Predictive Binding Affinity of Plant-Derived Natural Products Towards the Protein Kinase G Enzyme of Mycobacterium tuberculosis (MtPknG)
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Protein Preparation
4.2. Ligand Preparation
4.3. Binding Site Analysis and Prediction
4.4. Grid Box Preparation and Docking Studies
4.5. Protein–Ligand Interactions and Predictive Inhibition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Daniel, T.M. The history of tuberculosis. Respir. Med. 2006, 100, 1862–1870. [Google Scholar] [CrossRef]
- Global Tuberculosis Report 2018. World Health Organisation: Geneva, Switzerland, 2018. Available online: http://www.who.int/tb/publications/global_report/en/ (accessed on 1 May 2019).
- Janssen, S.; Jayachandran, R.; Khathi, L.; Zinsstag, J.; Grobusch, M.P.; Pieters, J. Exploring prospects of novel drugs for tuberculosis. Drug Des. Dev. Ther. 2012, 6, 217–224. [Google Scholar]
- Tiberi, S.; Muñoz-Torrico, M.; Duarte, R.; Dalcolmo, M.; D’Ambrosio, L.; Migliori, G.-B.; Zumla, A. New drugs and perspectives for new anti-tuberculosis regimens. Pulmonology 2018, 24, 86–98. [Google Scholar] [CrossRef]
- Singh, V.; Mizrahi, V. Identification and validation of novel drug targets in Mycobacterium tuberculosis. Drug Discov. Today 2017, 22, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.; Zhang, X. Protein targets for structure-based anti-Mycobacterium tuberculosis drug discovery. Protein Cell 2010, 1, 435–442. [Google Scholar] [CrossRef]
- Mdluli, K.; Kaneko, T.; Upton, A. Tuberculosis drug discovery and emerging targets. Ann. N. Y. Acad. Sci. 2014, 1323, 56–75. [Google Scholar] [CrossRef] [PubMed]
- Baugh, L.; Phan, I.; Begley, D.W.; Clifton, M.C.; Armour, B.; Dranow, D.M.; Taylor, B.M.; Muruthi, M.M.; Abendroth, J.; Fairman, J.W.; et al. Increasing the structural coverage of tuberculosis drug targets. Tuberculosis 2015, 95, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Av-Gay, Y.; Everett, M. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 2000, 8, 238–244. [Google Scholar] [CrossRef]
- Prisic, S.; Husson, R.N. Mycobacterium tuberculosis Serine/Threonine Protein Kinases. Microbiol. Spectr. 2014, 2, 681–708. [Google Scholar] [CrossRef] [PubMed]
- Walburger, A.; Koul, A.; Ferrari, G.; Nguyen, L.; Prescianotto-Baschong, C.; Huygen, K.; Klebl, B.; Thompson, C.; Bacher, G.; Pieters, J. Protein Kinase G from Pathogenic Mycobacteria Promotes Survival Within Macrophages. Science 2004, 304, 1800–1804. [Google Scholar] [CrossRef]
- Sundaramurthy, V.; Pieters, J. Interactions of pathogenic mycobacteria with host macrophages. Microbes Infect. 2007, 9, 1671–1679. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Wong, D.; Zheng, X.; Poirier, V.; Bach, H.; Hmama, Z.; Av-Gay, Y. Protein kinase and phosphatase signaling in Mycobacterium tuberculosis physiology and pathogenesis. Biochim. Biophys. Acta 2010, 1804, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Scherr, N.; Müller, P.; Perisa, D.; Combaluzier, B.; Jenö, P.; Pieters, J. Survival of Pathogenic Mycobacteria in Macrophages Is Mediated through Autophosphorylation of Protein Kinase G. J. Bacteriol. 2009, 191, 4546–4554. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Székely, R.; Waczek, F.; Szabadkai, I.; Németh, G.; Hegymegi-Barakonyi, B.; Erős, D.; Szokol, B.; Pató, J.; Hafenbradl, D.; Satchell, J.; et al. A novel drug discovery concept for tuberculosis: Inhibition of bacterial and host cell signalling. Immunol. Lett. 2008, 116, 225–231. [Google Scholar] [CrossRef]
- Kanehiro, Y.; Tomioka, H.; Pieters, J.; Tatano, Y.; Kim, H.; Iizasa, H.; Yoshiyama, H. Identification of Novel Mycobacterial Inhibitors Against Mycobacterial Protein Kinase, G. Front. Microbiol. 2018, 9, 1517. [Google Scholar] [CrossRef]
- Wehenkel, A.; Bellinzoni, M.; Graña, M.; Duran, R.; Villarino, A.; Fernández, P.; André-Leroux, G.; England, P.; Takiff, H.; Cerveñansky, C.; et al. Mycobacterial Ser/Thr protein kinases and phosphatases: Physiological roles and therapeutic potential. Biochim. Biophys. Acta 2008, 1784, 193–202. [Google Scholar] [CrossRef]
- Bellinzoni, M.; Wehenkel, A.M.; Durán, R.; Alzari, P.M. Novel mechanistic insights into physiological signaling pathways mediated by mycobacterial Ser/Thr protein kinases. Genes Immun. 2019, 20, 383–393. [Google Scholar] [CrossRef]
- Caballero, J.; Morales-Bayuelo, A.; Navarro-Retamal, C. Mycobacterium tuberculosis serine/threonine protein kinases: Structural information for the design of their specific ATP-competitive inhibitors. J. Comput. Mol. Des. 2018, 32, 1315–1336. [Google Scholar] [CrossRef]
- Gil, M.; Lima, A.; Rivera, B.; Rossello, J.; Urdániz, E.; Cascioferro, A.; Carrión, F.; Wehenkel, A.; Bellinzoni, M.; Batthyány, C.; et al. New substrates and interactors of the mycobacterial Serine/Threonine protein kinase PknG identified by a tailored interactomic approach. J. Proteom. 2019, 192, 321–333. [Google Scholar] [CrossRef]
- Khan, M.Z.; Kaur, P.; Nandicoori, V.K. Targeting the messengers: Serine/threonine protein kinases as potential targets for antimycobacterial drug development. IUBMB Life 2018, 70, 889–904. [Google Scholar] [CrossRef]
- Mori, M.; Sammartino, J.C.; Costantino, L.; Gelain, A.; Meneghetti, F.; Villa, S.; Chiarelli, L.R. An Overview on the Potential Antimycobacterial Agents Targeting Serine/Threonine Protein Kinases from Mycobacterium tuberculosis. Curr. Top. Med. Chem. 2019, 19, 646–661. [Google Scholar] [CrossRef] [PubMed]
- Wolff, K.A.; De La Peña, A.H.; Nguyen, H.T.; Pham, T.H.; Amzel, L.M.; Gabelli, S.B.; Nguyen, L. A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development. PLoS Pathog. 2015, 11, 1004839. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, J.; Salehi, B.; Stojanović-Radić, Z.Z.; Fokou, P.V.T.; Sharifi-Rad, M.; Mahady, G.B.; Sharifi-Rad, M.; Masjedi, M.-R.; Lawal, T.O.; Ayatollahi, S.A.; et al. Medicinal plants used in the treatment of tuberculosis-Ethnobotanical and ethnopharmacological approaches. Biotechnol. Adv. 2017. [Google Scholar] [CrossRef]
- Gautam, R.; Saklani, A.; Jachak, S.M. Indian medicinal plants as a source of antimycobacterial agents. J. Ethnopharmacol. 2007, 110, 200–234. [Google Scholar] [CrossRef]
- Newton, S.M.; Lau, C.; Gurcha, S.S.; Besra, G.S.; Wright, C.W. The evaluation of forty-three plant species for in vitro antimycobacterial activities; isolation of active constituents from Psoralea corylifolia and Sanguinaria canadensis. J. Ethnopharmacol. 2002, 79, 57–67. [Google Scholar] [CrossRef]
- Salomon, C.E.; Schmidt, L.E. Natural products as leads for tuberculosis drug development. Curr. Top. Med. Chem. 2012, 12, 735–765. [Google Scholar] [CrossRef]
- Guzman, J.D.; Gupta, A.; Bucar, F.; Gibbons, S.; Bhakta, S. Antimycobacterials from natural sources: Ancient times, antibiotic era and novel scaffolds. Front. Biosci. 2012, 17, 1861–1881. [Google Scholar] [CrossRef]
- Dashti, Y.; Grkovic, T.; Quinn, R.J. Predicting natural product value, an exploration of anti-TB drug space. Nat. Prod. Rep. 2014, 31, 990–998. [Google Scholar] [CrossRef]
- Santhosh, R.S.; Suriyanarayanan, B. Plants: A source for new antimycobacterial drugs. Planta Med. 2014, 80, 9–21. [Google Scholar] [CrossRef]
- Chinsembu, K.C. Tuberculosis and nature’s pharmacy of putative anti-tuberculosis agents. Acta Trop. 2016, 153, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Kolodziej, H. Traditionally used Pelargonium species: Chemistry and biological activity of umckaloabo extracts and their constituents. Curr. Top. Phytochem. 2000, 3, 77–93. [Google Scholar]
- Bladt, S.; Wagner, H. From the Zulu medicine to the European phytomedicine Umckaloabo®. Phytomedicine 2007, 14, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Helmstädter, A. Umckaloabo–Late vindication of a secret remedy. Pharm. Historian 1996, 26, 2–4. [Google Scholar]
- Newsom, S. Stevens’ cure: A secret remedy. J. R. Soc. Med. 2002, 95, 463–467. [Google Scholar]
- Sechehaye, A. The Treatment of Tuberculosis with Umckaloabo (Stevens’ Cure); B. Fraser & Co.: London, UK, 1930. [Google Scholar]
- An English Physician. Tuberculosis, Its Treatment and Cure with the Help of Umckaloabo (Stevens); B. Fraser & Co.: London, UK, 1931. [Google Scholar]
- Brendler, T.; Van Wyk, B.-E. A historical, scientific and commercial perspective on the medicinal use of Pelargonium sidoides (Geraniaceae). J. Ethnopharmacol. 2008, 119, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Sundarrajan, S.; Lulu, S.; Arumugam, M. Computational evaluation of phytocompounds for combating drug resistant tuberculosis by multi-targeted therapy. J. Mol. Model. 2015, 21, 247. [Google Scholar] [CrossRef]
- Appunni, S.; Rajisha, P.; Rubens, M.; Chandana, S.; Singh, H.N.; Swarup, V. Targeting PknB, an eukaryotic-like serine/threonine protein kinase of Mycobacterium tuberculosis with phytomolecules. Comput. Boil. Chem. 2017, 67, 200–204. [Google Scholar] [CrossRef]
- Scherr, N.; Honnappa, S.; Kunz, G.; Mueller, P.; Jayachandran, R.; Winkler, F.; Pieters, J.; Steinmetz, M.O. Structural basis for the specific inhibition of protein kinase G, a virulence factor of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2007, 104, 12151–12156. [Google Scholar] [CrossRef]
- Tiwari, D.; Singh, R.K.; Goswami, K.; Verma, S.K.; Prakash, B.; Nandicoori, V.K. Key Residues in Mycobacterium tuberculosis Protein Kinase G Play a Role in Regulating Kinase Activity and Survival in the Host*. J. Boil. Chem. 2009, 284, 27467–27479. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Tiwari, S.; Srivastava, K.K.; Siddiqi, M.I. Identification of Novel Inhibitors of Mycobacterium tuberculosis PknG Using Pharmacophore Based Virtual Screening, Docking, Molecular Dynamics Simulation, and Their Biological Evaluation. J. Chem. Inf. Model. 2015, 55, 1120–1129. [Google Scholar] [CrossRef] [PubMed]
- Santhi, N.; Aishwarya, S. Insights from the molecular docking of withanolide derivatives to the target protein PknG from Mycobacterium tuberculosis. Bioinformation 2011, 7, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Ma, S.; He, L.; Yuan, P.; She, Z.; Lu, Y. Sclerotiorin inhibits protein kinase G from Mycobacterium tuberculosis and impairs mycobacterial growth in macrophages. Tuberculosis 2017, 103, 37–43. [Google Scholar] [CrossRef] [PubMed]
- García-Sosa, A.T.; Hetényi, C.; Maran, U. Drug efficiency indices for improvement of molecular docking scoring functions. J. Comput. Chem. 2010, 31, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Kayser, O.; Kolodziej, H. Antibacterial Activity of Extracts and Constituents of Pelargonium sidoides and Pelargonium reniforme. Planta Med. 1997, 63, 508–510. [Google Scholar] [CrossRef] [PubMed]
- Mativandlela, S.; Meyer, J.; Hussein, A.; Lall, N. Antitubercular Activity of Compounds Isolated from Pelargonium sidoides. Pharm. Biol. 2007, 45, 645–650. [Google Scholar] [CrossRef]
- Seidel, V.; Taylor, P.W. In vitro activity of extracts and constituents of Pelagonium against rapidly growing mycobacteria. Int. J. Antimicrob. Agents 2004, 23, 613–619. [Google Scholar] [CrossRef]
- Kayser, O.; Kolodziej, H.; Kiderlen, A.F. Immunomodulatory principles of Pelargonium sidoides. Phytother. Res. 2001, 15, 122–126. [Google Scholar] [CrossRef]
- Koch, E.; Lanzendorfer-Goossens, H.; Whon, C. Stimulation of interferon (INF)-b-synthesis and natural killer (NK) cell activity by an aqueous-ethanolic extract from roots of Pelargonium sidoides (Umckaloabo®). Arch. Pharmacol. 2002, 365 (Suppl. 1), 288. [Google Scholar]
- Kolodziej, H. Antimicrobial, Antiviral and Immunomodulatory Activity Studies of Pelargonium sidoides (EPs((R)) 7630) in the Context of Health Promotion. Pharmaceuticals 2011, 4, 1295–1314. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Griffiths, W.; Taylor, P.; Griffiths, W. Components derived fromPelargoniumstimulate macrophage killing of Mycobacteriumspecies. J. Appl. Microbiol. 2009, 106, 1184–1193. [Google Scholar] [CrossRef] [PubMed]
- Akbay, P.; Başaran, A.A.; Ündeger, Ü.; Başaran, N.; Akbay, P. In vitro immunomodulatory activity of flavonoid glycosides from Urtica dioica L. Phytother. Res. 2003, 17, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Pragasam, S.J.; Venkatesan, V.; Rasool, M. Immunomodulatory and anti-inflammatory effect of p-coumaric acid, a common dietary polyphenol on experimental inflammation in rats. Inflammation 2013, 36, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Latté, K.P.; Ferreira, D.; Venkatraman, M.S.; Kolodziej, H. O-Galloyl-C-glycosylflavones from Pelargonium reniforme. Phytochemistry 2002, 59, 419–424. [Google Scholar] [CrossRef]
- Latté, K.P.; Kaloga, M.; Schäfer, A.; Kolodziej, H. An ellagitannin, n-butyl gallate, two aryltetralin lignans, and an unprecedented diterpene ester from Pelargonium reniforme. Phytochemistry 2008, 69, 820–826. [Google Scholar] [CrossRef]
- Hauer, H.; Germer, S.; Elsasser, J.; Ritter, T. Benzopyranones and their sulfate esters from Pelargonium sidoides. Planta Med. 2010, 76, 350–352. [Google Scholar] [CrossRef]
- Kolodziej, H. Fascinating metabolic pools of Pelargonium sidoides and Pelargonium reniforme, traditional and phytomedicinal sources of the herbal medicine Umckaloabo®. Phytomedicine 2007, 14, 9–17. [Google Scholar] [CrossRef]
- Allinger, N.L. Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. J. Am. Chem. Soc. 1977, 99, 8127–8134. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- Halgren, T. New Method for Fast and Accurate Binding-site Identification and Analysis. Chem. Boil. Drug Des. 2007, 69, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A. Identifying and Characterizing Binding Sites and Assessing Druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Lewu, F.; Grierson, D.; Afolayan, A.; Lewu, F. The leaves of Pelargonium sidoides may substitute for its roots in the treatment of bacterial infections. Boil. Conserv. 2006, 128, 582–584. [Google Scholar] [CrossRef]
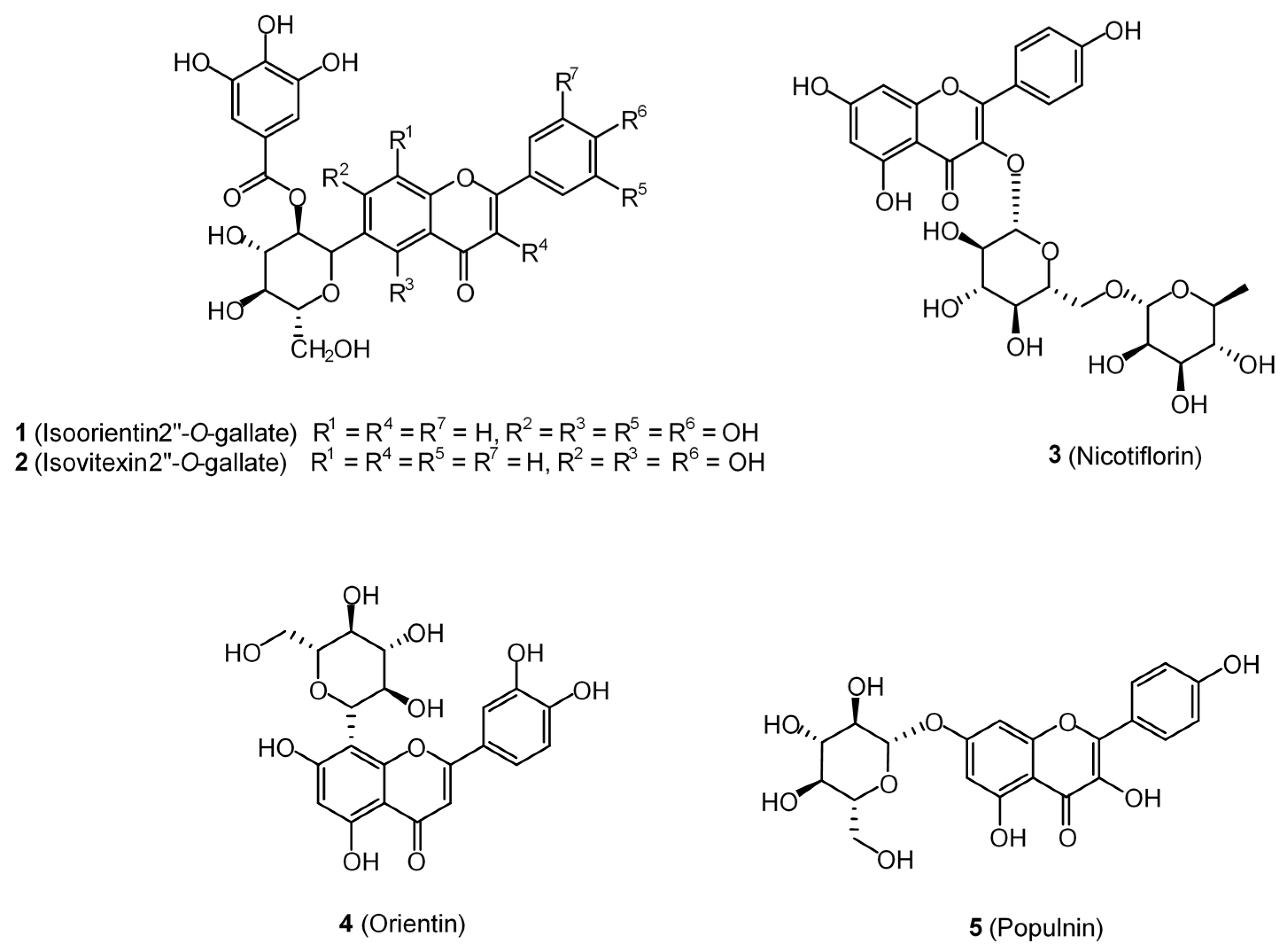
| Binding Site | SiteScore 1 | DScore 2 | Volume (Å) |
|---|---|---|---|
| 1 (AX20017-Co-crystallised site) | 1.138 | 1.174 | 271.31 |
| 2 | 1.027 | 1.034 | 1548.65 |
| 3 | 1.012 | 1.067 | 270.97 |
| 4 | 0.950 | 0.971 | 301.84 |
| 5 | 0.940 | 0.968 | 498.38 |
| Compound | P. reniforme | P. sidoides | Docking Score | Ligand Efficiency Indices | ||
|---|---|---|---|---|---|---|
| LE1 | LE2 | LE3 | ||||
| Isoorientin 2″-O-gallate (1) | AP | AP | −13.2 | 0.31 | 0.47 | 0.02 |
| Isovitexin 2″-O-gallate (2) | AP | −12.6 | 0.30 | 0.45 | 0.02 | |
| Nicotiflorin (3) | AP | −12.2 | 0.29 | 0.45 | 0.02 | |
| Orientin (4) | AP | AP | −11.8 | 0.37 | 0.56 | 0.03 |
| Populnin (5) | AP | −11.6 | 0.36 | 0.55 | 0.03 | |
| Rutin (6) | AP | −11.4 | 0.27 | 0.42 | 0.02 | |
| Vitexin (7) | AP | AP | −11.2 | 0.36 | 0.53 | 0.03 |
| Quercimeritrin (8) | AP | −11.2 | 0.34 | 0.53 | 0.02 | |
| Isoorientin (9) | AP | AP | −11.2 | 0.35 | 0.53 | 0.02 |
| Glucoluteolin (10) | AP | −11.1 | 0.35 | 0.53 | 0.02 | |
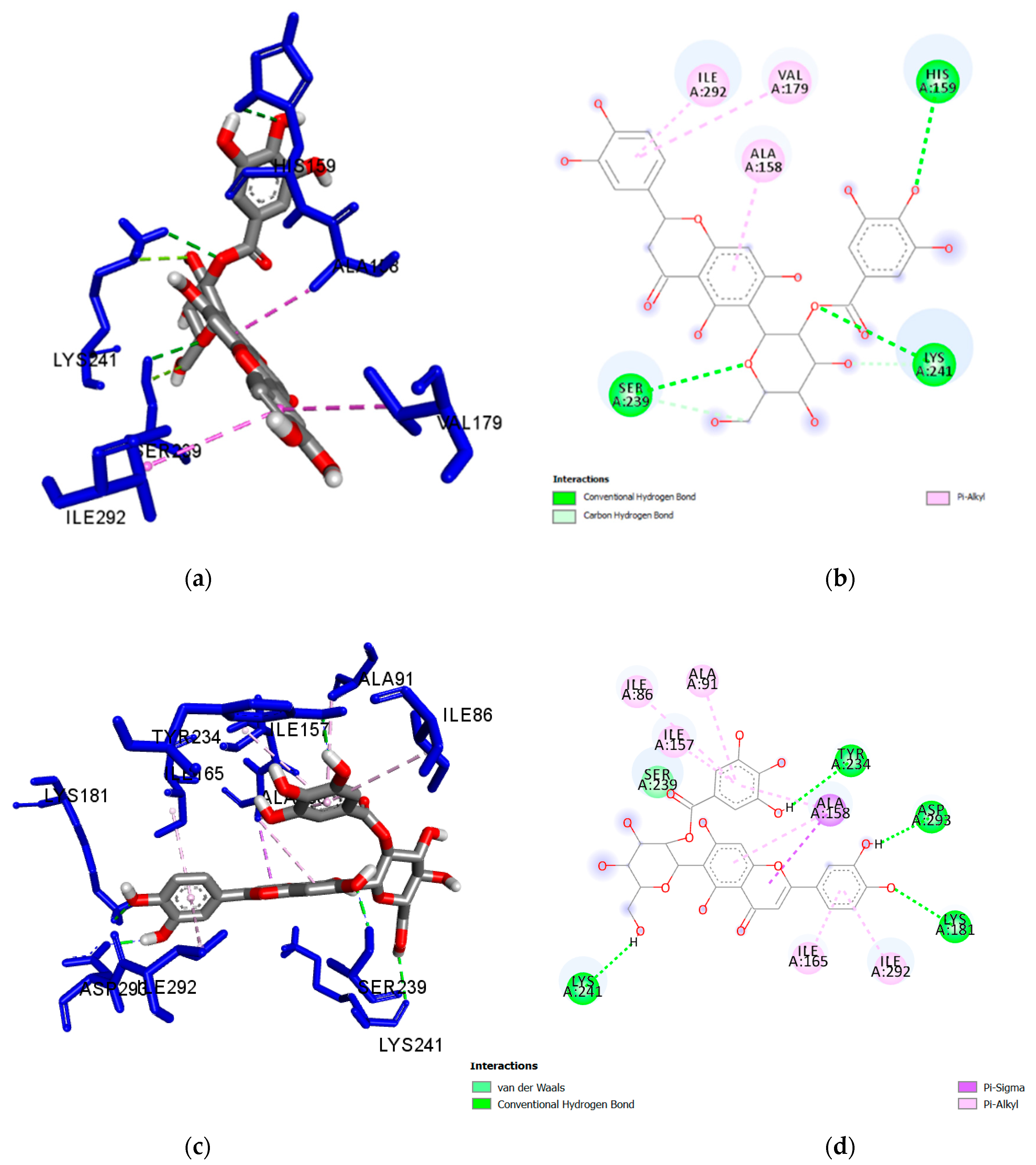
| Ligand | Interacting Residues | Distance (Å) | Category | Type |
|---|---|---|---|---|
| Isoorientin 2″-O-gallate (1) | Lys241 | 2.650 | H-Bond | Conventional |
| Ser239 | 2.825 | H-Bond | Conventional | |
| His159 | 3.063 | H-Bond | Conventional | |
| Lys241 | 3.140 | H-Bond | Carbon Hydrogen Bond | |
| Ser239 | 3.512 | H-Bond | Carbon Hydrogen Bond | |
| Ile292 | 4.701 | Hydrophobic | Pi-Alkyl | |
| Val179 | 4.893 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 4.195 | Hydrophobic | Pi-Alkyl | |
| Isovitexin 2″-O-gallate (2) | Lys241 | 2.168 | H-Bond | Conventional |
| Met232 | 2.903 | H-Bond | Conventional | |
| Ala158 | 3.898 | Hydrophobic | Pi-Sigma | |
| Ile292 | 4.811 | Hydrophobic | Pi-Alkyl | |
| Val235 | 5.002 | Hydrophobic | Pi-Alkyl | |
| Val179 | 4.317 | Hydrophobic | Pi-Alkyl | |
| Nicotiflorin (3) | Glu233 | 2.134 | H-Bond | Conventional |
| Glu280 | 2.286 | H-Bond | Conventional | |
| Gln238 | 2.290 | H-Bond | Conventional | |
| Ser239 | 2.357 | H-Bond | Conventional | |
| Ile86 | 5.025 | Hydrophobic | Alkyl | |
| Ile292 | 3.768 | Hydrophobic | Pi-Sigma | |
| Ile292 | 3.898 | Hydrophobic | Pi-Sigma | |
| Ile157 | 4.605 | Hydrophobic | Pi-Alkyl | |
| Ala91 | 4.608 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 4.846 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 5.218 | Hydrophobic | Pi-Alkyl | |
| Ile165 | 5.290 | Hydrophobic | Pi-Alkyl | |
| Met283 | 5.468 | Hydrophobic | Pi-Alkyl | |
| Val235 | 5.471 | Hydrophobic | Pi-Alkyl | |
| Orientin (4) | Lys181 | 2.248 | H-Bond | Conventional |
| Lys181 | 2.715 | H-Bond | Conventional | |
| Lys181 | 2.669 | H-Bond | Conventional | |
| Asp293 | 2.728 | H-Bond | Conventional | |
| Ala158 | 4.835 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 4.453 | Hydrophobic | Pi-Alkyl | |
| Ile157 | 4.846 | Hydrophobic | Pi-Alkyl | |
| Populnin (5) | Asp293 | 2.213 | H-Bond | Conventional |
| Gln238 | 2.278 | H-Bond | Conventional | |
| Lys181 | 2.498 | H-Bond | Conventional | |
| Gln238 | 3.455 | H-Bond | Carbon Hydrogen Bond | |
| Ile292 | 3.872 | Hydrophobic | Pi-Sigma | |
| Ala158 | 4.526 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 4.714 | Hydrophobic | Pi-Alkyl | |
| Ile165 | 4.817 | Hydrophobic | Pi-Alkyl | |
| Met283 | 5.127 | Hydrophobic | Pi-Alkyl | |
| Ile165 | 5.150 | Hydrophobic | Pi-Alkyl | |
| Ile292 | 5.159 | Hydrophobic | Pi-Alkyl | |
| Ile157 | 5.311 | Hydrophobic | Pi-Alkyl |
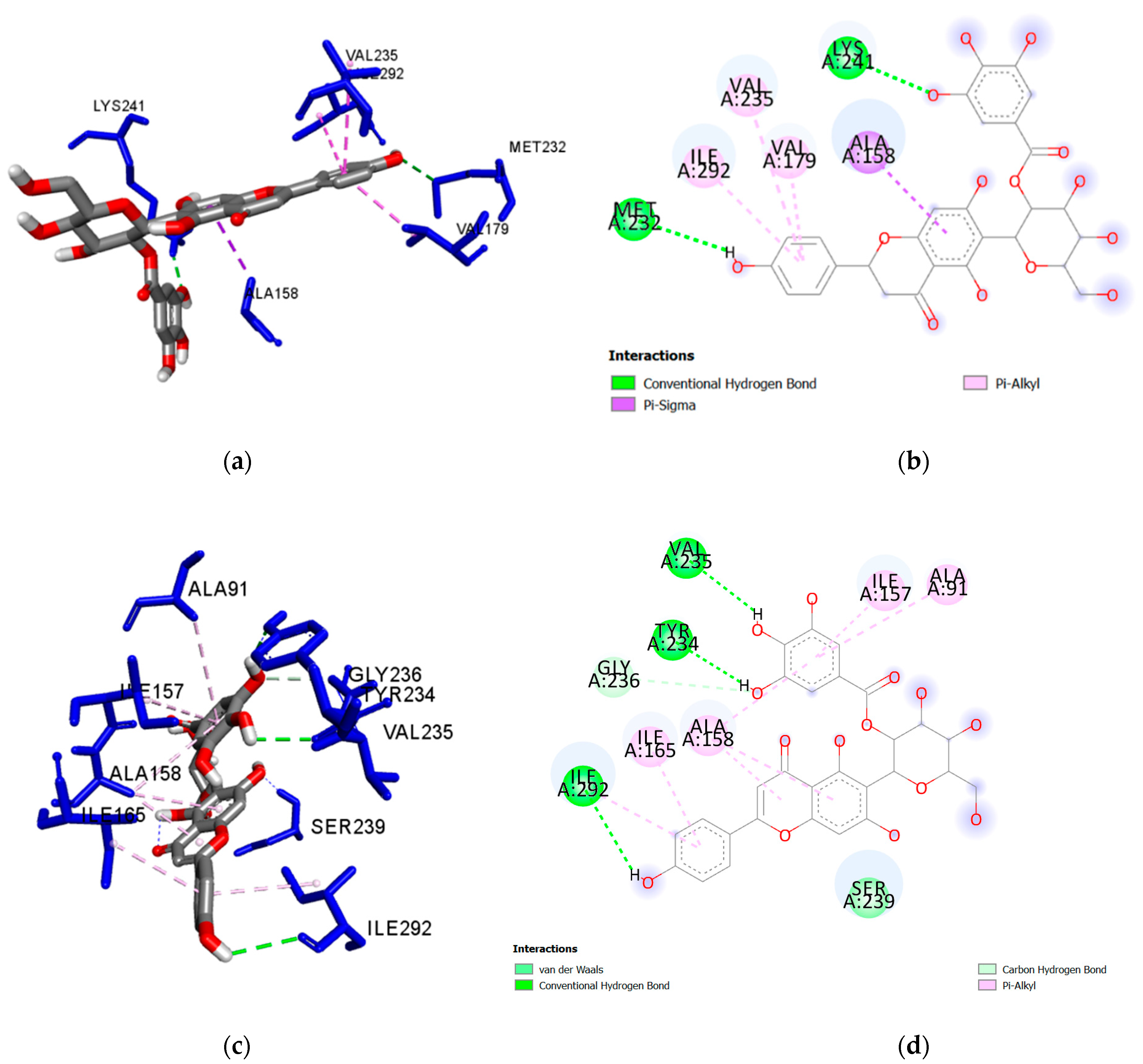
| Ligand | Interacting Residues | Distance (Å) | Category | Type |
|---|---|---|---|---|
| Isoorientin 2″-O-gallate (1) | Lys181 | 2.583 | H-Bond | Conventional |
| Lys241 | 2.657 | H-Bond | Conventional | |
| Ser239 | 2.086 | H-Bond | Conventional | |
| Tyr234 | 2.022 | H-Bond | Conventional | |
| Asp293 | 1.867 | H-Bond | Conventional | |
| Ile 86 | 5.361 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 3.929 | Hydrophobic | Pi-Sigma | |
| Ile292 | 5.263 | Hydrophobic | Pi-Alkyl | |
| Ala91 | 4.738 | Hydrophobic | Pi-Alkyl | |
| Ile165 | 4.592 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 4.984 | Hydrophobic | Pi-Alkyl | |
| Ile157 | 5.154 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 5.213 | Hydrophobic | Pi-Alkyl | |
| Isovitexin 2″-O-gallate (2) | Ser239 | 2.184 | H-Bond | Conventional |
| Tyr234 | 2.241 | H-Bond | Conventional | |
| Val235 | 2.699 | H-Bond | Conventional | |
| Ile292 | 3.044 | H-Bond | Conventional | |
| Gly236 | 3.376 | H-Bond | Carbon Hydrogen Bond | |
| Ala158 | 3.914 | Hydrophobic | Pi-Alkyl | |
| Ile292 | 4.878 | Hydrophobic | Pi-Alkyl | |
| Ile165 | 4.373 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 4.793 | Hydrophobic | Pi-Alkyl | |
| Ala91 | 4.847 | Hydrophobic | Pi-Alkyl | |
| Ile157 | 5.105 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 5.089 | Hydrophobic | Pi-Alkyl | |
| Nicotiflorin (3) | Lys181 | 3.005 | H-Bond | Conventional |
| Ser239 | 2.146 | H-Bond | Conventional | |
| Asn281 | 2.163 | H-Bond | Conventional | |
| Val235 | 2.174 | H-Bond | Conventional | |
| Ile292 | 3.747 | Hydrophobic | Pi-Sigma | |
| Ile86 | 4.966 | Hydrophobic | Alkyl | |
| Val235 | 5.072 | Hydrophobic | Pi-Alkyl | |
| Ile292 | 4.468 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 5.195 | Hydrophobic | Pi-Alkyl | |
| Ile165 | 4.364 | Hydrophobic | Pi-Alkyl | |
| Ile165 | 5.392 | Hydrophobic | Pi-Alkyl | |
| Orientin (4) | Ile157 | 2.477 | H-Bond | Conventional |
| Glu233 | 2.407 | H-Bond | Conventional | |
| Val235 | 2.155 | H-Bond | Conventional | |
| Val235 | 2.423 | H-Bond | Conventional | |
| Gly237 | 2.227 | H-Bond | Conventional | |
| Ser239 | 2.379 | H-Bond | Conventional | |
| Glu280 | 2.411 | H-Bond | Conventional | |
| Ala158 | 3.574 | Hydrophobic | Pi-Sigma | |
| Ala158 | 3.885 | Hydrophobic | Pi-Alkyl | |
| Ile165 | 4.567 | Hydrophobic | Pi-Alkyl | |
| Ile165 | 5.400 | Hydrophobic | Pi-Alkyl | |
| Val179 | 4.437 | Hydrophobic | Pi-Alkyl | |
| Ile292 | 5.460 | Hydrophobic | Pi-Alkyl | |
| Ile292 | 4.538 | Hydrophobic | Pi-Alkyl | |
| Populnin (5) | Gln238 | 2.130 | H-Bond | Conventional |
| Gln238 | 2.443 | H-Bond | Conventional | |
| Ser239 | 2.297 | H-Bond | Conventional | |
| Asn281 | 2.296 | H-Bond | Conventional | |
| Lys181 | 2.699 | H-Bond | Conventional | |
| Lys181 | 2.571 | H-Bond | Conventional | |
| Ala158 | 4.391 | Hydrophobic | Pi-Alkyl | |
| Ile165 | 5.080 | Hydrophobic | Pi-Alkyl | |
| Ile292 | 5.175 | Hydrophobic | Pi-Alkyl | |
| Ile157 | 4.571 | Hydrophobic | Pi-Alkyl | |
| Ala158 | 4.148 | Hydrophobic | Pi-Alkyl | |
| Ile165 | 4.783 | Hydrophobic | Pi-Alkyl | |
| Ile292 | 5.122 | Hydrophobic | Pi-Alkyl |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qasaymeh, R.M.; Rotondo, D.; Oosthuizen, C.B.; Lall, N.; Seidel, V. Predictive Binding Affinity of Plant-Derived Natural Products Towards the Protein Kinase G Enzyme of Mycobacterium tuberculosis (MtPknG). Plants 2019, 8, 477. https://doi.org/10.3390/plants8110477
Qasaymeh RM, Rotondo D, Oosthuizen CB, Lall N, Seidel V. Predictive Binding Affinity of Plant-Derived Natural Products Towards the Protein Kinase G Enzyme of Mycobacterium tuberculosis (MtPknG). Plants. 2019; 8(11):477. https://doi.org/10.3390/plants8110477
Chicago/Turabian StyleQasaymeh, Rana M., Dino Rotondo, Carel B. Oosthuizen, Namrita Lall, and Veronique Seidel. 2019. "Predictive Binding Affinity of Plant-Derived Natural Products Towards the Protein Kinase G Enzyme of Mycobacterium tuberculosis (MtPknG)" Plants 8, no. 11: 477. https://doi.org/10.3390/plants8110477
APA StyleQasaymeh, R. M., Rotondo, D., Oosthuizen, C. B., Lall, N., & Seidel, V. (2019). Predictive Binding Affinity of Plant-Derived Natural Products Towards the Protein Kinase G Enzyme of Mycobacterium tuberculosis (MtPknG). Plants, 8(11), 477. https://doi.org/10.3390/plants8110477







