Abstract
Leaves are initiated as lateral outgrowths from shoot apical meristems throughout the vegetative life of the plant. To achieve proper developmental patterning, cell-type specification and growth must occur in an organized fashion along the proximodistal (base-to-tip), mediolateral (central-to-edge), and adaxial–abaxial (top-bottom) axes of the developing leaf. Early studies of mutants with defects in patterning along multiple leaf axes suggested that patterning must be coordinated across developmental axes. Decades later, we now recognize that a highly complex and interconnected transcriptional network of patterning genes and hormones underlies leaf development. Here, we review the molecular genetic mechanisms by which leaf development is coordinated across leaf axes. Such coordination likely plays an important role in ensuring the reproducible phenotypic outcomes of leaf morphogenesis.
1. Introduction
The morphology of angiosperm leaves is extraordinarily diverse, and is a key component of natural variation in plant architecture. Nonetheless, all leaves share two ontogenetic features in common; they are (1) derived from the shoot apical meristem (SAM) and (2) asymmetrical from their inception [1]. The SAM is a stem cell reservoir at the growing tip(s) of the plant that ultimately supplies cells for all the above-ground organs. In this way, the SAM accounts for the continuous development of vegetative structures far beyond embryogenesis, which comprises a critical, strategic difference between plant and animal development. A leaf transitions from its origins as a small primordium to its mature form through the establishment and maintenance of three developmental axes upon which growth and differentiation proceed (Figure 1A,B) [2,3]. The leaf proximodistal axis (i.e., base-to-tip) is defined by the polarized growth of leaf initials away from the shoot, and later becomes elaborated by the establishment of proximal and distal cell and tissue types. For example, in many eudicot leaves, such as those of Arabidopsis, the leaf is subdivided into the proximal petiole and distal lamina. Leaf development also involves specialization of the upper and lower leaf surfaces, defining an adaxial–abaxial (top-bottom) axis of asymmetry. At its inception, the leaf primordium possesses inherent asymmetry along this axis due to the proximity of the adaxial leaf surface to the SAM relative to the abaxial leaf surface [1,4]. This early asymmetry drives the differentiation of diverging cell and tissue fates in the adaxial and abaxial domains of the leaf. Common anatomical differences between these domains include the formation of adaxial xylem and abaxial phloem, as well as differences in epidermal and mesophyll cell morphology. Finally, leaves grow to form a widened, flattened lamina from a marginal domain at the juxtaposition of the adaxial and abaxial leaf faces, thereby defining the mediolateral axis of the leaf. Development along the mediolateral axis is associated with the positioning of the medial leaf midvein and the proliferative transverse outgrowth of cells to form the leaf lamina, or blade.
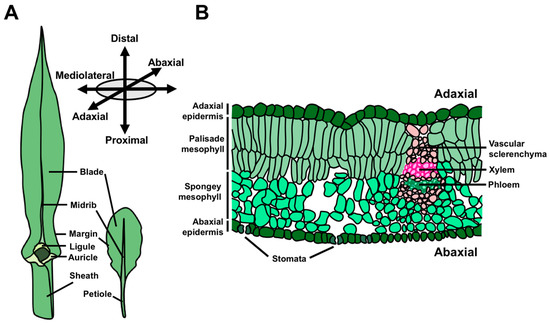
Figure 1.
Leaf anatomy and growth axes. (A) Leaves from maize, a monocot, and the two closely related eudicots, Cardamine hirsuta and Arabidopsis thaliana, highlighting major features of leaf anatomy. (B) Cross-section of a mature eudicot leaf illustrating the characteristic features of the adaxial and abaxial leaf domains.
A host of transcriptional regulators underlie the establishment and maintenance of the leaf developmental axes, often making use of inhibitory interactions to specify opposing cell fates. A classic example of this type of regulatory logic is seen in the context of the conserved antagonistic relationship between Class I KNOTTED-LIKE HOMEOBOX (KNOX), ASYMMETRIC LEAVES1 (AS1), and ASYMMETRIC LEAVES2 (AS2) transcription factors [5,6,7,8,9,10,11,12,13,14]. Class I KNOX genes are expressed in meristematic tissue and promote indeterminate growth, while AS1 and AS2 repress KNOX, an important first step in the transition from indeterminate to determinate cell identity along the proximodistal axis. Inhibitory interactions between adaxial and abaxial patterning regulators are also well-described. Along this axis, the adaxial cell fate-promoting HD-ZIP Class III (HD-ZIP III) transcription factors are repressed by abaxial cell fate-promoting KANADI (KAN) transcription factors and the HD-ZIPIII-targeting miR165/166 microRNAs [15,16,17,18,19,20]. In a parallel pathway, transcripts of the abaxial cell fate-promoting ETTIN/AUXIN RESPONSE FACTOR3 (ETT/ARF3) and ARF4 transcription factors are targeted by small-interfering RNAs (ta-siARFs) expressed in the adaxial domain [21,22,23]. Together, these mutually inhibitory interactions precisely specify and maintain the boundaries between the adaxial and abaxial faces of the leaf.
The reproducible phenotypic outcomes of leaf development may in part be ensured by multiple layers of redundancy built into the underlying genetic patterning network. Not only do multiple pathways act in parallel to regulate the same patterning processes, but gene duplication has expanded the number of redundantly acting pattering genes, potentially allowing for patterning robustness in the face of network perturbation or noise [24,25,26,27]. For example, the Arabidopsis genome contains four copies of the abaxially expressed KAN transcription-factor-encoding genes, which redundantly promote abaxial cell fate [28,29]. While genetic redundancy may allow for stable and predictable developmental outcomes along a given leaf axis, leaves develop simultaneously along multiple axes such that mechanisms controlling three-dimensional development must be synchronized to some extent. In the context of vertebrate limb bud development, evidence has emerged for molecular coordination across organ axes. For example, the transcription factors HAND2 and GLI3R, which promote anterior and posterior cell fate, respectively, have been shown to directly cross-regulate the expression of genes important for proximodistal patterning [30,31,32]. In plants, a model wherein adaxial–abaxial patterning establishes outgrowth of both the mediolateral and proximodistal leaf axes was proposed almost 25 years ago [33]. Here, we review several decades of developmental genetics research that test and extend this classical model of the mechanisms whereby leaves initiate from the SAM and undergo coordinated development along three axes. In general, coordinating factors sit at the top of complex gene regulatory networks, and work to organize and control the expression of well-characterized patterning modules. Their context-specific functions are frequently mediated by physical interaction and tightly regulated feedbacks with other patterning factors, to yield consistent morphological outcomes. Key patterning genes mentioned in this article are listed in Table 1.

Table 1.
Key genes mentioned in the text, their relevant homologs in other species, and the leaf axes they pattern.
2. Coordination of the Proximodistal and Adaxial–Abaxial Leaf Axes
2.1. ASYMMETRIC LEAVES1 and ASYMMETRIC LEAVES2
The Antirrhinum phantastica (phan) mutant was the first mutant described with impaired development along multiple leaf axes. Leaves of phan mutants exhibit a temperature-sensitive loss of adaxial–abaxial polarity, as well as ectopic KNOX gene expression in the leaf, indicating a role for PHAN in coordinating pattern formation along both the adaxial–abaxial and proximodistal axes [33,34]. The subsequent cloning of phan identified a loss-of-function mutation in a MYB-domain transcription factor-encoding gene as responsible for the phenotype [34]. PHAN orthologs are described in maize, Arabidopsis, tobacco, tomato, and pea [10,12,13,35,36,37,38,39]. Despite the high degree of sequence conservation among the PHAN orthologs, the phan phenotype differs across species. In maize, loss-of-function of the PHAN ortholog ROUGH SHEATH2 (RS2) yields leaves with ectopic KNOX expression, resulting in the intrusion of proximal tissue types, such as the auricle and ligule, into distal portions of the leaf blade [40,41,42]. This suggests that KNOX genes may act as proximal patterning factors in maize leaves and is consistent with detection of KN1 protein in the developing leaf base of wild-type leaves [43]. However, unlike phan mutants, leaf adaxial–abaxial polarity is retained in rs2 maize mutants [14]. In contrast, mutations in the PHAN orthologs of eudicots show proximodistal patterning defects, as well as varying degrees of adaxial–abaxial polarity defects, similar to the originally reported phan phenotype in Antirrhinum. In Arabidopsis, loss of function of the PHAN ortholog ASYMMETRIC LEAVES1 (AS1) results in leaves that are asymmetric, shortened along the proximodistal axis, and exhibit intrusion of petiole-like epidermal cells into the leaf lamina [10,44,45]. Similar to maize, these features are accompanied by ectopic Class I KNOX gene expression in the leaf [44,46]. Additionally, subtle defects in adaxial–abaxial polarity are observed [46,47]. More extreme patterning defects are observed in tobacco phan loss-of-function mutants [40]. Leaves lose much of their leaf blades and adaxial–abaxial patterning is perturbed throughout the petiole [48]. Phenotypic severity of phan is reduced under gibberellic acid (GA) treatment, a hormone that promotes differentiation and is biosynthetically inhibited by KNOX activity in undifferentiated tissues [49,50]. Finally, in pea phan mutants, proximal stipules are displaced distally and leaflet adaxial–abaxial polarity is perturbed, whereas in tomato phan mutants, the entire leaf is severely abaxialized [41,42,43]. Despite the differences in the phan phenotypes across taxa, there is a conserved role for PHAN in the downregulation of KNOX gene expression, and, to varying degrees, the regulation of adaxial–abaxial patterning in eudicots. Studies of phan phenotypes, therefore, set the stage for further investigation into cross-axes coordination of development in leaves.
Shortly after the cloning and characterization of AS1 in Arabidopsis, a second mutant was reported with perturbed leaf symmetry, resulting in the formation of highly asymmetric leaf lobes, as well as overgrowth of the adaxial leaf surface [11,51,52]. This mutant, named asymmetric leaves2 (as2), harbors a loss-of-function mutation in a gene encoding a LATERAL ORGAN BOUNDARIES (LOB)-domain transcription factor. AS2 is expressed uniformly throughout early-stage leaf primordia before becoming restricted to the adaxial domain, and similarly to AS1, negatively regulates the expression of Class I KNOX genes. In maize, the AS2 homolog INDETERMINATE GAMETOPHYTE1 (IG1) confers similar patterns of ectopic KNOX expression, as well as adaxial–abaxial polarity defects [53]. Overexpression of AS2 in Arabidopsis leads to leaf adaxialization [54]. Therefore, AS2, like AS1, links patterning along both the proximodistal and adaxial–abaxial leaf axes (Figure 2A). In fact, AS2 interacts physically with AS1, forming a complex that directly regulates the expression of BREVIPEDICELLUS (BP), an Arabidopsis Class I KNOX gene [55]. The repressive activity of the AS1–AS2 complex is mediated by its interaction with repressive chromatin remodeling factors, such as HIRA, in both Arabidopsis and maize [56]. In Arabidopsis, the AS1–AS2 complex can also recruit the POLYCOMB REPRESSIVE COMPLEX2 (PRC2) and LIKE HETEROCHROMATIN PROTEIN1 (LHP1) to repress KNOX gene expression [57,58]. Consistent with the role of the AS1–AS2 complex in the repression of proximal gene expression programs through downregulation of KNOX gene expression, the defects in midrib patterning, ectopic stipules, and leaf lobing—but not the defects in adaxial–abaxial patterning—are reduced by loss of class I KNOX gene function in the as1 as2 mutant background [59]. Treatment with GA also rescues the proximodistal patterning defects seen in the as1 as2 double mutants, consistent with previous work in tobacco [40].
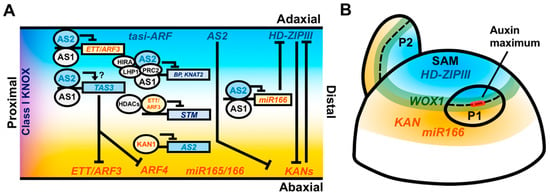
Figure 2.
Coordination of adaxial–abaxial and proximodistal leaf patterning factors. (A) Longitudinal model cross-section of the leaf with adaxial factors labeled in cyan, abaxial factors labeled in orange, and proximal factors labeled in purple. AS2 and HD-ZIPIII transcripts accumulate in the adaxial domain of the leaf, while ETT/ARF3, ARF4, miR165/166, and KAN transcripts accumulate in the abaxial domain. miR165/166 and the mature ta-siARF RNAs are capable of moving intercellularly and repressing HD-ZIPIII and ETT/ARF3 postranscriptionally. HD-ZIPIII and KAN genetically antagonize one another and AS2 can genetically repress KAN. The AS1–AS2 complex recruits the HIRA protein and the PRC2/LHP1 complex to repress expression of Class I KNOX genes and also directly binds and represses the expression of ETT/ARF3 and miR166 in the adaxial portion of the leaf. AS1–AS2 binds TAS3, which encodes the ta-siARF precursor transcript, possibly as a protective mechanism to exclude TAS3-antagonizing transcription factors from accessing their promoter binding sites. The abaxial factor ETT/ARF3 directly represses the expression of the Class I KNOX gene STM by recruiting HDACs. KAN restricts AS2 expression to the adaxial domain via a direct repressive interaction. (B) Radially organized expression domains of patterning factors within the meristem may prepattern the adaxial–abaxial axis of the leaf. HD-ZIPIII genes are expressed centrally while KAN and miR166 are expressed peripherally. WOX1 expression occurs at the juxtaposition of these domains. The auxin response is restricted to the boundary region where auxin promotes outgrowth of the leaf primordium, retaining the prepatterned domains of gene expression during the establishment of the proximodistal leaf axis.
Meanwhile, AS1 and AS2 coordinately regulate adaxial–abaxial patterning by repressing a distinct set of genes promoting abaxial cell fate. AS1–AS2 directly binds the abaxial identity gene ETT/ARF3, and represses its expression in the adaxial domain of the leaf via the recruitment of DNA methylating enzymes (Figure 2A) [60]. AS2 also binds ARF3, along with miR166a and TAS3A, two small-RNA encoding genes implicated in adaxial–abaxial patterning and AS2 is, in turn, repressed in the abaxial domain via direct binding by KAN1 (Figure 2A) [61,62]. AS2 represses KAN1 but it is unknown whether this interaction is direct. Taken together, AS1 and AS2 function in a highly conserved pathway to coordinately repress KNOX gene expression. The role of AS1 in leaf adaxial patterning in eudicots may largely be ascribed to its interaction with AS2, which regulates adaxial–abaxial patterning in both eudicots and monocots. The lack of adaxial–abaxial polarity defects in monocot as1 mutants could, therefore, reflect modifications in AS1–AS2 complex function and/or genetic redundancy (reviewed in [63]). Overall, the AS1–AS2 complex offers an example of developmental coordination across leaf axes through regulation of multiple downstream patterning modules and interactions with a diverse array of regulatory cofactors.
2.2. Auxin Controls Proximodistal and Adaxial–Abaxial Axis Formation
The plant hormone auxin also has key roles in coordinating the development of the proximodistal and mediolateral leaf axes, both downstream and upstream of key patterning factors. Polarized transport of auxin to foci at the flanks of the SAM predicts the site of leaf initiation and is critical for the initiation of the proximodistal leaf axis [64,65]. Auxin accumulation is driven by the polarization of PIN1 (PIN-FORMED1) auxin efflux carriers in the L1 layer of the meristem. Through an as yet undetermined pathway, the accumulation of auxin represses the expression of Class I KNOX gene expression at the site leaf primordium initiation, a necessary first step in proximodistal axis formation [66,67]. Interestingly, auxin application to the SAM is only able to promote primordial outgrowth at the SAM periphery, suggesting that an auxin response competency is required for the initiation of the proximodistal axis. This region of competency correlates with juxtaposed domains of cells expressing the HD-ZIPIII, KAN, and miR166a adaxial–abaxial patterning genes (Figure 2B) [4,5,68,69]. The expression of these genes within the SAM may reflect a prepattern that organizes the location of auxin-induced proximodistal outgrowth while simultaneously specifying the adaxial and abaxial domains of the initiating primordium. The homeobox transcription factor WUSCHEL (WUS), which is expressed in the SAM central zone and functions to maintain a stable population of meristematic stem cells, directly represses the expression of KAN1/2, suggesting a mechanism by which this prepattern is maintained [70]. How these adaxial–abaxial patterning factors mediate the restriction of auxin responsiveness is currently unknown. Nevertheless, it necessitates the coordination of proximodistal outgrowth with a predefined adaxial–abaxial prepattern, which likely contributes to stable patterning outcomes during leaf initiation.
Auxin may also play a role in coordinating the development of the proximodistal and adaxial–abaxial axes beyond the leaf initiation stage. High levels of auxin signaling are associated with the distal tips of the leaf and the developing vasculature [66,71] Meanwhile, treatment of tomato leaf primordia with exogenous auxin or the disruption of auxin transport and signaling is capable of abaxializing the leaf, suggesting that auxin functions to maintain the abaxial leaf domain [72]. However, contradictory evidence suggests a role for auxin in adaxial cell fate acquisition, as auxin promotes the expression of the HD-ZIPIII gene REV [68]. In fact, more recent analysis revealed no overall differences in auxin perception between the adaxial and abaxial leaf surfaces, arguing against a role for differential auxin accumulation along the adaxial–abaxial leaf axis [73]. Nonetheless, the observation of auxin-induced leaf abaxialization remains unexplained [72]. It is possible that polarized expression of ARFs, which mediate the transcriptional response to auxin, may control the differential effects of auxin within the abaxial and adaxial leaf domains downstream of auxin perception [21,74]. These genes may also serve as integrators of auxin-regulated adaxial–abaxial and proximodistal patterning in the leaf, since ETT/ARF3 directly binds and represses STM via the recruitment of histone deacetylases (Figure 2A) [75]. Further dissection of the role of ARFs in mediating the outcomes of auxin signaling during leaf development is clearly warranted and may reveal their activity in coordinating morphogenesis across developmental axes.
3. The Coordination of Adaxial–Abaxial and Mediolateral Patterning Underlies the Predictions of the Waites–Hudson Model
Waites and Hudson’s study of the Antirrhinum phan mutant first revealed the intimate relationship between the establishment of adaxial–abaxial polarity and the ability of the leaf to grow wide along the mediolateral axis [33,39]. This laid the foundation for the Waites–Hudson model, which predicts that adaxial–abaxial polarity is a prerequisite for mediolateral growth of the leaf. The study of numerous polarity mutants in both monocots and eudicots supports this model; when adaxial or abaxial cell fates are lost, leaves fail to grow wide and instead form radialized filamentous structures. In some cases, regions of blade outgrowth can occur between juxtaposed domains of adaxial or abaxial tissue, suggesting that mediolateral outgrowth can occur where these boundaries adjoin one another [17,28,29,76,77]. However, the presence of adaxial–abaxial polarity alone is not sufficient to drive mediolateral outgrowth. For example, the maize mutant ragged seedling2 (rgd2) possesses normal adaxial–abaxial patterning but often fails to form a blade [78].
3.1. PRESSED FLOWER and WOX1 Coordinate Mediolateral Outgrowth with Adaxial–Abaxial Patterning
Two important regulators of mediolateral development acting downstream of adaxial–abaxial patterning are the homeobox genes PRESSED FLOWER (PRS) and WUSCHEL-LIKE HOMEOBOX1 (WOX1). Loss of PRS function is associated with missing lateral sepals and stamens in the flower and deletion of lateral stipules at the leaf base [79,80]. The maize orthologs of PRS, NARROW SHEATH1 (NS1) and NARROW SHEATH1 (NS2), also have roles in patterning leaf outgrowth from the leaf margin; the width of the sheath and proximal leaf blade are severely reduced in ns1 ns2 double mutants [80,81]. Orthologs of NS1 and NS2 have been cloned in other monocot species, including rice (NARROW LEAF1 2/3 [NAL2/3]) and barley (NARROW LEAF DWARF1 [NLD1]), and yield comparable phenotypes when disrupted, although defects in margin patterning extend well into the leaf blade in these species [82,83]. While grass genomes lack a WOX1 paralog, WOX1 orthologs are present in the genomes of Arabidopsis, tobacco (LAMINA1 [LAM1]), petunia (MAEWEST [MAW]), and Medicago (STENEFOLIA [STF]) and have conserved roles in promoting the mediolateral outgrowth of the leaf blade [84,85,86]. The wox1 prs double mutant exhibits an even more extreme reduction in leaf width, suggesting that the two genes work together to promote mediolateral leaf development. Not only do these mutants have narrow leaves, but the adaxial–abaxial polarity of the epidermis and mesophyll is perturbed at the leaf margins, strongly indicating a role for these genes in axial coordination [85,87].
Prior to leaf initiation, the prepatterned adaxial–abaxial polarity within the meristem may organize the mediolateral domain of the primordium in which WOX1/PRS are expressed. Fluorescent reporter studies identified a circumferential band of WOX1 expression around the meristem at the boundary between HD-ZIPIII and KAN expression domains (Figure 2B) [68]. Subsequently, feedback regulation of WOX1/PRS is seen at later stages of leaf development. In kan1 kan2 double mutants, WOX1 and PRS are misexpressed in the abaxial domain of the leaf, suggesting that the KAN genes may function to negatively regulate the expression of WOX1/PRS [87]. Misregulation of WOX1/PRS may, therefore, explain the ectopic formation of abaxial margin-like outgrowths that occur in higher order kan mutants. AS2 also appears to negatively regulate the expression of WOX1/PRS in the adaxial domain of the leaf. AS1 orthologs in tomato and tobacco influence the extent of the adaxial domain in leaves and thereby, the positions where mediolateral growth can occur [41,48]. One possibility is that the AS1–AS2 complex contributes to the restriction of the WOX1/PRS domain in the leaf. Together, these interactions could help to restrict WOX1/PRS to the margin domain of the leaf, thereby rendering mediolateral outgrowth dependent on proper adaxial–abaxial patterning and lending a mechanistic basis to the predictions made by Waites and Hudson.
Given the adaxial–abaxial patterning defects in wox1 prs mutants, WOX1 and PRS are likely involved in feedback regulation of adaxial–abaxial patterning factors at later stages of development. Among the adaxial patterning factors, WOX1/PRS promote the expression of HD-ZIPIII genes in the developing leaf margins, likely in part through inhibition of miR165/166 (Figure 3A) [84] WOX1/PRS also repress the expression of AS2, restricting its expression to the adaxial domain of the leaf margin [87,88]. Meanwhile, WOX1/PRS influence the expression of abaxial-specific genes as well. For example, ARF4 expression is weakly promoted by WOX1 in Arabidopsis while ETT/ARF3, ARF4, and KAN paralogs in petunia are all upregulated by MAW (Figure 3A,B) [85]. ARF4 expression is also promoted by NAL2/3 in rice (Figure 3C) [82]. The means by which WOX1/PRS modulate expression of these adaxial–abaxial patterning genes remains unresolved, as well as the functional importance of these regulatory interactions. WOX genes appear to act in a majority of cases as transcriptional repressors, and STF is known to interact with the corepressor proteins LEUNIG (LUG) and TOPLESS (TPL) but also with AINTEGUMENTA3 (AN3), a transcriptional activator important for regulating cell proliferation (Figure 3D) [89,90,91,92]. Therefore, it is probable that both transcriptional activation and repressive functions of WOX1/PRS in the developing leaf margin maintain a stable boundary between the adaxial and abaxial leaf domains, thereby coordinating the patterning of the adaxial–abaxial and mediolateral leaf axes.
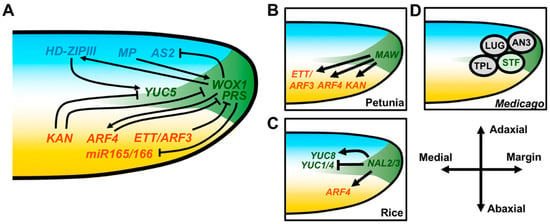
Figure 3.
Genetic interactions between marginal and adaxial–abaxial patterning genes at the leaf margin. (A) WOX1/PRS inhibits the expression of the adaxial patterning gene AS2 and the abaxial patterning genes miR165/166. Inhibition of miR165/166 indirectly promotes expression of HD-ZIPIII genes in the adaxial domain but may also do so via other direct/indirect mechanisms. KAN, ETT/ARF3 and ARF4 can repress WOX1/PRS and WOX1/PRS can promote ARF4. Auxin biosynthesis at the margin is promoted by expression of the auxin biosynthesis gene YUC5, which is inhibited by KAN and promoted by HD-ZIPIII. (B-D) WOX1 and PRS activity in other plant species. (B) The petunia homolog of WOX1, MAW, promotes the expression of ETT/ARF3, ARF4, and KAN homologs. (C) In rice, the PRS orthologs NAL2 and NAL3 repress the expression of ETT/ARF3 and ARF4 homologs as well as the auxin biosynthesis genes YUC1 and YUC4. YUC8 is instead upregulated by NAL2/3. (D) The Medicago WOX1 protein STF can physically interact with the TPL and LUG corepressor proteins but also with the transcriptional activator AN3.
3.2. Auxin Interconnects Adaxial–Abaxial and Mediolateral Patterning
Naturally, auxin also has a role in coordinating development along the mediolateral and adaxial–abaxial leaf axes. Local auxin biosynthesis, transport, and signaling all have a role in patterning the leaf margin and are under the direct transcriptional control of the opposing adaxial–abaxial patterning factors REV and KAN1 [93,94,95,96]. Auxin biosynthesis is necessary for marginal patterning, as higher-order loss-of-function mutations in the Arabidopsis YUCCA (YUC) auxin biosynthesis genes lead to defective margin development and REV directly promotes YUC5 expression, whereas KAN1 directly represses it [97]. In rice, NAL2/3 repress YUC1 and YUC4 but promote YUC8 expression [82]. Together, these data suggest that localized auxin biosynthesis may act downstream of adaxial–abaxial and/or mediolateral patterning to promote outgrowth at the margins (Figure 3A) [93,94,95,96].
Transport of auxin within the developing leaf is also crucial for proper development of the mediolateral and adaxial–abaxial leaf axes. In tomato, loss of PIN1 function results in occasional leaf radialization and disruption of auxin transport in lateral domains of the incipient leaf primordium [72]. Incisions separating the incipient leaf primordium from the SAM can also induce leaf radialization, and new data suggest that this phenomenon may be the consequence of perturbed auxin transport into the leaf [68,98,99].
Despite the apparent importance of spatiotemporal regulation of auxin transport and accumulation for leaf development, levels of auxin perception appear remarkably uniform across the adaxial–abaxial axis of the leaf, indicating potential polarization of the auxin response machinery [73]. ARF proteins mediate the transcriptional response to auxin and fall into two clades: type-A ARFs that possess transcriptional activation properties, and type-B ARFs, which act as transcriptional repressors [100,101,102]. The abaxial identity-promoting genes ARF3/ETT and ARF4 are both repressive Type-B ARFs. Other ARFs implicated in the control of adaxial cell fate include MONOPTEROS (MP), a Type-A ARF expressed in the adaxial domain of the leaf [103,104]. These abaxially and adaxially expressed ARFs appear to converge on the transcriptional regulation of WOX1 (Figure 3A). MP promotes the expression of WOX1, whereas ARF3/ETT and ARF4 repress it, consistent with their functional classifications [104]. Therefore, the complementary expression patterns of these Type-A and Type-B ARFs may translate auxin stimuli into instructions to pattern both the adaxial and abaxial faces of the leaf, thereby stably restricting the expression domain of WOX1 at the leaf margin. MP lacking the AUX/IAA-binding domain only conditions adaxial–abaxial patterning defects when expressed in the adaxial leaf domain, hinting that other auxin signaling components necessary for MP activity may be polarized along the adaxial–abaxial axis [103,105].
4. Control of Mediolateral Patterning along the Proximodistal Axis
4.1. Coordinating Indeterminate Growth and Growth Cessation across Axes
Leaf width can vary extensively along the proximodistal axis, suggesting that regulators of proximodistal patterning may influence mediolateral development. In eudicots, KNOX genes regulate patterning processes at the leaf margins. For example, the loss of BP, KNAT2, and KNAT6 function in the as1 mutant background can rescue many of the patterning defects triggered by ectopic Class I KNOX gene expression: the reduced petiole lengths and lobed margins conditioned by as1 are rescued [10,45,46,57]. In plants with compound leaves, KNOX genes have the additional role of promoting leaflet development at the leaf margins. For example, the STM gene is necessary for the formation of leaflets in the Arabidopsis relative Cardamine hirsuta, and overexpression of the STM ortholog KN1 is sufficient to yield highly dissected compound leaves in tomato [106,107,108]. STM acts by prolonging indeterminate, proliferative growth in the leaves of C. hirsuta, which is critical for the formation of leaflets [108,109] (Figure 4A). Leaflets are, in turn, patterned by the activity of auxin and CUP-SHAPED COTYLEDON (CUC) genes at the leaf margins. Marginal auxin maxima are generated by polarized auxin transport, thereby promoting leaflet outgrowth, whereas CUC expression is restricted to surrounding areas where it restricts growth [110,111]. This same mechanism is used to pattern the small serrations on the margins of Arabidopsis leaves, yet the lack of prolonged proliferative growth driven in part by STM in C. hirsuta prevents serrations from becoming fully fledged leaflets (Figure 4A). Unlike BP and KNAT2/6, STM is not subject to negative regulation by AS1, although as1 loss-of-function mutants in tomato have reduced numbers of leaflets. AS1 appears to affect leaflet number via its control of adaxial identity; leaves that fail to properly establish juxtaposed adaxial–abaxial leaf domains produce fewer leaflets in more distal positions [41,48]. Therefore, the regulation of cell indeterminacy by KNOX and AS1 along the proximodistal axis governs patterning outcomes at the margins.
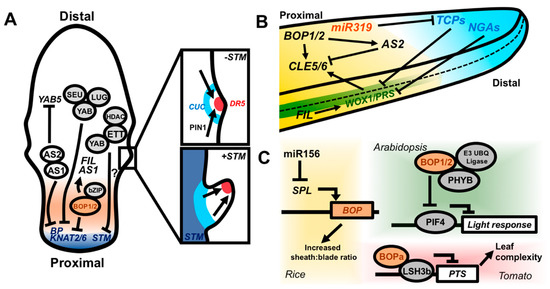
Figure 4.
Molecular coordination of the proximodistal and mediolateral leaf axes. (A) The Class I KNOX genes BP and KNAT2/6 are repressed by the AS1–AS2 complex, as well as by YAB transcription factors in complex with the corepressors SEU and LUG. The AS1–AS2 complex is, in turn, able to repress YAB5. YABs and ETT both repress STM expression and they can physically interact, potentially forming a repressive complex. BOP1/2 also repress BP and KNAT2/6 and promote expression of FIL and AS1. If Class I KNOX genes are not repressed in the leaf, growth is prolonged, which ultimately leads to the formation of leaflets via PIN1, auxin, and CUC2 activity (insets). Local auxin accumulation (indicated by expression of the DR5 promoter) driven by PIN1-mediated transport, drives margin outgrowth, while CUC genes inhibit growth in the surrounding sinus region. (B) Side-on view of a leaf illustrating the cross-regulation of proximodistal and mediolateral patterning factors in the context of the growth arrest front. The TCP and NGA genes promote cell differentiation and growth cessation in the distal portion of the leaf and are antagonized by proximally expressed miR319. TCPs and NGAs coordinately repress the expression of WOX1/PRS to allow for growth cessation at the margin. WOX1/PRS promote the proximal expression of CLE5/6, which affect leaf shape. BOP1/2 and AS2 also converge to promote and inhibit the expression of CLE5/6, respectively. BOP1/2 directly promote AS2 expression. FIL may promote mediolateral outgrowth by promoting WOX1 expression. (C) Genetic pathways by which BOPs may integrate internal and external cues to regulate patterning processes. In rice, miR166 represses SPL expression, which, in turn, inhibits BOP expression in the juvenile phase. Low BOP expression confers an increased sheath:blade ratio in juvenile leaves. Silencing of miR156 expression in adult leaves leads to increased BOP expression, yielding reduced blade:sheath ratios. BOP1/2 likely also function in light signaling. Arabidopsis BOP1/2 interact with PHYB and an E3 ubiquitin ligase complex to degrade PIF4 and attenuate light responsive gene expression. BOPa in tomato interacts with the light response regulator LSH3b and LSH3b represses PTS expression with downstream KNOX-mediated promotion of leaf complexity.
Growth cessation is also tightly coordinated between the proximodistal and mediolateral leaf axes. As the leaf matures, a wave of reduced cell-proliferation migrates basipetally across the leaf. Members of the TEOSINTE BRANCHED1/CINCINNATA/PROLIFERATING CELL FACTOR (TCP) transcription factor family, along with the NGATHA (NGA) transcription factors, regulate the movement of this arrest front, thereby promoting the cessation of growth in the leaf [112,113,114,115]. The proximally expressed microRNA miR319 counters TCP activity by post-transcriptionally silencing TCP mRNAs (Figure 4B). As growth ceases along both the proximodistal and mediolateral axes, it is reasonable to infer that these inhibitors of cell proliferation may also act to control the expression of mediolateral patterning genes. In fact, the combined overexpression of miR319a and knockout of NGA1,2,3 and 4 induces prolonged expression of WOX1/PRS, leading to severe overproliferation of the leaf margin (Figure 4B) [116]. This places WOX1 and PRS downstream of the TCP-NGA-miR319 proximodistal patterning module and provides a potential mechanism by which growth cessation along both the proximodistal and mediolateral axes is synchronized, yielding properly proportioned leaves.
4.2. YABBY Genes Regulate Proximodistal and Mediolateral Leaf Development
Members of the plant-specific YABBY (YAB) family of transcription factors are also important coordinators of mediolateral and proximodistal leaf development. Originally, YAB genes were linked to the control of abaxial cell identity in the leaf [117,118,119,120]. In Arabidopsis, FIL and other YABs are expressed in the abaxial domain of the leaf primordium; however, in other plant species, the spatial expression patterns of YAB genes diverge from the situation in Arabidopsis. Namely, maize YAB genes are expressed adaxially, whereas in rice, YAB genes exhibit no apparent polarity in developing leaf primordia [18,76,121,122]. Furthermore, overexpression of the abaxializing factor KAN2 in Arabidopsis does not condition ectopic expression of FIL, suggesting that the spatial regulation of FIL expression is in some sense independent of leaf adaxial–abaxial polarity [76]. Instead, YAB genes appear to at least in part regulate the growth of the leaf mediolateral axis. In support of this, the ectopic laminar outgrowths found on the abaxial side of kan1 kan2 kan3 mutants fail to develop in the yab3 background [76]. Furthermore, quadruple yab mutants show severely perturbed expression of margin patterning factors, yet adaxial–abaxial patterning genes are only mildly affected [123]. The PRS/WOX1 patterning module may have a conserved role in regulating the expression of YAB genes. In Arabidopsis, WOX1 promotes the expression of FIL and, in rice, NAL2/3 positively and negatively regulates the expression of multiple YAB genes [82,87,124]. Therefore, YAB genes likely act, to an extent, downstream of mediolateral patterning processes at the leaf margin.
Meanwhile, alongside their roles in mediolateral patterning, YAB genes antagonize the activity of proximal patterning factors. The KNOX genes KNAT2, BP, and KNAT6 are upregulated in the yab3 fil mutant background, and yab3 fil partially suppresses the stm mutant phenotype [125]. Consistent with these findings, ectopic meristems form on the adaxial leaf face in yab3 fil mutants. Transcriptomic analysis of leaves from higher-order yab mutants identified expression of WUS in leaf tissue, again indicating that meristem and/or axillary meristem (i.e., proximal) gene expression programs are reactivated in the leaf [123]. In Arabidopsis, FIL, YAB3, and YAB5 repress gene expression by forming a complex with the transcriptional repressor proteins LEUNIG and SEUSS (Figure 4A) [126]. Genetic disruption of this complex is associated with larger and more disorganized SAMs, suggesting that this complex may act non-cell autonomously to repress meristematic gene expression. Effects on SAM size are also observed in loss-of-function mutants of two maize YAB genes, DROOPING LEAF1/2 (DRL1/2); however, the effect is reversed compared to Arabidopsis, as drl1 drl2 mutant meristems are smaller than wild-type [121]. In rice, YAB1 works in a negative regulatory circuit with the hormone GA, which promotes leaf growth and differentiation. YAB1 is upregulated by GA and, in turn, represses expression of the GA biosynthesis gene GA3OX2 [122]. A lack of GA homeostasis in monocot yab mutants may explain the smaller SAM phenotypes of drl1/2 maize mutants, as meristematic cell identity is in part dependent on GA biosynthesis repression via KN1 [49,50]. Clearly, if this regulatory network exists in eudicots, its importance for regulating SAM size is diminished. Finally, YABs can also interact with ETT/ARF3 in vitro, raising the possibility that this interaction may contribute to the known downregulation of Class I KNOX genes by ETT/ARF3 via their interactions with histone deacetylases (Figure 4A) [75]. Together, these findings place YAB genes as key regulators of mediolateral and proximodistal patterning in leaves, yet the precise roles of YAB function is rendered ambiguous owing to the species-specific differences in phenotypes. For example, the characterized YAB genes of maize and rice control differentiation of the leaf midrib: no defects in mediolateral or adaxial-abaxial polarity are observed [121,127]. A better understanding of YAB gene function in the context of proximodistal and mediolateral patterning will likely emerge as reverse genetics overcomes the high levels of genetic redundancy in the YAB family.
4.3. BLADE-ON-PETIOLE Genes Integrate Developmental Patterning with Environmental Sensing
Genes that coordinate the development of multiple leaf axes simultaneously may also be poised to mediate broad developmental changes in response to endogenous or exogenous cues. Such roles are emerging for the BLADE ON PETIOLE (BOP) genes. BOPs encode BTB/POZ domain transcription factors that harbor an ankyrin repeat. In Arabidopsis, bop1 and bop2 mutants develop ectopic outgrowths of blade tissue along the petiole [128,129,130]. Thus, BOP genes may restrict mediolateral outgrowth to the appropriate position along the proximodistal axis of the leaf [131]. The BOP genes repress the expression of the Class I KNOX genes BP, KNAT2, and KNAT6 in Arabidopsis [128]. Furthermore, in cauline leaves and floral lateral organs the BOP genes are required for the development of the proximal abscission zone by regulating the expression of ATH1 (ARABIDOPSIS THALIANA HOMEOBOX1) [132,133]. BOP1/2 possess transcriptional activation activity, and may repress Class I KNOX gene expression via their direct upregulation of AS2 [134,135]. BOP1/2 can also physically interact with bZip proteins, an interaction involved in controlling ATH1 expression [136]. Meanwhile, BOP1/2 may regulate leaf width by controlling the expression of mediolateral patterning factors. BOP1/2 promote the expression of FIL, which could stimulate the mediolateral expansion of the leaf blade at specific positions along the proximodistal axis [134]. In addition, BOP1/2 promote the expression of two proximally expressed CLE (CLAVATA-LIKE EMBRYO SURROUNDING REGION) peptide-encoding genes that are simultaneously upregulated by WOX1 and repressed by AS2 [137]. CLEs typically have intercellular signaling roles and bind to leucine-rich receptor-like kinases. In Arabidopsis, cle5/6 mutants exhibit subtle changes in leaf width and petiole angle. BOP1/2 may, in part, regulate the mediolateral development of the leaf by converging with WOX1 and AS2 on CLE5/6 regulation (Figure 4B).
Recently, BOPs have also been identified as orchestrating developmental responses to endogenous and external cues. For example, BOP gene function in rice intersects with the vegetative phase change pathway. Rice contains three BOP-like genes and higher-order mutant combinations lead to progressively reduced blade:sheath ratios, with this effect becoming more pronounced as leaves transition from juvenile to adult stages [138]. Indeed, the juvenile leaf 1 has a much higher level of BOP expression than do subsequent leaves and is almost entirely composed of sheath tissue. Phase-specific regulation of BOP expression is driven by the highly conserved miR156-SQUAMOSA PROMOTER BINDING LIKE (SPL) pathway in which reduction in miR156 expression relieves the repression of SPL, thereby promoting the juvenile-to-adult phase transition (Figure 4C) [139]. BOP genes are likely downstream of this pathway but, it is unknown whether BOPs are direct targets of SPL. BOPs also mediate the developmental outcomes of light signaling. PHYTOCHROME B physically interacts with both BOP1 and BOP2 in Arabidopsis, and BOP2 can mediate the degradation of the light-response transcriptional regulator PHYTOCHROME INTERACTING FACTOR4 (PIF4) through a physical interaction with an E3 ubiquitin ligase complex under red light (Figure 4C) [140]. BOPs also interact with the LIGHT-DEPENDENT SHORT HYPOCOTYLS3b (LSH3b) protein of tomato [141]. In Arabidopsis, LSH genes promote cell growth in response to light and, in tomato, LSH3b binds and negatively regulates PETROSELINUM, which enhances leaf complexity through its posttranscriptional modification of KNOX activity [141,142,143]. Therefore, BOP control of development along the proximodistal and mediolateral axes may act downstream of both internal and external cues to coordinately pattern the leaf.
5. Conclusions
Plants are capable of producing consistently patterned leaves throughout their lifespan, a feat that requires highly robust control of underlying genetic patterning factors. One way that such robustness may be achieved is through the close coordination of factors patterning the three growth axes of leaves, a prediction realized by the work of Waites and Hudson almost twenty five years ago. Here, we have described several genetic and hormonal factors that simultaneously regulate developmental patterning along multiple leaf axes simultaneously. Such control mechanisms likely ensure that growth along these axes proceeds in a synchronized and organized fashion. Perhaps most striking is the highly redundant repression of Class I KNOX genes by factors regulating development along all three axes. Clearly, KNOX repression is an essential and tightly regulated developmental program of the leaf. Additionally, coordinating factors frequently cross-regulate multiple patterning modules, as well as interact with a host of co-regulatory proteins to carry out their functions. Looking forward, advances in live imaging methods will allow for enhanced analysis of leaf development and its associated spatiotemporal gene expression patterns in real time. This, coupled with rapdily advancing genomics technologies, will provide a major boon to understanding the complex and interwoven leaf axial patterning pathways. Through such work, we will come closer to understanding the remarkable consistency of leaf patterning in three dimensions.
Funding
J. Satterlee is supported by NSF IOS-PGRP grant #1238142.
Acknowledgments
We thank J. Strable and B. Conlon for comments on the manuscript and figures.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kaplan, D.R. Fundamental Concepts of Leaf Morphology and Morphogenesis: A Contribution to the Interpretation of Molecular Genetic Mutants. Int. J. Plant Sci. 2001, 162, 465–474. [Google Scholar] [CrossRef]
- Steeves, T.A.; Sussex, I.M.B. Patterns in plant development. Nord. J. Bot. 1989, 11, 204. [Google Scholar]
- Sinha, N. Leaf Development in Angiosperms. Annu. Rev. Plant Phys. Plant Mol. Biol. 1999, 50, 419–446. [Google Scholar] [CrossRef]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef]
- Vollbrecht, E.; Veit, B.; Sinha, N.; Hake, S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 1991, 350, 241–243. [Google Scholar] [CrossRef]
- Sinha, N.R.; Williams, R.E.; Hake, S. Overexpression of the maize homeobox gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 1993, 7, 787–795. [Google Scholar] [CrossRef]
- Vollbrecht, E.; Reiser, L.; Hake, S. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 2000, 127, 3161–3172. [Google Scholar]
- Smith, L.G.; Greene, B.; Veit, B.; Hake, S. A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 1992, 116, 21–30. [Google Scholar]
- Chuck, G.; Lincoln, C.; Hake, S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 1996, 8, 1277–1289. [Google Scholar]
- Byrne, M.E.; Barley, R.; Curtis, M.; Arroyo, J.M.; Dunham, M.; Hudson, A.; Martienssen, R.A. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 2000, 408, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Semiarti, E.; Ueno, Y.; Tsukaya, H.; Iwakawa, H.; Machida, C.; Machida, Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 2001, 128, 1771–1783. [Google Scholar] [PubMed]
- Timmermans, M.C.; Hudson, A.; Becraft, P.W.; Nelson, T. ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 1999, 284, 151–153. [Google Scholar] [CrossRef] [PubMed]
- Tsiantis, M.; Schneeberger, R.; Golz, J.F.; Freeling, M.; Langdale, J.A. The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 1999, 284, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Schneeberger, R.; Tsiantis, M.; Freeling, M.; Langdale, J.A. The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 1998, 125, 2857–2865. [Google Scholar] [PubMed]
- McConnell, J.R.; Emery, J.; Eshed, Y.; Bao, N.; Bowman, J.; Barton, M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001, 411, 709–713. [Google Scholar] [CrossRef]
- Otsuga, D.; DeGuzman, B.; Prigge, M.J.; Drews, G.N.; Clark, S.E. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001, 25, 223–236. [Google Scholar] [CrossRef]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef]
- Juarez, M.T.; Twigg, R.W.; Timmermans, M.C.P. Specification of adaxial cell fate during maize leaf development. Development 2004, 131, 4533–4544. [Google Scholar] [CrossRef]
- Juarez, M.T.; Kui, J.S.; Thomas, J.; Heller, B.A.; Timmermans, M.C.P. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 2004, 428, 84–88. [Google Scholar] [CrossRef]
- Kidner, C.A.; Martienssen, R.A. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 2004, 428, 81–84. [Google Scholar] [CrossRef]
- Pekker, I.; Alvarez, J.P.; Eshed, Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 2005, 17, 2899–2910. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, D.H.; Nogueira, F.T.S.; Howell, M.D.; Montgomery, T.A.; Carrington, J.C.; Timmermans, M.C.P. Pattern formation via small RNA mobility. Genes Dev. 2009, 23, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Skopelitis, D.S.; Benkovics, A.H.; Husbands, A.Y.; Timmermans, M.C.P. Boundary Formation through a Direct Threshold-Based Readout of Mobile Small RNA Gradients. Dev. Cell 2017, 43, 265–273.e6. [Google Scholar] [CrossRef] [PubMed]
- Kafri, R.; Springer, M.; Pilpel, Y. Genetic Redundancy: New Tricks for Old Genes. Cell 2009, 136, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Leal, D.; Xu, C.; Kwon, C.-T.; Soyars, C.; Demesa-Arevalo, E.; Man, J.; Liu, L.; Lemmon, Z.H.; Jones, D.S.; Van Eck, J.; et al. Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 2019, 51, 786–792. [Google Scholar] [CrossRef]
- El-Brolosy, M.A.; Kontarakis, Z.; Rossi, A.; Kuenne, C.; Günther, S.; Fukuda, N.; Kikhi, K.; Boezio, G.L.M.; Takacs, C.M.; Lai, S.-L.; et al. Genetic compensation triggered by mutant mRNA degradation. Nature 2019, 568, 193–197. [Google Scholar] [CrossRef]
- Ma, Z.; Zhu, P.; Shi, H.; Guo, L.; Zhang, Q.; Chen, Y.; Chen, S.; Zhang, Z.; Peng, J.; Chen, J. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 2019, 568, 259–263. [Google Scholar] [CrossRef]
- Kerstetter, R.A.; Bollman, K.; Taylor, R.A.; Bomblies, K.; Poethig, R.S. KANADI regulates organ polarity in Arabidopsis. Nature 2001, 411, 706–709. [Google Scholar] [CrossRef]
- Eshed, Y.; Baum, S.F.; Perea, J.V.; Bowman, J.L. Establishment of polarity in lateral organs of plants. Curr. Biol. 2001, 11, 1251–1260. [Google Scholar] [CrossRef]
- Vokes, S.A.; Ji, H.; Wong, W.H.; McMahon, A.P. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008, 22, 2651–2663. [Google Scholar] [CrossRef]
- Osterwalder, M.; Speziale, D.; Shoukry, M.; Mohan, R.; Ivanek, R.; Kohler, M.; Beisel, C.; Wen, X.; Scales, S.J.; Christoffels, V.M.; et al. HAND2 Targets Define a Network of Transcriptional Regulators that Compartmentalize the Early Limb Bud Mesenchyme. Dev. Cell 2014, 31, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Zuniga, A. Next generation limb development and evolution: old questions, new perspectives. Development 2015, 142, 3810–3820. [Google Scholar] [CrossRef] [PubMed]
- Waites, R.; Hudson, A. phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 1995, 121, 2143. [Google Scholar]
- Waites, R.; Selvadurai, H.R.N.; Oliver, I.R.; Hudson, A. The PHANTASTICA Gene Encodes a MYB Transcription Factor Involved in Growth and Dorsoventrality of Lateral Organs in Antirrhinum. Cell 1998, 93, 779–789. [Google Scholar] [CrossRef]
- McHale, N.A.; Koning, R.E. PHANTASTICA Regulates Development of the Adaxial Mesophyll in Nicotiana Leaves. Plant Cell 2004, 16, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; McCormick, S.; Timmermans, M.; Sinha, N. The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 2003, 424, 438–443. [Google Scholar] [CrossRef]
- Kim, M.; Pham, T.; Hamidi, A.; McCormick, S.; Kuzoff, R.K.; Sinha, N. Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves. Development 2003, 130, 4405–4415. [Google Scholar] [CrossRef]
- Tattersall, A.D.; Turner, L.; Knox, M.R.; Ambrose, M.J.; Ellis, T.H.N.; Hofer, J.M.I. The Mutant crispa Reveals Multiple Roles for PHANTASTICA in Pea Compound Leaf Development. Plant Cell 2005, 17, 1046. [Google Scholar] [CrossRef]
- Theodoris, G.; Inada, N.; Freeling, M. Conservation and molecular dissection of ROUGH SHEATH2 and ASYMMETRIC LEAVES1 function in leaf development. Proc. Natl. Acad. Sci.USA 2003, 100, 6837–6842. [Google Scholar] [CrossRef]
- Jackson, D.; Veit, B.; Hake, S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 1994, 120, 405. [Google Scholar]
- Hake, S.; Smith, H.M.S.; Holtan, H.; Magnani, E.; Mele, G.; Ramirez, J. The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 2004, 20, 125–151. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Tsiantis, M. KNOX genes: versatile regulators of plant development and diversity. Development 2010, 137, 3153–3165. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D. Double Labeling of KNOTTED1 mRNA and Protein Reveals Multiple Potential Sites of Protein Trafficking in the Shoot Apex. Plant Physiol. 2002, 129, 1423–1429. [Google Scholar] [CrossRef] [PubMed]
- Ori, N.; Eshed, Y.; Chuck, G.; Bowman, J.L.; Hake, S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 2000, 127, 5523–5532. [Google Scholar]
- Sun, Y.; Zhou, Q.; Zhang, W.; Fu, Y.; Huang, H. ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 2002, 214, 694–702. [Google Scholar] [CrossRef]
- Long, J.A.; Moan, E.I.; Medford, J.I.; Barton, M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 1996, 379, 66–69. [Google Scholar] [CrossRef]
- Xu, L.; Xu, Y.; Dong, A.; Sun, Y.; Pi, L.; Xu, Y.; Huang, H. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 2003, 130, 4097–4107. [Google Scholar] [CrossRef]
- Zoulias, N.; Koenig, D.; Hamidi, A.; McCormick, S.; Kim, M. A role for PHANTASTICA in medio-lateral regulation of adaxial domain development in tomato and tobacco leaves. Ann. Bot. 2012, 109, 407–418. [Google Scholar] [CrossRef]
- Bolduc, N.; Hake, S. The Maize Transcription Factor KNOTTED1 Directly Regulates the Gibberellin Catabolism Gene ga2ox1. Plant Cell 2009, 21, 1647–1658. [Google Scholar] [CrossRef]
- Bolduc, N.; Yilmaz, A.; Mejia-Guerra, M.K.; Morohashi, K.; O’Connor, D.; Grotewold, E.; Hake, S. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 2012, 26, 1685–1690. [Google Scholar] [CrossRef]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C.; et al. The ASYMMETRIC LEAVES2 Gene of Arabidopsis thaliana, Required for Formation of a Symmetric Flat Leaf Lamina, Encodes a Member of a Novel Family of Proteins Characterized by Cysteine Repeats and a Leucine Zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, H.; Iwasaki, M.; Kojima, S.; Ueno, Y.; Soma, T.; Tanaka, H.; Semiarti, E.; Machida, Y.; Machida, C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves: AS2 represses adaxial cell proliferation of leaves. Plant J. 2007, 51, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.M.S. The indeterminate gametophyte1 Gene of Maize Encodes a LOB Domain Protein Required for Embryo Sac and Leaf Development. Plant Cell 2007, 19, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Shuai, B.; Springer, P.S. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 2003, 15, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C.P. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef]
- Phelps-Durr, T.L.; Thomas, J.; Vahab, P.; Timmermans, M.C.P. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 2005, 17, 2886–2898. [Google Scholar] [CrossRef]
- Lodha, M.; Marco, C.F.; Timmermans, M.C.P. The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 2013, 27, 596–601. [Google Scholar] [CrossRef]
- Li, Z.; Li, B.; Liu, J.; Guo, Z.; Liu, Y.; Li, Y.; Shen, W.-H.; Huang, Y.; Huang, H.; Zhang, Y.; et al. Transcription factors AS1 and AS2 interact with LHP1 to repress KNOX genes in Arabidopsis. J. Integr. Plant Biol. 2016, 58, 959–970. [Google Scholar] [CrossRef]
- Ikezaki, M.; Kojima, M.; Sakakibara, H.; Kojima, S.; Ueno, Y.; Machida, C.; Machida, Y. Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 2010, 61, 70–82. [Google Scholar] [CrossRef]
- Iwasaki, M.; Takahashi, H.; Iwakawa, H.; Nakagawa, A.; Ishikawa, T.; Tanaka, H.; Matsumura, Y.; Pekker, I.; Eshed, Y.; Vial-Pradel, S.; et al. Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development 2013, 140, 1958–1969. [Google Scholar] [CrossRef]
- Husbands, A.Y.; Benkovics, A.H.; Nogueira, F.T.S.; Lodha, M.; Timmermans, M.C.P. The ASYMMETRIC LEAVES Complex Employs Multiple Modes of Regulation to Affect Adaxial-Abaxial Patterning and Leaf Complexity. Plant Cell 2015, 27, 3321–3335. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Lin, W.-C.; Huang, T.; Poethig, R.S.; Springer, P.S.; Kerstetter, R.A. KANADI1 regulates adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc. Natl. Acad. Sci. USA 2008, 105, 16392–16397. [Google Scholar] [CrossRef] [PubMed]
- Conklin, P.A.; Strable, J.; Li, S.; Scanlon, M.J. On the mechanisms of development in monocot and eudicot leaves. New Phytol. 2019, 221, 706–724. [Google Scholar] [CrossRef]
- Reinhardt, D.; Mandel, T.; Kuhlemeier, C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 2000, 12, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, D.; Pesce, E.-R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Traas, J.; Friml, J.; Kuhlemeier, C. Regulation of phyllotaxis by polar auxin transport. Nature 2003, 426, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Barkoulas, M.; Tsiantis, M. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 2006, 133, 3955–3961. [Google Scholar] [CrossRef] [PubMed]
- Scanlon, M.J. The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol. 2003, 133, 597–605. [Google Scholar] [CrossRef]
- Caggiano, M.P.; Yu, X.; Bhatia, N.; Larsson, A.; Ram, H.; Ohno, C.K.; Sappl, P.; Meyerowitz, E.M.; Jönsson, H.; Heisler, M.G. Cell type boundaries organize plant development. eLife 2017, 6. [Google Scholar] [CrossRef]
- Yu, T.; Guan, C.; Wang, J.; Sajjad, M.; Ma, L.; Jiao, Y. Dynamic patterns of gene expression during leaf initiation. J. Genet. Genom. 2017, 44, 599–601. [Google Scholar] [CrossRef]
- Yadav, R.K.; Perales, M.; Gruel, J.; Ohno, C.; Heisler, M.; Girke, T.; Jonsson, H.; Reddy, G.V. Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 2014, 9, 654. [Google Scholar] [CrossRef]
- Bar, M.; Ori, N. Leaf development and morphogenesis. Development 2014, 141, 4219–4230. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Wang, Y.; Yu, T.; Cunha, A.; Wu, B.; Vernoux, T.; Meyerowitz, E.; Jiao, Y. Auxin depletion from leaf primordia contributes to organ patterning. Proc. Natl. Acad. Sci. USA 2014, 111, 18769–18774. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, N.; Åhl, H.; Jönsson, H.; Heisler, M.G. Quantitative analysis of auxin sensing in leaf primordia argues against proposed role in regulating leaf dorsoventrality. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Sessions, A.; Nemhauser, J.L.; McColl, A.; Roe, J.L.; Feldmann, K.A.; Zambryski, P.C. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 1997, 124, 4481–4491. [Google Scholar]
- Chung, Y.; Zhu, Y.; Wu, M.-F.; Simonini, S.; Kuhn, A.; Armenta-Medina, A.; Jin, R.; Østergaard, L.; Gillmor, C.S.; Wagner, D. Auxin Response Factors promote organogenesis by chromatin-mediated repression of the pluripotency gene SHOOTMERISTEMLESS. Nat. Commun. 2019, 10, 886. [Google Scholar] [CrossRef]
- Eshed, Y.; Izhaki, A.; Baum, S.F.; Floyd, S.K.; Bowman, J.L. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 2004, 131, 2997–3006. [Google Scholar] [CrossRef]
- Candela, H.; Johnston, R.; Gerhold, A.; Foster, T.; Hake, S. The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell 2008, 20, 2073–2087. [Google Scholar] [CrossRef]
- Henderson, D.C.; Muehlbauer, G.J.; Scanlon, M.J. Radial leaves of the maize mutant ragged seedling2 retain dorsiventral anatomy. Dev. Biol. 2005, 282, 455–466. [Google Scholar] [CrossRef][Green Version]
- Matsumoto, N.; Okada, K. A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 2001, 15, 3355–3364. [Google Scholar] [CrossRef]
- Nardmann, J.; Ji, J.; Werr, W.; Scanlon, M.J. The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 2004, 131, 2827–2839. [Google Scholar] [CrossRef]
- Scanlon, M.J.; Schneeberger, R.G.; Freeling, M. The maize mutant narrow sheath fails to establish leaf margin identity in a meristematic domain. Development 1996, 122, 1683–1691. [Google Scholar]
- Cho, S.-H.; Yoo, S.-C.; Zhang, H.; Pandeya, D.; Koh, H.-J.; Hwang, J.-Y.; Kim, G.-T.; Paek, N.-C. The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol. 2013, 198, 1071–1084. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Tanaka, S.-Y.; Masumoto, Y.; Nobori, N.; Ishii, H.; Hibara, K.-I.; Itoh, J.-I.; Tanisaka, T.; Taketa, S. Barley NARROW LEAFED DWARF1 encoding a WUSCHEL-RELATED HOMEOBOX 3 (WOX3) regulates the marginal development of lateral organs. Breed. Sci. 2016, 66, 416–424. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McHale, N.A.; Marcotrigiano, M. LAM1 is required for dorsoventrality and lateral growth of the leaf blade in Nicotiana. Development 1998, 125, 4235–4243. [Google Scholar] [PubMed]
- Vandenbussche, M.; Horstman, A.; Zethof, J.; Koes, R.; Rijpkema, A.S.; Gerats, T. Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell 2009, 21, 2269–2283. [Google Scholar] [CrossRef]
- Tadege, M.; Lin, H.; Bedair, M.; Berbel, A.; Wen, J.; Rojas, C.M.; Niu, L.; Tang, Y.; Sumner, L.; Ratet, P.; et al. STENOFOLIA regulates blade outgrowth and leaf vascular patterning in Medicago truncatula and Nicotiana sylvestris. Plant Cell 2011, 23, 2125–2142. [Google Scholar] [CrossRef]
- Nakata, M.; Matsumoto, N.; Tsugeki, R.; Rikirsch, E.; Laux, T.; Okada, K. Roles of the Middle Domain–Specific WUSCHEL-RELATED HOMEOBOX Genes in Early Development of Leaves in Arabidopsis. Plant Cell 2012, 24, 519–535. [Google Scholar] [CrossRef]
- Zhang, F.; Tadege, M. Repression of AS2 by WOX family transcription factors is required for leaf development in Medicago and Arabidopsis. Plant Signal. Behav. 2015, 10, e993291. [Google Scholar]
- Lin, H.; Niu, L.; McHale, N.A.; Ohme-Takagi, M.; Mysore, K.S.; Tadege, M. Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proc. Natl. Acad. Sci. USA 2013, 110, 366–371. [Google Scholar] [CrossRef]
- Lin, H.; Niu, L.; Tadege, M. STENOFOLIA acts as a repressor in regulating leaf blade outgrowth. Plant Signal. Behav. 2013, 8, e24464. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, H.; Kalve, S.; Wolabu, T.W.; Nakashima, J.; Golz, J.F.; Tadege, M. Control of leaf blade outgrowth and floral organ development by LEUNIG, ANGUSTIFOLIA3 and WOX transcriptional regulators. New Phytol. 2019, 223, 2024–2038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, Y.; Li, G.; Tang, Y.; Kramer, E.M.; Tadege, M. STENOFOLIA Recruits TOPLESS to Repress ASYMMETRIC LEAVES2 at the Leaf Margin and Promote Leaf Blade Outgrowth in Medicago truncatula. Plant Cell 2014, 26, 650–664. [Google Scholar] [CrossRef] [PubMed]
- Brandt, R.; Salla-Martret, M.; Bou-Torrent, J.; Musielak, T.; Stahl, M.; Lanz, C.; Ott, F.; Schmid, M.; Greb, T.; Schwarz, M.; et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 2012, 72, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Liu, T.; Newell, N.R.; Magnani, E.; Huang, T.; Kerstetter, R.; Michaels, S.; Barton, M.K. Establishing a framework for the Ad/abaxial regulatory network of Arabidopsis: ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell 2013, 25, 3228–3249. [Google Scholar] [CrossRef]
- Merelo, P.; Xie, Y.; Brand, L.; Ott, F.; Weigel, D.; Bowman, J.L.; Heisler, M.G.; Wenkel, S. Genome-Wide Identification of KANADI1 Target Genes. PLoS ONE 2013, 8, e77341. [Google Scholar] [CrossRef]
- Huang, T.; Harrar, Y.; Lin, C.; Reinhart, B.; Newell, N.R.; Talavera-Rauh, F.; Hokin, S.A.; Barton, M.K.; Kerstetter, R.A. Arabidopsis KANADI1 Acts as a Transcriptional Repressor by Interacting with a Specific cis -Element and Regulates Auxin Biosynthesis, Transport, and Signaling in Opposition to HD-ZIPIII Factors. Plant Cell 2014, 26, 246–262. [Google Scholar] [CrossRef]
- Wang, W.; Xu, B.; Wang, H.; Li, J.; Huang, H.; Xu, L. YUCCA genes are expressed in response to leaf adaxial-abaxial juxtaposition and are required for leaf margin development. Plant Physiol. 2011, 157, 1805–1819. [Google Scholar] [CrossRef]
- Sussex, I.M. Experiments on the cause of dorsiventrality in leaves. Nature 1951, 167, 651–652. [Google Scholar] [CrossRef]
- Shi, J.; Dong, J.; Xue, J.; Wang, H.; Yang, Z.; Jiao, Y.; Xu, L.; Huang, H. Model for the role of auxin polar transport in patterning of the leaf adaxial-abaxial axis. Plant J. 2017, 92, 469–480. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Roosjen, M.; Paque, S.; Weijers, D. Auxin Response Factors: output control in auxin biology. J. Exp. Bot. 2018, 69, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5844–5849. [Google Scholar] [CrossRef] [PubMed]
- Krogan, N.T.; Berleth, T. A dominant mutation reveals asymmetry in MP/ARF5 function along the adaxial-abaxial axis of shoot lateral organs. Plant Signal. Behav. 2012, 7, 940–943. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.; Wu, B.; Yu, T.; Wang, Q.; Krogan, N.T.; Liu, X.; Jiao, Y. Spatial Auxin Signaling Controls Leaf Flattening in Arabidopsis. Curr. Biol. 2017, 27, 2940–2950.e4. [Google Scholar] [CrossRef] [PubMed]
- Simonini, S.; Deb, J.; Moubayidin, L.; Stephenson, P.; Valluru, M.; Freire-Rios, A.; Sorefan, K.; Weijers, D.; Friml, J.; Østergaard, L. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes Dev. 2016, 30, 2286–2296. [Google Scholar] [CrossRef]
- Hareven, D.; Gutfinger, T.; Parnis, A.; Eshed, Y.; Lifschitz, E. The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 1996, 84, 735–744. [Google Scholar] [CrossRef]
- Hay, A.; Tsiantis, M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 2006, 38, 942–947. [Google Scholar] [CrossRef]
- Barkoulas, M.; Hay, A.; Kougioumoutzi, E.; Tsiantis, M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 2008, 40, 1136–1141. [Google Scholar] [CrossRef]
- Kierzkowski, D.; Runions, A.; Vuolo, F.; Strauss, S.; Lymbouridou, R.; Routier-Kierzkowska, A.-L.; Wilson-Sánchez, D.; Jenke, H.; Galinha, C.; Mosca, G.; et al. A Growth-Based Framework for Leaf Shape Development and Diversity. Cell 2019, 177, 1405–1418.e17. [Google Scholar] [CrossRef]
- Nikovics, K.; Blein, T.; Peaucelle, A.; Ishida, T.; Morin, H.; Aida, M.; Laufs, P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 2006, 18, 2929–2945. [Google Scholar] [CrossRef]
- Bilsborough, G.D.; Runions, A.; Barkoulas, M.; Jenkins, H.W.; Hasson, A.; Galinha, C.; Laufs, P.; Hay, A.; Prusinkiewicz, P.; Tsiantis, M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 2011, 108, 3424–3429. [Google Scholar] [CrossRef] [PubMed]
- Nath, U.; Crawford, B.C.W.; Carpenter, R.; Coen, E. Genetic Control of Surface Curvature. Science 2003, 299, 1404. [Google Scholar] [CrossRef] [PubMed]
- Efroni, I.; Blum, E.; Goldshmidt, A.; Eshed, Y. A Protracted and Dynamic Maturation Schedule Underlies Arabidopsis Leaf Development. Plant Cell 2008, 20, 2293. [Google Scholar] [CrossRef] [PubMed]
- Trigueros, M.; Navarrete-Gómez, M.; Sato, S.; Christensen, S.K.; Pelaz, S.; Weigel, D.; Yanofsky, M.F.; Ferrándiz, C. The NGATHA Genes Direct Style Development in the Arabidopsis Gynoecium. Plant Cell 2009, 21, 1394–1409. [Google Scholar] [CrossRef]
- Alvarez, J.P.; Goldshmidt, A.; Efroni, I.; Bowman, J.L.; Eshed, Y. The NGATHA Distal Organ Development Genes Are Essential for Style Specification in Arabidopsis. Plant Cell 2009, 21, 1373–1393. [Google Scholar] [CrossRef]
- Alvarez, J.P.; Furumizu, C.; Efroni, I.; Eshed, Y.; Bowman, J.L. Active suppression of a leaf meristem orchestrates determinate leaf growth. eLife 2016, 5. [Google Scholar] [CrossRef]
- Siegfried, K.R.; Eshed, Y.; Baum, S.F.; Otsuga, D.; Drews, G.N.; Bowman, J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 1999, 126, 4117–4128. [Google Scholar]
- Sawa, S.; Watanabe, K.; Goto, K.; Kanaya, E.; Morita, E.H.; Okada, K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 1999, 13, 1079–1088. [Google Scholar] [CrossRef]
- Eshed, Y.; Baum, S.F.; Bowman, J.L. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 1999, 99, 199–209. [Google Scholar] [CrossRef]
- Golz, J.F.; Roccaro, M.; Kuzoff, R.; Hudson, A. GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development 2004, 131, 3661–3670. [Google Scholar] [CrossRef]
- Strable, J.; Wallace, J.G.; Unger-Wallace, E.; Briggs, S.; Bradbury, P.J.; Buckler, E.S.; Vollbrecht, E. Maize YABBY Genes drooping leaf1 and drooping leaf2 Regulate Plant Architecture. Plant Cell 2017, 29, 1622–1641. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Zhao, Y.; Ma, Q.; Hu, Y.; Hedden, P.; Zhang, Q.; Zhou, D.X. The Rice YABBY1 Gene Is Involved in the Feedback Regulation of Gibberellin Metabolism. Plant Phys. 2007, 144, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Sarojam, R.; Sappl, P.G.; Goldshmidt, A.; Efroni, I.; Floyd, S.K.; Eshed, Y.; Bowman, J.L. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 2010, 22, 2113–2130. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Hu, Y.; Zhao, Y.; Liu, H.; Zhou, D.X. A WUSCHEL-LIKE HOMEOBOX Gene Represses a YABBY Gene Expression Required for Rice Leaf Development. Plant Phys. 2007, 144, 380–390. [Google Scholar] [CrossRef]
- Kumaran, M.K.; Bowman, J.L.; Sundaresan, V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 2002, 14, 2761–2770. [Google Scholar] [CrossRef]
- Stahle, M.I.; Kuehlich, J.; Staron, L.; von Arnim, A.G.; Golz, J.F. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 2009, 21, 3105–3118. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Nagasawa, N.; Kawasaki, S.; Matsuoka, M.; Nagato, Y.; Hirano, H.-Y. The YABBY Gene DROOPING LEAF Regulates Carpel Specification and Midrib Development in Oryza sativa. Plant Cell 2004, 16, 500–509. [Google Scholar] [CrossRef]
- Ha, C.M.; Kim, G.T.; Kim, B.C.; Jun, J.H.; Soh, M.S.; Ueno, Y.; Machida, Y.; Tsukaya, H.; Nam, H.G. The BLADE-ON-PETIOLE1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 2003, 130, 161–172. [Google Scholar] [CrossRef]
- Ha, C.M.; Jun, J.H.; Nam, H.G.; Fletcher, J.C. BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 1361–1370. [Google Scholar] [CrossRef]
- Hepworth, S.R.; Zhang, Y.; McKim, S.; Li, X.; Haughn, G.W. BLADE-ON-PETIOLE–Dependent Signaling Controls Leaf and Floral Patterning in Arabidopsis. Plant Cell 2005, 17, 1434–1448. [Google Scholar] [CrossRef]
- Ichihashi, Y.; Kawade, K.; Usami, T.; Horiguchi, G.; Takahashi, T.; Tsukaya, H. Key Proliferative Activity in the Junction between the Leaf Blade and Leaf Petiole of Arabidopsis. Plant Phys. 2011, 157, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- McKim, S.M.; Stenvik, G.E.; Butenko, M.A.; Kristiansen, W.; Cho, S.K.; Hepworth, S.R.; Aalen, R.B.; Haughn, G.W. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 2008, 135, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Mena, C.; Sablowski, R. ARABIDOPSIS THALIANA HOMEOBOX GENE1 Establishes the Basal Boundaries of Shoot Organs and Controls Stem Growth. Plant Cell 2008, 20, 2059–2072. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.M.; Jun, J.H.; Nam, H.G.; Fletcher, J.C. BLADE-ON-PETIOLE1 and 2 Control Arabidopsis Lateral Organ Fate through Regulation of LOB Domain and Adaxial-Abaxial Polarity Genes. Plant Cell 2007, 19, 1809–1825. [Google Scholar] [CrossRef]
- Jun, J.H.; Ha, C.M.; Fletcher, J.C. BLADE-ON-PETIOLE1 Coordinates Organ Determinacy and Axial Polarity in Arabidopsis by Directly Activating ASYMMETRIC LEAVES2. Plant Cell 2010, 22, 62–76. [Google Scholar] [CrossRef]
- Wang, Y.; Salasini, B.C.; Khan, M.; Devi, B.; Bush, M.; Subramaniam, R.; Hepworth, S.R. Clade I TGACG-Motif Binding Basic Leucine Zipper Transcription Factors Mediate BLADE-ON-PETIOLE-Dependent Regulation of Development. Plant Physiol. 2019, 180, 937–951. [Google Scholar] [CrossRef]
- DiGennaro, P.; Grienenberger, E.; Dao, T.Q.; Jun, J.H.; Fletcher, J.C. Peptide signaling molecules CLE5 and CLE6 affect Arabidopsis leaf shape downstream of leaf patterning transcription factors and auxin. Plant Direct 2018, 2, e00103. [Google Scholar] [CrossRef]
- Toriba, T.; Tokunaga, H.; Shiga, T.; Nie, F.; Naramoto, S.; Honda, E.; Tanaka, K.; Taji, T.; Itoh, J.-I.; Kyozuka, J. BLADE-ON-PETIOLE genes temporally and developmentally regulate the sheath to blade ratio of rice leaves. Nat. Commun. 2019, 10, 619. [Google Scholar] [CrossRef]
- Poethig, R.S. Vegetative phase change and shoot maturation in plants. Curr. Top. Dev. Biol. 2013, 105, 125–152. [Google Scholar]
- Zhang, B.; Holmlund, M.; Lorrain, S.; Norberg, M.; Bakó, L.; Fankhauser, C.; Nilsson, O. BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. eLife 2017, 6. [Google Scholar] [CrossRef]
- Ichihashi, Y.; Aguilar-Martinez, J.A.; Farhi, M.; Chitwood, D.H.; Kumar, R.; Millon, L.V.; Peng, J.; Maloof, J.N.; Sinha, N.R. Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc. Natl. Acad. Sci. USA 2014, 111, E2616–E2621. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Nakazawa, M.; Takase, T.; Manabe, K.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Matsui, M. Overexpression of LSH1, a member of an uncharacterised gene family, causes enhanced light regulation of seedling development. Plant J. 2004, 37, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Koenig, D.; Kang, J.; Yoong, F.Y.; Sinha, N. Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 2008, 18, 672–677. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).