virus X is the mushroom Phytophthora infestans (Mont.) De Bary. Agron. Livest. 2010, 3, 18–23. [Google Scholar]
- Desoignies, N.; Schramme, F.; Ongena, M.; Legrève, A. Systemic resistance induced by Bacillus lipopeptides in Beta vulgaris reduces infection by the rhizomania disease vector Polymyxa betae. Mol. Plant Pathol. 2013, 14, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Andika, I.B.; Wei, S.; Cao, C.; Salaipeth, L.; Kondo, H.; Sun, L. Phytopathogenic fungus hosts a plant virus: A naturally occurring cross-kingdom viral infection. Proc. Natl. Acad. Sci. USA 2017, 114, 12267–12272. [Google Scholar] [CrossRef] [PubMed]
- Mascia, T.; Labarile, R.; Doohan, F.; Gallitelli, D. Tobacco mosaic virus infection triggers an RNAi-based response in Phytophthora infestans. Sci. Rep. 2019, 9, 2657. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://www.redoxagro.com/viricide-3639616.html (accessed on 4 December 2019).
- Available online: https://onzelivre.nl/disease-management.php (accessed on 4 December 2019).
- De Fazio, G.; Caner, J.; Vicente, M. Effect of virazole (ribavirin) on tomato spotted wilt virus in two systemic hosts, tomato and tobacco. Arch. Virol. 1980, 63, 305–309. [Google Scholar] [CrossRef]
- Kondakova, V.G.; Schuster, G. Elimination of Strawberry Mottle Virus and Strawberry Crinkle Virus from Isolated Apices of Three Strawberry Varieties by the Addition of 2,4-Dioxohexahydro-1,3,5-triazine (5-Azadihydrouracil) to the Nutrient Medium. J. Phytopathol. 1991, 132, 84–86. [Google Scholar] [CrossRef]
- Palukaitis, P.; Yoon, J.-Y.; Choi, S.-K.; Carr, J.P. Manipulation of induced resistance to viruses. Curr. Opin. Virol. 2017, 26, 141–148. [Google Scholar] [CrossRef]
- Kavino, M.; Harish, S.; Kumar, N.; Saravanakumar, D.; Samiyappan, R. Induction of systemic resistance in banana (Musa spp.) against Banana bunchy top virus (BBTV) by combining chitin with root-colonizing Pseudomonas fluorescens strain CHA0. Eur. J. Plant Pathol. 2008, 120, 353–362. [Google Scholar] [CrossRef]
- Mishra, S.; Jagadeesh, K.S.; Krishnaraj, P.U.; Prem, S. Biocontrol of tomato leaf curl virus (ToLCV) in tomato with chitosan supplemented formulations of Pseudomonas sp. Under field conditions. Aust. J. Crop Sci. 2014, 8, 347–355. [Google Scholar]
- Firmansyah, D.; Widodo Hidayat, S.H. Chitosan and Plant Growth Promoting Rhizobacteria Application to Control Squash mosaic virus on Cucumber Plants. Asian J. Plant Pathol. 2017, 11, 148–155. [Google Scholar]
- Yi, H.-S.; Yang, J.W.; Ryu, C.-M. ISR meets SA Routside: Additive action of the endophyte Bacillus pumilus INR7 and the chemical inducer, benzothiadiazole, on induced resistance against bacterial spotin field-grown pepper. Front. Plant Sci. 2013, 4, 122. [Google Scholar] [CrossRef]
- Beris, D.; Theologidis, I.; Skandalis, N.; Vassilakos, N. Bacillus amyloliquefaciens strain MBI600 induces salicylic acid-dependent resistance in tomato plants against Tomato spotted wilt virus and Potato virus Y. Sci. Rep. 2018, 8, 10320. [Google Scholar] [CrossRef] [PubMed]
- Clay, K. Fungal endophytes of grasses: A defensive mutualism between plants and fungi. Ecology 1988, 69, 10–16. [Google Scholar] [CrossRef]
- Siegel, M.R.; Latch, G.C.M. Expression of antifungal activity in agar culture by isolates of grass endophytes. Mycologia 1991, 83, 529–537. [Google Scholar] [CrossRef]
- Le Cocq, K.; Gurr, S.J.; Hirsch, P.R.; Mauchline, T.H. Exploitation of endophytes for sustainable agricultural intensification. Mol. Plant Pathol. 2017, 18, 469–473. [Google Scholar] [CrossRef]
- Bouizgarne, B. Bacteria for Plant Growth Promotion and Disease Management. In Bacteria in Agrobiology: Disease Management; Maheshwari, D.K., Ed.; Chapter 2; Springer: Berlin/Heidelberg, Germany, 2012; pp. 15–46. [Google Scholar]
- Yue, Q.; Miller, C.J.; White, J.F., Jr.; Richardson, M.D. Isolation and characterization of fungal inhibitors from Epichloë festucae. J. Agric. Food Chem. 2000, 48, 4687–4692. [Google Scholar] [CrossRef]
- Saritha, M.; Prasad, N.V.; Tollamadugu, K.V. The Status of Research and Application of Biofertilizers and Biopesticides: Global Scenario. In Recent Developments in Applied Microbiology and Biochemistry; Academic Press: Cambridge, MA, USA, 2019; pp. 195–207. [Google Scholar]
- Available online: https://agrow.agribusinessintelligence.informa.com/-/media/agri/agrow/ag-market-reviews-pdfs/supplements/agrow-biopesticides_2013.pdf (accessed on 05 December 2019).
- Available online: http://en.sibbio.ru/catalog/crop/bitoksibatsilin/ (accessed on 4 December 2019).
- Sudhakar, N.; Thajuddin, N.; Murugesana, K. Plant growth-promoting rhizobacterial mediated protection of tomato in the field against cucumber mosaic virus and its vector Aphis gossypii. Biocontrol Sci. Technol. 2011, 21, 367–386. [Google Scholar] [CrossRef]
- Muvea, A.M.; Subramanian, S.; Maniania, N.K.; Poehling, H.-M.; Ekesi, S.; Meyhöfer, R. Endophytic Colonization of Onions Induces Resistance Against Viruliferous Thrips and Virus Replication. Front. Plant Sci. 2018, 9, 1785. [Google Scholar] [CrossRef]
- Loebenstein, G.; Lovrekovich, L. Interference with tobacco mosaic virus local lesion formation in tobacco by injection heat-killed cells of Pseudomonas syringae. Virology 1966, 30, 587–591. [Google Scholar] [CrossRef]
- Mann, E.W. Inhibition of tobacco mosaic virus by a bacterial extract. Phytopathology 1969, 59, 658–662. [Google Scholar]
- Bergstrom, G.C.; Johnson, M.C.; Kuc, J. Effects of local infection of cucumber by Colletotrichum lagenarium, Pseudomonas lachrymans, or tobacco necrosis virus on systemic resistance to cucumber mosaic virus. Phytopathology 1982, 72, 922–926. [Google Scholar] [CrossRef]
- Vinodkumar, S.; Nakkeeran, S.; Renukadevi, P.; Mohankumar, S. Diversity and antiviral potential of rhizospheric and endophytic Bacillus species and phyto-antiviral principles against tobacco streak virus in cotton. Agric. Ecosyst. Environ. 2018, 267, 42–51. [Google Scholar]
- Wang, S.; Wu, H.; Qiao, J.; Ma, L.; Liu, J.; Xia, Y.; Gao, X. Molecular Mechanism of Plant Growth Promotion and Induced Systemic Resistance to Tobacco Mosaic Virus by Bacillus spp. J. Microbiol. Biotechnol. 2009, 19, 1250–1258. [Google Scholar] [CrossRef] [PubMed]
- Lian, L.; Xie, L.; Zheng, L.; Lin, Q. Induction of systemic resistance in tobacco against Tobacco mosaic virus by Bacillus spp. Biocontrol Sci. Technol. 2011, 21, 281–292. [Google Scholar] [CrossRef]
- Zeyruk, V.N.; Barkalov, A.G.; Tikhonova, L.V.; Bessonov, A.S.; Nazarov, N.M.; Paremsky, I.Y.; Korshunov, A.V.; Chernikov, V.I.; Masyuk, Y.; Marianovskaya, M.V.; et al. Method of Reproduction of Healthy Potato Plants. Patent of the Russian Federation No. 2001123454/13, 23 August 2001. [Google Scholar]
- Shankar, A.C.; Udaya, N.S.; Chandra, N.-R.S.; Kumar, H.B.; Reddy, M.S.; Niranjana, S.R.; Prakash, H.S. Rhizobacteria mediated resistance against the blackeye cowpea mosaic strain of bean common mosaic virus in cowpea (Vigna unguiculata). Pest Manag. Sci. 2009, 65, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.F.; Zehnder, G.W.; Schuster, D.J.; Sikora, E.J.; Polston, J.E.; Kloepper, J.W. Plant Growth-Promoting Rhizobacterial Mediated Protection in Tomato Against Tomato mottle virus. Plant Dis. 2000, 84, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Maurhofer, M.; Hase, C.; Meuwly, P.; Métraux, J.P.; Défago, G. Induction of systemic resistance of tobacco to tobacco necrosis virus by the root-colonizing Pseudomonas fluorescens strain CHA0: Influence of the gacA gene and of pyoverdine production. Phytopathology 1994, 84, 139–146. [Google Scholar] [CrossRef]
- Karthikeyan, G.; Doraisamy, S.; Rabindran, R. Pseudomonas fluorescens mediated systemic resistance against urdbean leaf crinkle virus in blackgram (Vigna mungo). Arch. Phytopathol. Plant Protect. 2009, 42, 201–212. [Google Scholar] [CrossRef]
- Maurhofer, M.; Reimmann, C.; Sacherer, S.P.; Heeb, S.; Haas, D.; Defago, G. Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against Tobacco necrosis virus. Phytopathology 1998, 88, 678–684. [Google Scholar] [CrossRef]
- Damayanti, T.A.; Katerina, T. Protection of hot pepper against multiple infections of viruses by utilizing root colonizing bacteria. J. ISSAAS 2008, 14, 92–100. [Google Scholar]
- Lee, G.; Lee, S.H.; Kim, K.M.; Ryu, C.M. Foliar application of the leaf-colonizing yeast Pseudozyma churashimaensis elicits systemic defense of pepper against bacterial and viral pathogens. Sci. Rep. 2017, 10, 39432. [Google Scholar] [CrossRef]
- Kumar, S.; Chauhan, P.S.; Agrawal, L.; Raj, R.; Srivastava, A.; Gupta, S.; Mishra, S.K.; Yadav, S.; Singh, P.C.; Raj, S.K.; et al. Paenibacillus lentimorbus Inoculation Enhances Tobacco Growth and Extenuates the Virulence of Cucumber mosaic virus. PLoS ONE 2016, 11, e0149980. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://minsemlab.ru/agrobakter/biogran (accessed on 4 December 2019).
- Yang, J.; Guo, C.; Zhai, X.; Shen, L.; Qian, Y.; Wang, F. Inactivation of Tobacco mosaic virus in soil by Pseudomonas putida A3-m strain to prevent virus mosaic disease. Afr. J. Microbiol. Res. 2012, 6, 6300–6307. [Google Scholar] [CrossRef]
- Sharipova, M.; Rockstroh, A.; Balaban, N.; Mardanova, A.; Toymentseva, A.; Tikhonova, A.; Vologin, S.; Stashevsky, Z. Antiviral Effect of Ribonuclease from Bacillus pumilus against phytopathogenic RNA-Viruses. Agric. Sci. 2015, 6, 1357–1366. [Google Scholar]
- Zehnder, G.W.; Yao, C.; Murphy, J.F.; Sikora, E.J.; Kloepper, J.W. Induction of resistance in tomato against Cucumber mosaic cucumovirus by plant growth-promoting rhizobacteria. Biolcontrol 2000, 45, 127–137. [Google Scholar] [CrossRef]
- Su, P.; Tan, X.; Li, C.; Zhang, D.; Cheng, J.; Zhang, S.; Zhou, X.; Yan, Q.; Peng, J.; Zhang, Z.; et al. Photosynthetic bacterium Rhodopseudomonas palustris GJ-22 induces systemic resistance against viruses. Microb. Biotechnol. 2017, 10, 612–624. [Google Scholar] [CrossRef]
- Ryu, C.M.; Murphy, J.F.; Mysore, K.S.; Kloepper, J.W. Plant growth-promoting rhizobacteria systemically protect Arabidopsis thaliana against Cucumber mosaic virus by a salicylic acid and NPR1-independent and jasmonic acid-dependent signaling pathway. Plant J. 2004, 39, 381–392. [Google Scholar] [CrossRef]
- Lee, G.H.; Ryu, C.M. Spraying of Leaf-Colonizing Bacillus amyloliquefaciens Protects Pepper from Cucumber mosaic virus. Plant Dis. 2016, 100, 2099–2105. [Google Scholar] [CrossRef]
- Jetiyanon, K.; Fowler, W.; Kloepper, J.W. Broad-spectrum protection against several pathogens by PGPR mixtures under field conditions in Thailand. Plant Dis. 2003, 87, 1390–1394. [Google Scholar] [CrossRef]
- Raupach, G.S.; Liu, L.; Murphy, J.F.; Tuzun, S.; Kloepper, J.W. Induced systemic resistance in cucumber and tomato against cucumber mosaic cucumovirus using plant growth-promoting rhizobacteria (PGPR). Plant Dis. 1996, 80, 891–894. [Google Scholar] [CrossRef]
- Murphy, J.F.; Reddy, M.S.; Ryu, C.M.; Kloepper, J.W.; Li, R. Rhizobacteria mediated growth promotion of tomato leads to protection against Cucumber mosaic virus. Phytopathology 2003, 93, 1301–1307. [Google Scholar] [CrossRef]
- Srinivasan, K.; Mathivanan, N. Biological control of sunflower necrosis virus disease with powder and liquid formulations of plant growth promoting microbial consortia under field conditions. Biol. Control 2009, 51, 395–402. [Google Scholar] [CrossRef]
- Abdalla, O.A.; Bibi, S.; Zhang, S. Application of plant growth-promoting rhizobacteria to control Papaya ringspot virus and Tomato chlorotic spot virus. Arch. Phytopathol. Plant Prot. 2017, 50, 584–597. [Google Scholar] [CrossRef]
- Harish, S.; Kavino, M.; Kumar, N.; Saravanakumar, D.; Soorianathasundaram, K.; Samiyappan, R. Biohardening with plant growth promoting rhizosphere and endophytic bacteria induces systemic resistance against Banana bunchy top virus. Appl. Soil Ecol. 2008, 39, 187–200. [Google Scholar] [CrossRef]
- Ulyanova, V.; Vershinina, V.; Ilinskaya, O. Barnase and binase: Twins with distinct fates. FEBS J. 2011, 278, 3633–3643. [Google Scholar] [CrossRef]
- Ilinskaya, O.; Ulyanova, V.; Lisevich, I.; Dudkina, E.; Zakharchenko, N.; Kusova, A.; Faizullin, D.; Zuev, Y. The Native Monomer of Bacillus pumilus Ribonuclease Does Not Exist Extracellularly. BioMed Res. Int. 2018, 2018, 4837623. [Google Scholar] [CrossRef]
- Khalaf, E.M.; Raizada, M.N. Bacterial Seed Endophytes of Domesticated Cucurbits Antagonize Fungal and Oomycete Pathogens Including Powdery Mildew. Front Microbiol. 2018, 9, 42. [Google Scholar] [CrossRef]
- Nakamura, A.; Koide, Y.; Miyazaki, H.; Kitamura, A.; Masaki, H.; Beppu, T.; Uozumi, T. Gene cloning and characterization of a novel extracellular ribonuclease of Bacillus subtilis. Eur. J. Biochem. 1992, 209, 121–127. [Google Scholar] [CrossRef]
- Hahnen, E.; Znamenskaya, L.; Koczan, D.; Leshchinskaya, I.; Hobom, G. A novel secreted ribonuclease from Bacillus intermedius: Gene structure and regulatory control. Mol. Gen. Genet. 2000, 263, 571–580. [Google Scholar] [CrossRef]
- Trifonova, E.A.; Ibragimova, S.M.; Volkova, O.A.; Shumny, V.K.; Kochetov, A.V. Ribonuclease activity as a new prospective disease resistance marker in potato. Vavilov J. Genet. Breed. 2018, 22, 987–991. [Google Scholar] [CrossRef]
- Fedorova, A.A.; Azzami, K.; Ryabchikova, E.I.; Spitsyna, Y.E.; Silnikov, V.N.; Ritter, W.; Gross, H.J.; Tautz, J.; Vlassov, V.V.; Beier, H.; et al. Inactivation of a non-enveloped RNA virus by artificial ribonucleases: Honey bees and acute bee paralysis virus as a new experimental model for in vivo antiviral activity assessment. Antivir. Res. 2011, 91, 267–277. [Google Scholar] [CrossRef]
- Zhou, W.W.; Niu, T.G. Purification and some properties of an extracellular ribonuclease withantiviral activity against tobacco mosaic virus from Bacillus cereus. Biotechnol. Lett. 2009, 31, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Trifonova, E.A.; Komarova, M.L.; Leonova, N.S.; Shcherban’, A.B.; Kochetov, A.V.; Malinovskii, V.I.; Shumnyi, V.K. Transgenic Potato (Solanum tuberosum L.) Plants Expressing the Gene of Secretory Nuclease from Serratia marcescens. Dokl. Biochem. Biophys. 2004, 394, 39–41. [Google Scholar] [PubMed]
- Zhirnov, I.V.; Trifonova, E.A.; Romanova, A.V.; Filipenko, E.A.; Kochetov, A.V.; Shumny, V.K.; Sapotsky, M.V.; Malinovsky, V.I. Induced expression of Serratia marcescens ribonuclease III gene in transgenic Nicotiana tabacum L. cv. SR1 tobacco plants. Russ. J. Genet. 2016, 52, 1137–1141. [Google Scholar] [CrossRef]
- Cao, X.; Lu, Y.; Di, D.; Zhang, Z.; Liu, H.; Tian, L.; Zhang, A.; Zhang, Y.; Shi, L.; Guo, B.; et al. Enhanced virus resistance in transgenic maize expressing a dsRNA-specific endoribonuclease gene from E. coli. PLoS ONE 2013, 8, e60829. [Google Scholar] [CrossRef]
- Zhang, L.; French, R.; Langenberg, W.G.; Mitra, A. Accumulation of barley stripe mosaic virus is significantly reduced in transgenic wheat plants expressing a bacterial ribonuclease. Transgenic Res. 2001, 10, 13–19. [Google Scholar] [CrossRef]
- Yang, X.; Niu, L.; Zhang, W.; He, H.; Yang, J.; Xing, G.; Guo, D.; Zhao, Q.; Zhong, X.; Li, H.; et al. Increased multiple virus resistance in transgenic soybean overexpressing the double-strand RNA-specific ribonuclease gene PAC1. Transgenic Res. 2019, 28, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1. [Google Scholar] [CrossRef]
- Pakniat-Jahromy, A.; Behjatnia, S.A.; Dry, I.B.; Izadpanah, K.; Rezaian, M.A. A new strategy for generating geminivirus resistant plants using a DNA betasatellite/split barnase construct. J. Virol. Methods. 2010, 170, 57–66. [Google Scholar] [CrossRef]
- Chikh-Ali, M.; Rowley, J.S.; Kuhl, J.; Gray, S.M.; Karasev, A.V. Evidence of a Monogenic Nature of the Nz Gene Conferring Resistance Against Potato virus Y Strain Z (PVYZ) in Potato. Am. J. Potato Res. 2014, 91, 649. [Google Scholar] [CrossRef]
- Glais, L.; Bellstedt, D.U.; Lacomme, C. Diversity, Characterization and Classification of PVY. In Potato Virus Y: Biodiversity, Pathogenicity, Epidemiology and Management; Lacomme, C., Glais, L., Bellstedt, D., Dupuis, B., Karasev, A., Jacquot, E., Eds.; Springer: Cham, Switzerland, 2017; pp. 43–76. [Google Scholar]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef]
- Zvereva, A.S.; Golyaev, V.; Turco, S.; Gubaeva, E.G.; Rajeswaran, R.; Schepetilnikov, M.V.; Srour, O.; Ryabova, L.A.; Boller, T.; Pooggin, M.M. Viral protein suppresses oxidative burst and salicylic acid dependent autophagy and facilitates bacterial growth on virus-infected plants. New Phytol. 2016, 211, 1020–1034. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.G.; Wang, C.H.; Guo, H.S. Application of RNA silencing to plant disease resistance. Silence 2012, 3, 5. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, D.K.; Johri, B.N. Interactions of Bacillus spp. and plants—with special reference to induced systemic resistance (ISR). Microbiol. Res. 2009, 68, 1754–1759. [Google Scholar] [CrossRef] [PubMed]
- Maksimov, I.V.; Veselova, S.V.; Nuzhnaya, T.V.; Sarvarova, E.R.; Khairullin, R.M. Plant growth-promoting bacteria in the regulation of plant resistance to stress factors. Russ. J. Plant Physiol. 2015, 62, 715–726. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The Chemistry of Plant–Microbe Interactions in the Rhizosphere and the Potential for Metabolomics to Reveal Signaling Related to Defense Priming and Induced Systemic Resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef]
- Zvereva, A.S.; Pooggin, M.M. Silencing and Innate Immunity in Plant Defense against Viral and Non-Viral Pathogens. Viruses 2012, 4, 2578–2597. [Google Scholar] [CrossRef]
- Boris, K.V.; Kochieva, E.Z. NBS-LRR Resistance Genes to Potato Virus X. Biol. Bull. Rev. 2013, 133, 124–132. [Google Scholar] [CrossRef]
- Sorokina, E.V. Toll-like receptors and primary pathogen recognition in infectious and non-infectious cutaneous pathology. Int. J. Immunopathol. Allergol. Infectol. 2012, 2, 6–15. [Google Scholar]
- Guevara-Morato, M.A.; de Lacoba, M.G.; García-Luque, I.; Serra, M.T. Characterization of a pathogenesis-related protein 4 (PR-4) induced in Capsicum chinense L3 plants with dual RNA-ase and DNA-ase activities. J. Exp. Bot. 2010, 61, 3259–3271. [Google Scholar] [CrossRef]
- Bai, S.; Dong, C.; Li, B.; Dai, H. A PR-4 gene identified from Malus domestica is involved in the defense responses against Botryosphaeria dothidea. Plant Physiol. Biochem. 2013, 62, 23–32. [Google Scholar] [CrossRef]
- Kandan, A.; Radja Commare, R.; Nandakumar, R.; Ramiah, M.; Raguchander, T.; Samiyappan, R. Induction of phenylpropanoid metabolism by Pseudomonas fluorescens against Tomato spotted wilt virus in tomato. Folia Microbiol. 2002, 47, 121–129. [Google Scholar] [CrossRef]
- Zarate, S.; Kempema, L.; Walling, L.L. Silverleaf Whitefly Induces Salicylic Acid Defenses and Suppresses Effectual Jasmonic Acid Defenses. Plant Physiol. 2007, 143, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Yun, D.C.; Yang, S.Y.; Kim, Y.C.; Kim, I.S.; Kim, Y.H. Identification of surfactin as an aphicidal metabolite produced by Bacillus amyloliquefaciens G1. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 751–753. [Google Scholar] [CrossRef]
- Hussein, W.; Awad, H.; Fahim, S. Systemic Resistance Induction of Tomato Plants against ToMV Virus by Surfactin Produced from Bacillus subtilis BMG02. Am. J. Microbiol. Res. 2016, 4, 153–158. [Google Scholar]
- Djavaheri, M.; Mercado-Blanco, J.; Versluis, C.; Meyer, J.M.; Loon, L.C.; Bakker, P.A. Iron-regulated metabolites produced by Pseudomonas fluorescens WCS374r are not required for eliciting induced systemic resistance against Pseudomonas syringae pv. tomato in Arabidopsis. Microbiol. Open. 2012, 1, 311–325. [Google Scholar] [CrossRef]
- Xie, S.; Jiang, H.; Ding, T.; Xu, Q.; Chai, W.; Cheng, B. Bacillus amyloliquefaciens FZB42 represses plant miR846 to induce systemic resistance via a jasmonic acid-dependent signalling pathway. Mol. Plant Pathol. 2018, 19, 1612–1623. [Google Scholar] [CrossRef]
- Nazari, F.; Safaie, N.; Soltani, B.M.; Shams-Bakhsh, M.; Sharifi, M. Bacillus subtilis affects miRNAs and flavanoids production in Agrobacterium-Tobacco interaction. Plant Physiol Biochem. 2017, 118, 98–106. [Google Scholar] [CrossRef]
- Niu, D.; Xia, J.; Jiang, C.; Qi, B.; Ling, X.; Lin, S.; Zhang, W.; Guo, J.; Jin, H.; Zhao, H. Bacillus cereus AR156 primes induced systemic resistance by suppressing miR825/825* and activating defense-related genes in Arabidopsis. J. Integr. Plant Biol. 2016, 58, 426–439. [Google Scholar] [CrossRef]
- Kong, H.G.; Shin, T.S.; Kim, T.H.; Ryu, C.-M. Stereoisomers of the Bacterial Volatile Compound 2,3-Butanediol Differently Elicit Systemic Defense Responses of Pepper against Multiple Viruses in the Field. Front. Plant Sci. 2018, 9, 90. [Google Scholar] [CrossRef]
- Pumplin, N.; Voinnet, O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 2013, 11, 745–760. [Google Scholar] [CrossRef]
- Zhan, X.; Zhang, F.; Zhong, Z.; Chen, R.; Wang, Y.; Chang, L.; Bock, R.; Nie, B.; Zhang, J. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 2019, 17, 1814–1822. [Google Scholar] [CrossRef]
- Fuentes, A.; Carlos, N.; Ruiz, Y.; Callard, D.; Sánchez, Y.; Ochagavía, M.E.; Seguin, J.; Malpica-López, N.; Hohn, T.; Lecca, M.R. Field trial and molecular characterization of RNAi-transgenic tomato plants that exhibit resistance to tomato yellow leaf curl geminivirus. Mol. Plant Microbe Interact. 2016, 29, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Huvenne, H.; Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 2010, 56, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Whitten, M.M.A.; Facey, P.D.; Sol, R.D.; Fernández-Martínez, L.T.; Evans, M.C.; Mitchell, J.J.; Bodger, O.G.; Dyson, P.J. Symbiont-mediated RNA interference in insects. Proc. R. Soc. B. 2016, 283, 20160042. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://www.motherjones.com/tom-philpott/2015/08/coming-farm-field-near-you-gene-silencing-pesticides-rna-rnai (accessed on 4 December 2019).
- Available online: http://gogreenpestcontrol.ca/rna-insecticide-could-target-specific-pests (accessed on 4 December 2019).
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).