Transcription Factors Associated with Leaf Senescence in Crops
Abstract
1. Introduction
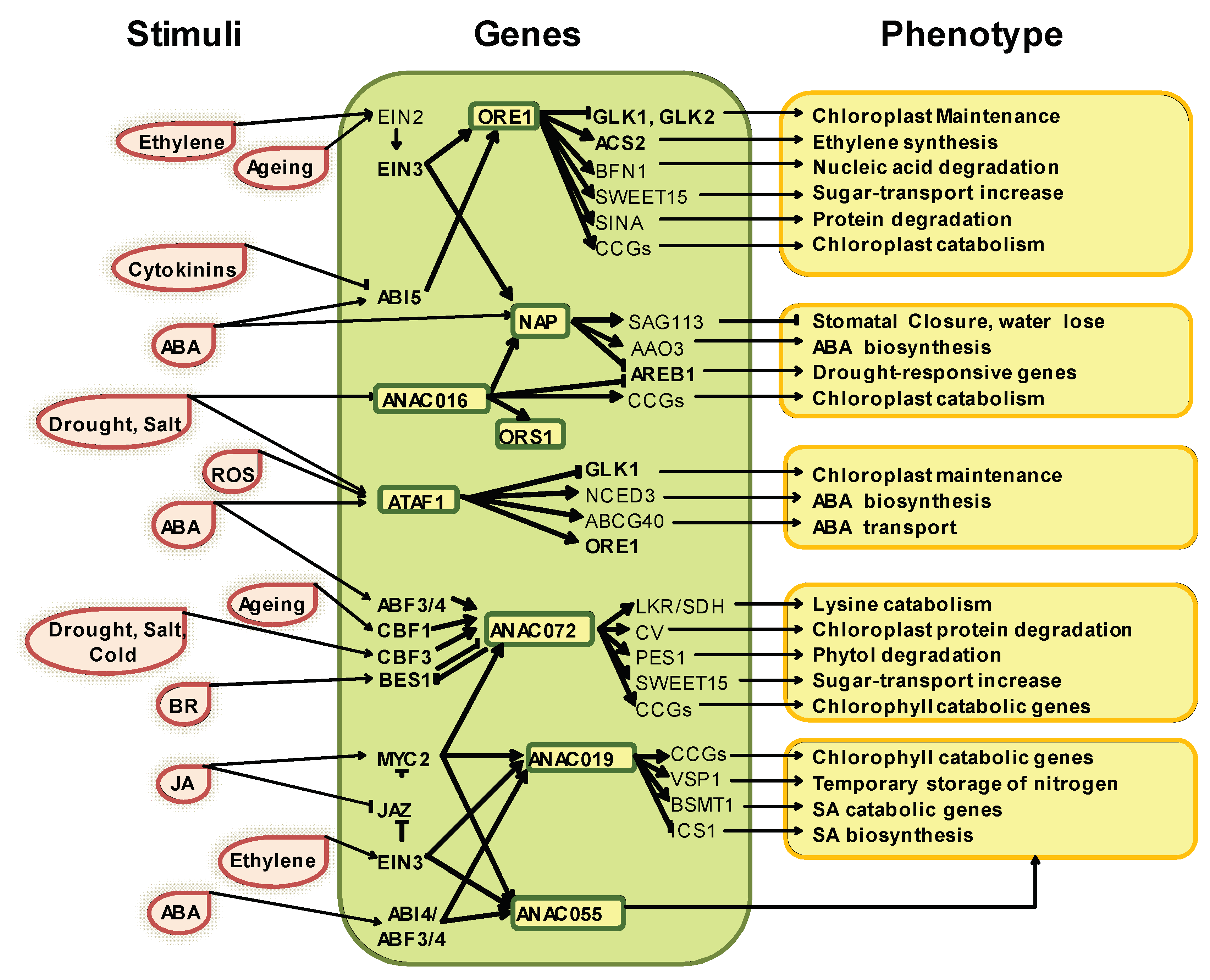
2. Senescence Regulation Network in Arabidopsis and NAC TF Contribution
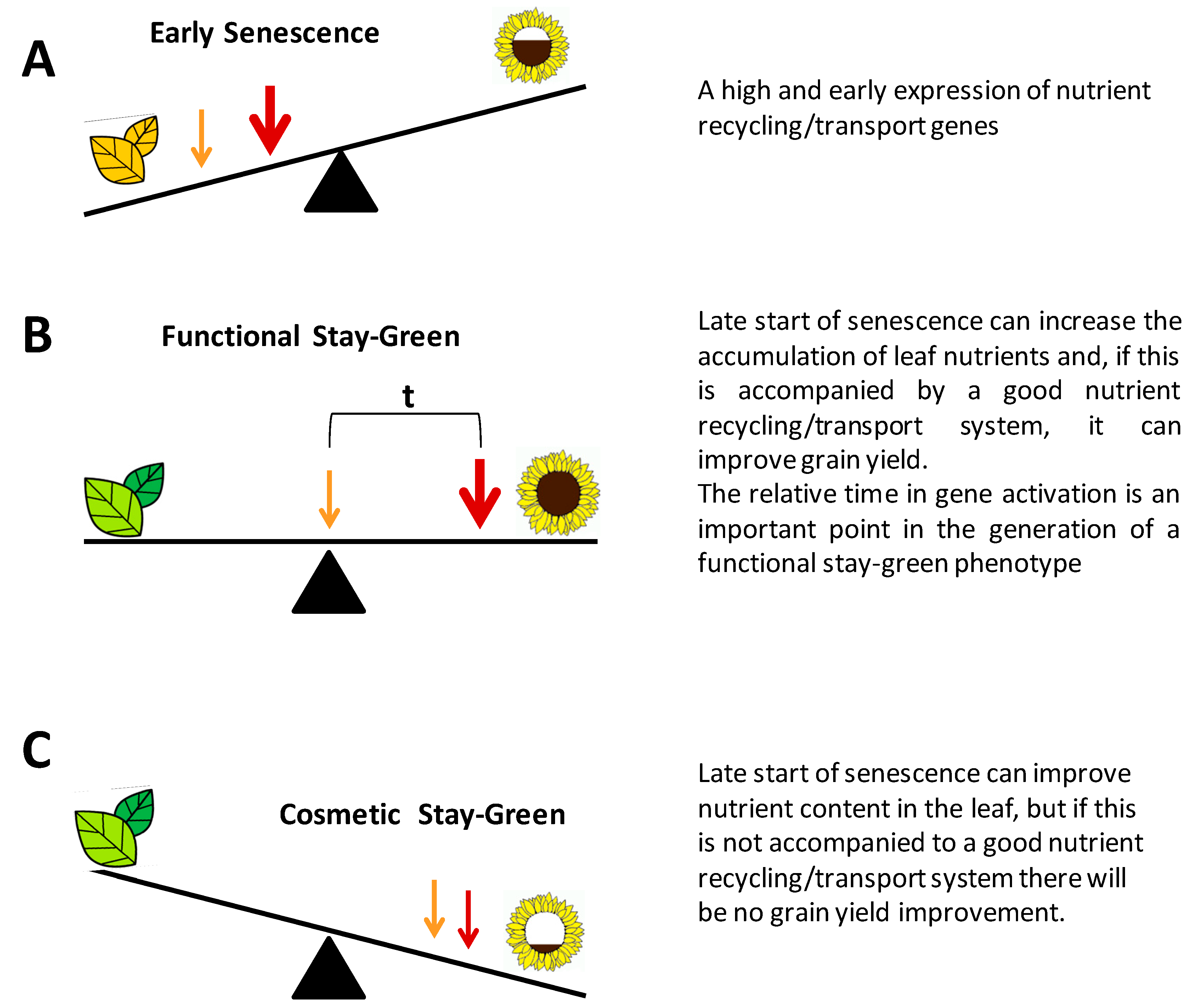
3. The Stay-Green Trait in Crops
4. Main TFs Associated with Leaf Senescence in Crops
4.1. Rice
4.2. Wheat
4.3. Barley
4.4. Maize
4.5. Sorghum
4.6. Soybean
4.7. Sunflower
4.8. Cotton
4.9. Grapevine
4.10. Ornamental Species
5. Expression Pattern and Function Integration of NACs TFs Involved in Leaf Senescence across Species
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Buchanan-Wollaston, V.; Earl, S.; Harrison, E.; Mathas, E.; Navabpour, S.; Page, T.; Pink, D. The molecular analysis of leaf senescence—A genomics approach. Plant Biotechnol. J. 2003, 1, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Wu, A.; Mueller-Roeber, B. Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal. Behav. 2010, 5, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Stoddart, J.L. Leaf senescence. Annu. Rev. Plant Physiol. 1980, 5, 83–111. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf Senescence: Systems and Dynamics Aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Huang, L.; Young, M.; Ougham, H. Evolution of plant senescence. BMC Evol. Biol. 2009, 9, 163. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M.; Rentsch, D. Uptake and Partitioning of Amino Acids and Peptides. Mol. Plant 2010, 3, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Tabbita, F.; Pearce, S.; Barneix, A.J. Breeding for increased grain protein and micronutrient content in wheat: Ten years of the GPC-B1 gene. J. Cereal Sci. 2017, 73, 183–191. [Google Scholar] [CrossRef]
- Yamaji, N.; Ma, J.F. A Transporter at the Node Responsible for Intervascular Transfer of Silicon in Rice. Plant Cell 2009, 21, 2878–2883. [Google Scholar] [CrossRef]
- Patrick, J.W.; Offler, C.E. Compartmentation of transport and transfer events in developing seeds. J. Exp. Bot. 2001, 52, 551–564. [Google Scholar] [CrossRef]
- Pottier, M.; Daubresse, C.M.; Yoshimoto, K.; Thomine, S. Autophagy as a possible mechanism for micronutrient remobilization from leaves to seeds. Front. Plant Sci. 2014, 5, 11. [Google Scholar] [CrossRef]
- Maillard, A.; DiquÃlou, S.; Billard, V.; LaÃnÃ, P.; Garnica, M.; Prudent, M.; Garcia-Mina, J.M.; Yvin, J.C.; Ourry, A. Leaf mineral nutrient remobilization during leaf senescence and modulation by nutrient deficiency. Front. Plant Sci. 2015, 6, 317. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Seo, H.S.; Koh, H.J.; et al. The Senescence-Induced Staygreen Protein Regulates Chlorophyll Degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [PubMed]
- Carrión, C.A.; Costa, M.L.; Martínez, D.E.; Mohr, C.; Humbeck, K.; Guiamet, J.J. In vivo inhibition of cysteine proteases provides evidence for the involvement of ‘senescence-associated vacuoles’ in chloroplast protein degradation during dark-induced senescence of tobacco leaves. J. Exp. Bot. 2013, 64, 4967–4980. [Google Scholar] [CrossRef] [PubMed]
- Martínez, D.E.; Costa, M.L.; Gomez, F.M.; Otegui, M.S.; Guiamet, J.J. ‘Senescence-associated vacuoles’ are involved in the degradation of chloroplast proteins in tobacco leaves. Plant J. 2008, 56, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.S.; Penfold, C.A.; Jenkins, D.; et al. High-Resolution Temporal Profiling of Transcripts during Arabidopsis Leaf Senescence Reveals a Distinct Chronology of Processes and Regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Riaño-Pachón, D.M.; Mueller-Roeber, B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 2008, 10, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Köhler, B.; Mueller-Roeber, B.; Zanor, M.I. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef]
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef]
- Gil Nam, H.; Woo, H.R.; Kim, J.H.; Lim, P.O. The Delayed Leaf Senescence Mutants of Arabidopsis, ore1, ore3, and ore9 are Tolerant to Oxidative Stress. Plant Cell Physiol. 2004, 45, 923–932. [Google Scholar]
- Matallana-Ramirez, L.P.; Rauf, M.; Farage-Barhom, S.; Dortay, H.; Xue, G.P.; Dröge-Laser, W.; Lers, A.; Balazadeh, S.; Mueller-Roeber, B. NAC Transcription Factor ORE1 and Senescence-Induced BIFUNCTIONAL NUCLEASE1 (BFN1) Constitute a Regulatory Cascade in Arabidopsis. Mol. Plant 2013, 6, 1438–1452. [Google Scholar] [CrossRef]
- Moschen, S.; Luoni, S.B.; Paniego, N.B.; Hopp, H.E.; Dosio, G.A.A.; Fernandez, P.; Heinz, R.A. Identification of candidate genes associated with leaf senescence in cultivated sunflower (Helianthus annuus L.). PLoS ONE 2014, 9, e104379. [Google Scholar] [CrossRef] [PubMed]
- Moschen, S.; Higgins, J.; Di Rienzo, J.A.; Heinz, R.A.; Paniego, N.; Fernandez, P. Network and biosignature analysis for the integration of transcriptomic and metabolomic data to characterize leaf senescence process in sunflower. BMC Bioinform. 2016, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Moschen, S.; Di Rienzo, J.A.; Higgins, J.; Tohge, T.; Watanabe, M.; Rivarola, M.; Dopazo, J.; Hopp, H.E.; Hoefgen, R.; Fernie, A.R.; et al. Integration of transcriptomic and metabolic data reveals hub transcription factors involved in drought stress response in sunflower (Helianthus annuus L.). Plant Mol. Biol. 2017, 94, 549–564. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Hong, S.H.; Kim, Y.W.; Lee, I.H.; Jun, J.H.; Phee, B.K.; Rupak, T.; Jeong, H.; Lee, Y.; Hong, B.S.; et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 4023–4036. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Nam, H.G.; Lim, P.O. ScienceDirect Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Cai, Z.; Gan, S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004, 27, 521–549. [Google Scholar] [CrossRef]
- Pruneda-Paz, J.L.; Breton, G.; Nagel, D.H.; Kang, S.E.; Bonaldi, K.; Doherty, C.J.; Ravelo, S.; Galli, M.; Ecker, J.R.; Kay, S.A. A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Rep. 2014, 8, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Podzimska-Sroka, D.; O’Shea, C.; Gregersen, P.L.; Skriver, K. NAC Transcription Factors in Senescence: From Molecular Structure to Function in Crops. Plants 2015, 4, 412–448. [Google Scholar] [CrossRef]
- Zentgraf, U.; Jobst, J.; Kolb, D.; Rentsch, D. Senescence-Related Gene Expression Profiles of Rosette Leaves of Arabidopsis thaliana: Leaf Age Versus Plant Age. Plant Biol. 2004, 6, 178–183. [Google Scholar] [CrossRef]
- Balazadeh, S.; Kwasniewski, M.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Xue, G.P.; Mueller-Roeber, B. ORS1, an H2O2-Responsive NAC Transcription Factor, Controls Senescence in Arabidopsis thaliana. Mol. Plant 2011, 4, 346–360. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive Analysis of NAC Domain Transcription Factor Gene Family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.K.; Hye, R.W.; Kim, J.; Pyung, O.L.; In, C.L.; Seung, H.C.; Hwang, D.; Hong, G.N. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef]
- Jaradat, M.R.; Feurtado, J.A.; Huang, D.; Lu, Y.; Cutler, A.J. Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 2013, 13, 192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, G.; Jia, J.; Liu, X.; Kong, X. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J. Exp. Bot. 2012, 63, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. Regulation of Apetala2/Ethylene Response Factors in Plants. Front. Plant Sci. 2017, 8, 150. [Google Scholar] [CrossRef]
- Koyama, T. The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front. Plant Sci. 2014, 5, 650. [Google Scholar] [CrossRef]
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, M.; Song, Q.; Zhao, L.; Wang, G.; Zhou, C. Identification and function analyses of senescence-associated WRKYs in wheat. Biochem. Biophys. Res. Commun. 2016, 474, 761–767. [Google Scholar] [CrossRef]
- Miao, Y.; Smykowski, A.; Zentgraf, U. A novel upstream regulator ofWRKY53transcription during leaf senescence in Arabidopsis thaliana. Plant Biol. 2008, 10, 110–120. [Google Scholar] [CrossRef]
- Ulker, B.; Mukhtar, M.S.; Somssich, I.E. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 2007, 226, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Sakuraba, Y.; Han, S.H.; Yoo, S.C.; Paek, N.C. Mutation of the Arabidopsis NAC016 Transcription Factor Delays Leaf Senescence. Plant Cell Physiol. 2013, 54, 1660–1672. [Google Scholar] [CrossRef] [PubMed]
- Garapati, P.; Xue, G.P.; Munné-Bosch, S.; Balazadeh, S. Transcription Factor ATAF1 in Arabidopsis Promotes Senescence by Direct Regulation of Key Chloroplast Maintenance and Senescence Transcriptional Cascades. Plant Physiol. 2015, 168, 1122–1139. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.I.; Asensi-Fabado, M.A.; Munne-Bosch, S.; Antonio, C.; Tohge, T.; et al. JUNGBRUNNEN1, a Reactive Oxygen Species-Responsive NAC Transcription Factor, Regulates Longevity in Arabidopsis. Plant Cell Online 2012, 24, 482–506. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.D.; Seo, P.J.; Yoon, H.K.; Park, C.M. The Arabidopsis NAC Transcription Factor VNI2 Integrates Abscisic Acid Signals into Leaf Senescence via the COR/RD Genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef]
- Hunter, D.A.; Yoo, S.D.; Butcher, S.M.; McManus, M.T. Expression of 1-Aminocyclopropane-1-Carboxylate Oxidase during Leaf Ontogeny in White Clover. Plant Physiol. 2002, 120, 131–142. [Google Scholar] [CrossRef]
- Grbić, V.; Bleecker, A.B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995, 8, 595–602. [Google Scholar] [CrossRef]
- Jibran, R.; Hunter, D.A.; Dijkwel, P.P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013, 82, 547–561. [Google Scholar] [CrossRef]
- Jing, H.C.; Schippers, J.H.M.; Hille, J.; Dijkwel, P.P. Ethylene-induced leaf senescence depends on age-related changes and OLD genes in Arabidopsis. J. Exp. Bot. 2005, 56, 2915–2923. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, S.J.; Lee, I.H.; Chu, H.; Penfold, C.A.; Kim, J.H.; Buchanan-Wollaston, V.; Gil Nam, H.; Woo, H.R.; Lim, P.O. Comparative transcriptome analysis in Arabidopsis ein2/ore3 and ahk3/ore12 mutants during dark-induced leaf senescence. J. Exp. Bot. 2018, 69, 3023–3036. [Google Scholar] [CrossRef] [PubMed]
- Rauf, M.; Arif, M.; Dortay, H.; Matallana-Ramírez, L.P.; Waters, M.T.; Gil Nam, H.; Lim, P.O.; Mueller-Roeber, B.; Balazadeh, S. ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Rep. 2013, 14, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.N.; Zhong, S.; Weirauch, M.T.; Hon, G.; Pelizzola, M.; Li, H.; Huang, S.S.C.; Schmitz, R.J.; Urich, M.; Kuo, D.; et al. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2013, 2, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Hirayama, T.; Roman, G.; Nourizadeh, S.; Ecker, J.R. EIN2, a Bifunctional Transducer of Ethylene and Stress Responses in Arabidopsis. Science 1999, 284, 2148–2152. [Google Scholar] [CrossRef] [PubMed]
- Lei, G.; Shen, M.; Li, Z.-G.; Zhang, B.; Duan, K.-X.; Wang, N.; Cao, Y.-R.; Zhang, W.-K.; Ma, B.; Ling, H.-Q.; et al. EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ. 2011, 34, 1678–1692. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.L.; Chen, N.Z.; An, R.; Su, Z.; Qi, B.S.; Ren, F.; Chen, J.; Wang, X.C. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. 2007, 63, 289–305. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Li, K.; Sun, F.; Hu, H.; Zhao, Y.; Han, C.; Zhang, W.; Duan, Y.; Liu, M.; et al. Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol. Biol. 2007, 64, 633–644. [Google Scholar] [CrossRef]
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of Abscisic Acid Synthesis and Signaling Mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Asad, M.A.U.; Zakari, S.A.; Zhao, Q.; Zhou, L.; Ye, Y.; Cheng, F. Abiotic Stresses Intervene with ABA Signaling to Induce Destructive Metabolic Pathways Leading to Death: Premature Leaf Senescence in Plants. Int. J. Mol. Sci. 2019, 20, 256. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Kim, Y.S.; Han, S.H.; Lee, B.D.; Paek, N.C. The Arabidopsis Transcription Factor NAC016 Promotes Drought Stress Responses by Repressing AREB1 Transcription through a Trifurcate Feed-Forward Regulatory Loop Involving NAP. Plant Cell 2015, 27, 1771–1787. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Worley, E.; Udvardi, M. A NAP-AAO3 Regulatory Module Promotes Chlorophyll Degradation via ABA Biosynthesis in Arabidopsis Leaves. Plant Cell 2014, 26, 4862–4874. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Aoki, H.; Koiwai, H.; Kamiya, Y.; Nambara, E.; Koshiba, T. Comparative Studies on the Arabidopsis Aldehyde Oxidase (AAO) Gene Family Revealed a Major Role of AAO3 in ABA Biosynthesis in Seeds. Plant Cell Physiol. 2004, 45, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Gan, S.S. An Abscisic Acid-AtNAP Transcription Factor-SAG113 Protein Phosphatase 2C Regulatory Chain for Controlling Dehydration in Senescing Arabidopsis Leaves. Plant Physiol. 2011, 158, 961–969. [Google Scholar] [CrossRef]
- Guan, C.; Wang, X.; Feng, J.; Hong, S.; Liang, Y.; Ren, B.; Zuo, J. Cytokinin Antagonizes Abscisic Acid-Mediated Inhibition of Cotyledon Greening by Promoting the Degradation of ABSCISIC ACID INSENSITIVE5 Protein in Arabidopsis. Plant Physiol. 2014, 164, 1515–1526. [Google Scholar] [CrossRef]
- Schippers, J.H.M. Science Direct Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 2015, 27, 77–83. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Han, S.H.; Lee, S.H.; Hörtensteiner, S.; Paek, N.C. Arabidopsis NACO16 promotes chlorophyll breakdown by directly upregulating STAYGREEN1 transcription. Plant Cell Rep. 2016, 35, 155–166. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Seok, H.Y.; Woo, D.H.; Nguyen, L.V.; Tran, H.T.; Tarte, V.N.; Mehdi SM, M.; Lee, S.Y.; Moon, Y.H. Arabidopsis AtNAP functions as a negative regulator via repression of AREB1 in salt stress response. Planta 2017, 245, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Lindemose, S.; De Masi, F.; Reimer, J.J.; Nielsen, M.; Perera, V.; Workman, C.T.; Turck, F.; Grant, M.R.; Mundy, J.; et al. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 2013, 3, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Hickman, R.; Hill, C.; Penfold, C.A.; Breeze, E.; Bowden, L.; Moore, J.D.; Zhang, P.; Jackson, A.; Cooke, E.; Bewicke-Copley, F.; et al. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 2013, 75, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Lindemose, S.; Jensen, M.K.; Van De Velde, J.; O’Shea, C.; Heyndrickx, K.S.; Workman, C.T.; Vandepoele, K.; Skriver, K.; De Masi, F. A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res. 2014, 42, 7681–7693. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; Jiang, H.; Li, C.B.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Li, C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008, 18, 756–767. [Google Scholar] [CrossRef]
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015, 84, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Zhou, X.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Spivey, N.W.; Zeng, W.; Liu, P.P.; Fu, Z.Q.; Klessig, D.F.; He, S.Y.; Dong, X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef]
- Schweizer, F.; Bodenhausen, N.; Lassueur, S.; Masclaux, F.G.; Reymond, P. Differential Contribution of Transcription Factors to Arabidopsis thaliana Defense Against Spodoptera littoralis. Front. Plant Sci. 2013, 4, 1–12. [Google Scholar] [CrossRef]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.S.P.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef]
- Tran, L.S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and Functional Analysis of Arabidopsis Stress-Inducible NAC Transcription Factors That Bind to a Drought-Responsive cis-Element in the early responsive to dehydration stress 1 Promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, M.; Yan, Y.; Liu, X.; Li, L. Dual Function of NAC072 in Gene Regulation in Arabidopsis. Front. Plant Sci. 2016, 7, 1075. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.X.; Sun, Y.; Burlingame, A.L.; Wang, Z.Y. Brassinosteroid signal transduction from cell surface receptor kinases to nuclear transcription factors. Nature 2009, 11, 1254–1260. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.; Kwon, S.I.; Choe, S. Antagonistic Regulation of Arabidopsis Growth by Brassinosteroids and Abiotic Stresses. Mol. Cells 2014, 37, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.Y.; Li, L.; et al. RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 2017, 8, 14573. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 2016, 33, 147–156. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Hickman, R.; Van Verk, M.C.; Van Dijken, A.J.H.; Mendes, M.P.; Vroegop-Vos, I.A.; Caarls, L.; Steenbergen, M.; Van der Nagel, I.; Wesselink, G.J.; Jironkin, A.; et al. Architecture and Dynamics of the Jasmonic Acid Gene Regulatory Network. Plant Cell 2017, 29, 2086–2105. [Google Scholar] [CrossRef]
- Kim, J.; Chang, C.; Tucker, M.L. To grow old: Regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 2015, 6, 1–7. [Google Scholar] [CrossRef]
- Li, Z.; Peng, J.; Wen, X.; Guo, H. ETHYLENE-INSENSITIVE3 Is a Senescence-Associated Gene That Accelerates Age-Dependent Leaf Senescence by Directly Repressing miR164 Transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gao, J.; Yao, L.; Ren, G.; Zhu, X.; Gao, S.; Qiu, K.; Zhou, X.; Kuai, B. The role of ANAC072 in the regulation of chlorophyll degradation during age-and dark-induced leaf senescence. Plant Cell Rep. 2016, 35, 1729–1741. [Google Scholar] [CrossRef] [PubMed]
- Kamranfar, I.; Xue, G.P.; Tohge, T.; Sedaghatmehr, M.; Fernie, A.R.; Balazadeh, S.; Mueller-Roeber, B. Transcription factor RD26 is a key regulator of metabolic reprogramming during dark-induced senescence. New Phytol. 2018, 218, 1543–1557. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wei, J.; Wang, M.; Shi, Z.; Gong, W.; Zhang, M. The Crystal Structure of Arabidopsis VSP1 Reveals the Plant Class C-Like Phosphatase Structure of the DDDD Superfamily of Phosphohydrolases. PLoS ONE 2012, 7, e49421. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, J.H.; Kim, J.; Kim, J.J.; Hong, S.; Kim, J.; Kim, J.H.; Woo, H.R.; Hyeon, C.; Lim, P.O.; et al. Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4930–E4939. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, C.M. Signaling linkage between environmental stress resistance and leaf senescence in Arabidopsis. Plant Signal. Behav. 2011, 6, 1564–1566. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, Zn, and Fe content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef]
- Thomas, H.; Smart, C.M. Crops that stay green. Ann. Appl. Biol. 1993, 123, 193–219. [Google Scholar] [CrossRef]
- Moschen, S.; Bengoa Luoni, S.; Di Rienzo, J.A.; Caro, M.D.; Tohge, T.; Watanabe, M.; Hollmann, J.; González, S.; Rivarola, M.; García-García, F.; et al. Integrating transcriptomic and metabolomic analysis to understand natural leaf senescence in sunflower. Plant Biotechnol. J. 2016, 14, 719–734. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wang, K.J.; Jiang, G.M.; Liu, M.Z.; Gao, L.M. Photosynthetic rate and yield formation in different maize hybrids. Biol. Plant. 2007, 51, 165–168. [Google Scholar] [CrossRef]
- Zhao, D.; Derkx, A.P.; Liu, D.C.; Buchner, P.; Hawkesford, M.J. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 2015, 17, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Cruickshank, A.W.; Borrell, A.K.; Henzell, R.; Jordan, D.R.; Hunt, C.H. The Relationship between the Stay-Green Trait and Grain Yield in Elite Sorghum Hybrids Grown in a Range of Environments. Crop. Sci. 2012, 52, 1153–1161. [Google Scholar]
- Gustafson, P.; Lee, S.H.; Fu, J.D.; Yan, Y.F.; Kim, M.Y.; Lee, B.W. Population-specific quantitative trait loci mapping for functional stay-green trait in rice (Oryza sativa L.). Genome 2011, 54, 235–243. [Google Scholar]
- Hafsi, M.; Mechmeche, W.; Bouamama, L.; Djekoune, A.; Zaharieva, M.; Monneveux, P. Flag Leaf Senescence, as Evaluated by Numerical Image Analysis, and its Relationship with Yield under Drought in Durum Wheat. J. Agron. Crop. Sci. 2000, 185, 275–280. [Google Scholar] [CrossRef]
- Kusaba, M.; Tanaka, A.; Tanaka, R. Stay-green plants: What do they tell us about the molecular mechanism of leaf senescence. Photosynth. Res. 2013, 117, 221–234. [Google Scholar] [CrossRef]
- De La Vega, A.; Cantore, M.; Sposaro, M.; Trapani, N.; Pereira, M.L.; Hall, A. Canopy stay-green and yield in non-stressed sunflower. Field Crop. Res. 2011, 121, 175–185. [Google Scholar] [CrossRef]
- Luquez, V.M.; Guiamét, J.J. The stay green mutations d1 and d2 increase water stress susceptibility in soybeans. J. Exp. Bot. 2002, 53, 1421–1428. [Google Scholar] [CrossRef]
- Dohleman, F.G.; Long, S.P. More Productive Than Maize in the Midwest: How Does Miscanthus Do It? Plant Physiol. 2009, 150, 2104–2115. [Google Scholar] [CrossRef]
- Zhao, Y.; Qiang, C.; Wang, X.; Chen, Y.; Deng, J.; Jiang, C.; Sun, X.; Chen, H.; Li, J.; Piao, W.; et al. New alleles for chlorophyll content and stay-green traits revealed by a genome wide association study in rice (Oryza sativa). Sci. Rep. 2019, 9, 2541. [Google Scholar] [CrossRef]
- Wang, N.; Zheng, Y.; Xin, H. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013, 32, 61–75. [Google Scholar] [CrossRef]
- Borrill, P.; Harrington, S.A.; Simmonds, J.; Uauy, C. Identification of transcription factors regulating senescence in wheat through gene regulatory network modelling. bioRxiv 2018, 180, 456749. [Google Scholar] [CrossRef] [PubMed]
- Leng, Y.; Ye, G.; Zeng, D. Genetic Dissection of Leaf Senescence in Rice. Int. J. Mol. Sci. 2017, 18, 2686. [Google Scholar] [CrossRef]
- Khush, G.S. What it will take to Feed 5.0 Billion Rice consumers in 2030. Plant Mol. Biol. 2005, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Y.; Lv, B.; Li, J.; Luo, L.; Lu, S.; Zhang, X.; Ma, H.; Ming, F. The NAC Family Transcription Factor OsNAP Confers Abiotic Stress Response Through the ABA Pathway. Plant Cell Physiol. 2014, 55, 604–619. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Huang, W.; Liu, L.; Chen, T.; Zhou, F.; Lin, Y. Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 2013, 13, 132. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, R.; Chen, X.; Wu, X.; Ming, F. Characterization of OsNAP from Oryza sativa L. and its application in molecular breeding. J. Fudan Univ. Nat. Sci. 2014, 51, 507–514. [Google Scholar]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef]
- Ma, B.; He, S.J.; Duan, K.X.; Yin, C.C.; Chen, H.; Yang, C.; Xiong, Q.; Song, Q.X.; Lu, X.; Chen, H.W.; et al. Identification of Rice Ethylene-Response Mutants and Characterization of MHZ7/OsEIN2 in Distinct Ethylene Response and Yield Trait Regulation. Mol. Plant 2013, 6, 1830–1848. [Google Scholar] [CrossRef]
- Lee, S.H.; Sakuraba, Y.; Lee, T.; Kim, K.W.; An, G.; Lee, H.Y.; Paek, N.C. Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 2015, 57, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Ricachenevsky, F.K.; Menguer, P.K.; Sperotto, R.A. kNACking on heaven’s door: How important are NAC transcription factors for leaf senescence and Fe/Zn remobilization to seeds? Front. Plant Sci. 2013, 4, 226. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.J.; Jung, H.; Choi, Y.D.; Kim, J.-K. Genome-wide analyses of direct target genes of four rice NAC-domain transcription factors involved in drought tolerance. BMC Genom. 2018, 19, 40. [Google Scholar] [CrossRef] [PubMed]
- El Mannai, Y.; Akabane, K.; Hiratsu, K.; Satoh-Nagasawa, N.; Wabiko, H. The NAC Transcription Factor Gene OsY37 (ONAC011) Promotes Leaf Senescence and Accelerates Heading Time in Rice. Int. J. Mol. Sci. 2017, 18, 2165. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Piao, W.; Lim, J.H.; Han, S.H.; Kim, Y.S.; An, G.; Paek, N.C. Rice ONAC106 Inhibits Leaf Senescence and Increases Salt Tolerance and Tiller Angle. Plant Cell Physiol. 2015, 56, 2325–2339. [Google Scholar] [CrossRef]
- Sperotto, R.A.; Ricachenevsky, F.K.; de Abreu Waldow, V.; Fett, J.P. Iron biofortification in rice: It’s a long way to the top. Plant Sci. 2012, 190, 24–39. [Google Scholar] [CrossRef] [PubMed]
- Kutman, U.B.; Yildiz, B.; Cakmak, I. Effect of nitrogen on uptake, remobilization and partitioning of zinc and iron throughout the development of durum wheat. Plant Soil 2011, 342, 149–164. [Google Scholar] [CrossRef]
- Ortiz, R.; Braun, H.-J.; Crossa, J.; Crouch, J.H.; Davenport, G.; Dixon, J.; Dreisigacker, S.; Duveiller, E.; He, Z.; Huerta, J.; et al. Wheat genetic resources enhancement by the International Maize and Wheat Improvement Center (CIMMYT). Genet. Resour. Crop Evol. 2008, 55, 1095–1140. [Google Scholar] [CrossRef]
- Aksoy, M.A.; Beghin, J.C. Global Agricultural Trade and Developing Countries; The World Bank: Washington, DC, USA, 2004; ISBN 978-0-8213-5863-4. [Google Scholar]
- Zörb, C.; Langenkämper, G.; Betsche, T.; Niehaus, K.; Barsch, A. Metabolite Profiling of Wheat Grains (Triticum aestivum L.) from Organic and Conventional Agriculture. J. Agric. Food Chem. 2006, 54, 8301–8306. [Google Scholar] [CrossRef]
- Langenkämper, G.; Zörb, C.; Seifert, M.; Mäder, P.; Fretzdorff, B.; Betsche, T. Nutritional quality of organic and conventional wheat. J. Appl. Bot. Food Qual. 2006, 80, 150–154. [Google Scholar]
- Shewry, P.R. Wheat. J. Exp. Bot. 2009, 60, 1537–1553. [Google Scholar] [CrossRef] [PubMed]
- Saidi, M.N.; Mergby, D.; Brini, F. Identification and expression analysis of the NAC transcription factor family in durum wheat (Triticum turgidum L. ssp. durum). Plant Physiol. Biochem. 2017, 112, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Brevis, J.C.; Uauy, C.; Dubcovsky, J. The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. J. Exp. Bot. 2006, 57, 2785–2794. [Google Scholar]
- Pearce, S.; Tabbita, F.; Cantu, D.; Buffalo, V.; Avni, R.; Vazquez-Gross, H.; Zhao, R.; Conley, C.J.; Distelfeld, A.; Dubcovksy, J. Regulation of Zn and Fe transporters by the GPC1 gene during early wheat monocarpic senescence. BMC Plant Biol. 2014, 14, 368. [Google Scholar] [CrossRef] [PubMed]
- Avni, R.; Zhao, R.; Pearce, S.; Jun, Y.; Uauy, C.; Tabbita, F.; Fahima, T.; Slade, A.; Dubcovsky, J.; Distelfeld, A. Functional characterization of GPC-1 genes in hexaploid wheat. Planta 2014, 239, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, M.W.; Gregersen, P.L. Members of the barley NAC transcription factor gene family show differential co-regulation with senescence-associated genes during senescence of flag leaves. J. Exp. Bot. 2014, 65, 4009–4022. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, M.W.; Holm, P.B.; Gregersen, P.L. Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggests conserved functions compared to both monocots and dicots. BMC Res. Notes 2011, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, M.W.; Matthewman, C.; Podzimska-Sroka, D.; O’Shea, C.; Lindemose, S.; Møllegaard, N.E.; Holme, I.B.; Hebelstrup, K.; Skriver, K.; Gregersen, P.L. Barley plants over-expressing the NAC transcription factor gene HvNAC005 show stunting and delay in development combined with early senescence. J. Exp. Bot. 2016, 67, 5259–5273. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, J.; Gregersen, P.L.; Krupinska, K. Identification of predominant genes involved in regulation and execution of senescence-associated nitrogen remobilization in flag leaves of field grown barley. J. Exp. Bot. 2014, 65, 3963–3973. [Google Scholar] [CrossRef]
- Foyer, C.H.; Karpinska, B.; Krupinska, K. The functions of WHIRLY1 and REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 in cross tolerance responses in plants: A hypothesis. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130226. [Google Scholar] [CrossRef]
- Janack, B.; Sosoi, P.; Krupinska, K.; Humbeck, K. Knockdown of WHIRLY1 Affects Drought Stress-Induced Leaf Senescence and Histone Modifications of the Senescence-Associated Gene HvS40. Plants 2016, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Kucharewicz, W.; Distelfeld, A.; Bilger, W.; Müller, M.; Munné-Bosch, S.; Hensel, G.; Krupinska, K. Acceleration of leaf senescence is slowed down in transgenic barley plants deficient in the DNA/RNA-binding protein WHIRLY1. J. Exp. Bot. 2017, 68, 983–996. [Google Scholar] [CrossRef] [PubMed]
- FAO. FAOSTAT, Production. Available online: http://faostat3.fao.org/browse/Q/QC/E (accessed on 18 January 2019).
- Borrell, A.; Hammer, G.; Oosterom, E. Stay-green: A consequence of the balance between supply and demand for nitrogen during grain filling? Ann. Appl. Biol. 2001, 138, 91–95. [Google Scholar] [CrossRef]
- Nasielski, J.; Earl, H.; Deen, B.; Deen, B. Luxury Vegetative Nitrogen Uptake in Maize Buffers Grain Yield under Post-silking Water and Nitrogen Stress: A Mechanistic Understanding. Front. Plant Sci. 2019, 10, 318. [Google Scholar] [CrossRef] [PubMed]
- Woli, K.P.; Sawyer, J.E.; Boyer, M.J.; Abendroth, L.J.; Elmore, R.W. Corn Era Hybrid Nutrient Concentration and Accumulation of Secondary and Micronutrients. Agron. J. 2019, 111, 1604–1620. [Google Scholar] [CrossRef]
- Tollenaar, M.; Ahmadzadeh, A.; Lee, E.A. Physiological Basis of Heterosis for Grain Yield in Maize. Crop. Sci. 2004, 44, 2086–2094. [Google Scholar] [CrossRef]
- Ma, B.; Dwyer, L. Nitrogen uptake and use of two contrasting maize hybrids differing in leaf senescence. Plant Soil 1998, 199, 283–291. [Google Scholar] [CrossRef]
- Bekavac, G.; Purar, B.; Stojaković, M.; Jocković, D.; Ivanović, M.; Nastasić, A. Genetic analysis of stay-green trait in broad-based maize populations. Cereal Res. Commun. 2007, 35, 31–41. [Google Scholar] [CrossRef]
- Rajcan, I.; Tollenaar, M. Source: Sink ratio and leaf senescence in maize: II. Nitrogen metabolism during grain filling. Field Crop. Res. 1999, 60, 255–265. [Google Scholar] [CrossRef]
- Mu, X.; Chen, Q.; Chen, F.; Yuan, L.; Mi, G. Dynamic remobilization of leaf nitrogen components in relation to photosynthetic rate during grain filling in maize. Plant Physiol. Biochem. 2018, 129, 27–34. [Google Scholar] [CrossRef]
- Li, P.; Cao, W.; Fang, H.; Xu, S.; Yin, S.; Zhang, Y.; Lin, D.; Wang, J.; Chen, Y.; Xu, C.; et al. Transcriptomic Profiling of the Maize (Zea mays L.) Leaf Response to Abiotic Stresses at the Seedling Stage. Front. Plant Sci. 2017, 8, 290. [Google Scholar] [CrossRef]
- Shiriga, K.; Sharma, R.; Kumar, K.; Yadav, S.K.; Hossain, F.; Thirunavukkarasu, N. Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene 2014, 2, 407–417. [Google Scholar] [CrossRef]
- Zhang, W.Y.; Xu, Y.C.; Li, W.L.; Yang, L.; Yue, X.; Zhang, X.S.; Zhao, X.Y. Transcriptional Analyses of Natural Leaf Senescence in Maize. PLoS ONE 2014, 9, e115617. [Google Scholar] [CrossRef]
- Zhang, J.; Fengler, K.A.; Van Hemert, J.L.; Gupta, R.; Mongar, N.; Sun, J.; Allen, W.B.; Wang, Y.; Weers, B.; Mo, H.; et al. Identification and characterization of a novel stay-green QTL that increases yield in maize. Plant Biotechnol. J. 2019, pbi.13139. [Google Scholar] [CrossRef]
- Campos, H.; Cooper, M.; Habben, J.; Edmeades, G.; Schussler, J. Improving drought tolerance in maize: A view from industry. Field Crop. Res. 2004, 90, 19–34. [Google Scholar] [CrossRef]
- Calviño, M.; Messing, J. Sweet sorghum as a model system for bioenergy crops. Curr. Opin. Biotechnol. 2012, 23, 323–329. [Google Scholar] [CrossRef]
- Wu, X.Y.; Hu, W.J.; Xia, Y.; Zhao, Y.; Wang, L.D.; Zhang, L.M.; Luo, H.; Luo, J.C.; Jing, H.C. Transcriptome profiling of developmental leaf senescence in sorghum (Sorghum bicolor). Plant Mol. Biol. 2016, 92, 555–580. [Google Scholar] [CrossRef]
- Pettersson, D.; Pontoppi, K. Soybean Meal and The Potential for Upgrading Its Feeding Value by Enzyme Supplementation. Soybean Bio Act. Compd. 2013, 13, 288–307. [Google Scholar]
- Patil, G.; Mian, R.; Vuong, T.; Pantalone, V.; Song, Q.; Chen, P.; Shannon, G.J.; Carter, T.C.; Nguyen, H.T. Molecular mapping and genomics of soybean seed protein: A review and perspective for the future. Theor. Appl. Genet. 2017, 130, 1975–1991. [Google Scholar] [CrossRef]
- American Soybean Association. SoyStats. Available online: http://www.soystats.com (accessed on 1 March 2019).
- Warrington, C.V.; Abdel-Haleem, H.; Hyten, D.L.; Cregan, P.B.; Orf, J.H.; Killam, A.S.; Bajjalieh, N.; Li, Z.; Boerma, H.R. QTL for seed protein and amino acids in the Benning × Danbaekkong soybean population. Theor. Appl. Genet. 2015, 128, 839–850. [Google Scholar] [CrossRef]
- Melo, B.P.; Fraga, O.T.; Silva, J.C.; Ferreira, D.O.; Brustolini, O.J.; Carpinetti, P.A.; Machado, J.P.; Reis, P.A.; Fontes, E.P. Revisiting the Soybean GmNAC Superfamily. Front. Plant Sci. 2018, 9, 1–22. [Google Scholar] [CrossRef]
- Meng, Q.; Zhang, C.; Gai, J.; Yu, D. Molecular cloning, sequence characterization and tissue-specific expression of six NAC-like genes in soybean (Glycine max (L.) Merr.). J. Plant Physiol. 2007, 164, 1002–1012. [Google Scholar] [CrossRef]
- Pinheiro, G.L.; Marques, C.S.; Costa, M.D.; Reis, P.A.; Alves, M.S.; Carvalho, C.M.; Fietto, L.G.; Fontes, E.P. Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 2009, 444, 10–23. [Google Scholar] [CrossRef]
- Faria, J.A.; Ab Reis, P.; Reis, M.T.; Rosado, G.L.; Pinheiro, G.L.; Mendes, G.C.; Fontes, E.P. The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress- and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol. 2011, 11, 129. [Google Scholar] [CrossRef]
- Reis, P.A.B.; Carpinetti, P.A.; Freitas, P.P.; Santos, E.G.; Camargos, L.F.; Oliveira, I.H.; Silva, J.C.F.; Carvalho, H.H.; Dal-Bianco, M.; Soares-Ramos, J.R.; et al. Functional and regulatory conservation of the soybean ER stress-induced DCD/NRP-mediated cell death signaling in plants. BMC Plant Biol. 2016, 16, 156. [Google Scholar] [CrossRef]
- Irsigler, A.S.; Costa, M.D.; Zhang, P.; Ab Reis, P.; Dewey, R.E.; Boston, R.S.; Fontes, E.P. Expression profiling on soybean leaves reveals integration of ER- and osmotic-stress pathways. BMC Genom. 2007, 8, 431. [Google Scholar] [CrossRef]
- Pimenta, M.R.; Alves, R.; Silva, P.A.; Mendes, G.C.; Brustolini, B.; Caetano, D.N.; Paulo, J.; Machado, B.; Jose, O.; Silva, C.F.; et al. The Stress-Induced Soybean NAC Transcription Factor GmNAC81 Plays a Positive Role in Developmentally Programmed Leaf Senescence. Plant Cell Physiol. 2016, 57, 1098–1114. [Google Scholar] [CrossRef]
- Mendes, G.C.; Reis, P.A.B.; Calil, I.P.; Carvalho, H.H.; Aragão, F.J.L.; Fontes, E.P.B. GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc. Natl. Acad. Sci. USA 2013, 110, 19627–19632. [Google Scholar] [CrossRef]
- Carvalho, H.H.; Silva, P.A.; Mendes, G.C.; Brustolini, O.J.; Pimenta, M.R.; Gouveia, B.C.; Valente, M.A.; Ramos, H.J.; Soares-Ramos, J.R.; Fontes, E.P. The Endoplasmic Reticulum Binding Protein BiP Displays Dual Function in Modulating Cell Death Events. Plant Cell Physiol. 2014, 164, 654–670. [Google Scholar] [CrossRef]
- Fernandez, P.; Di Rienzo, J.; Fernandez, L.; Hopp, H.E.; Paniego, N.; Heinz, R.A. Transcriptomic identification of candidate genes involved in sunflower responses to chilling and salt stresses based on cDNA microarray analysis. BMC Plant Biol. 2008, 8, 11. [Google Scholar] [CrossRef]
- Lavaud, Y.; Dosio, G.A.A.; González, L.M.; Aguirrezábal, L.A.N.; Izquierdo, N.G.; Andrade, F.H. Intercepted Solar Radiation during Seed Filling Determines Sunflower Weight per Seed and Oil Concentration. Crop. Sci. 2003, 43, 152–161. [Google Scholar]
- Dosio, G.A.; Aguirreza´bal, L.A.; Pereyra, V.R.; Andrade, F.H. Solar Radiation Intercepted during Seed Filling and Oil Production in Two Sunflower Hybrids. Crop. Sci. 2000, 40, 1637–1644. [Google Scholar] [CrossRef]
- Hall, A.J.; Connor, D.J.; Whitfield, D.M. Contribution of pre-anthesis assimilates to grain-filling in irrigated and water-stressed sunflower crops I. Estimates using labelled carbon. F. Crop. Res. 1989, 20, 95–112. [Google Scholar] [CrossRef]
- Fernandez, P.; Soria, M.; Blesa, D.; DiRienzo, J.; Moschen, S.; Rivarola, M.; Clavijo, B.J.; Gonzalez, S.; Peluffo, L.; Príncipi, D.; et al. Development, Characterization and Experimental Validation of a Cultivated Sunflower (Helianthus annuus L.) Gene Expression Oligonucleotide Microarray. PLoS ONE 2012, 7, e45899. [Google Scholar] [CrossRef]
- Agüera, E.; Cabello, P.; De La Haba, P. Induction of leaf senescence by low nitrogen nutrition in sunflower (Helianthus annuus) plants. Physiol. Plant. 2010, 138, 256–267. [Google Scholar] [CrossRef]
- De La Mata, L.; Cabello, P.; De La Haba, P.; Agüera, E. Growth under elevated atmospheric CO2 concentration accelerates leaf senescence in sunflower (Helianthus annuus L.) plants. J. Plant Physiol. 2012, 169, 1392–1400. [Google Scholar] [CrossRef]
- Manavella, P.A.; Arce, A.L.; Dezar, C.A.; Bitton, F.; Renou, J.-P.; Crespi, M.; Chan, R.L. Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb-4 transcription factor. Plant J. 2006, 48, 125–137. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Page, T.; Harrison, E.; Breeze, E.; Lim, P.O.; Gil Nam, H.; Lin, J.F.; Wu, S.H.; Swidzinski, J.; Ishizaki, K.; Leaver, C.J.; et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005, 42, 567–585. [Google Scholar]
- Gialdi, A.L.; Moschen, S.; Villán, C.; Fernández, M.L.; Maldonado, S.; Paniego, N.; Heinz, R.; Fernandez, P.; López, A. Identification and characterization of contrasting sunflower genotypes to early leaf senescence process combining molecular and physiological studies (Helianthus annuus L.). Plant Sci. 2016, 250, 40–50. [Google Scholar] [CrossRef][Green Version]
- Dong, H.; Li, W.J.; Tang, W.; Zhang, D.M. Research Progress in Physiological Premature Senescence in Cotton. Cotton Sci. 2010, 3, 15, (In Chinese with English Abstract). [Google Scholar]
- Liu, W.; Zhang, W.; Zheng, N.; Zhai, W.; Qi, F. Study of Cotton Leaf Senescence Induced by Alternaria Alternata Infection. In Advanced Structural Safety Studies; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Lin, M.; Pang, C.; Fan, S.; Song, M.; Wei, H.; Yu, S. Global analysis of the Gossypium hirsutum L. Transcriptome during leaf senescence by RNA-Seq. BMC Plant Biol. 2015, 15, 43. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Ma, J.; Li, L.; Fan, S.; Guo, Y.; Song, M.; Wei, H.; Pang, C.; Yu, S. GhNAC12, a neutral candidate gene, leads to early aging in cotton (Gossypium hirsutum L). Gene 2016, 576, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Demura, T.; Ye, Z.H. SND1, a NAC Domain Transcription Factor, Is a Key Regulator of Secondary Wall Synthesis in Fibers of Arabidopsis. Plant Cell Online 2006, 18, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Bibi, N.; Gan, S.; Li, F.; Yuan, S.; Ni, M.; Wang, M.; Shen, H.; Wang, X. A novel NAP member GhNAP is involved in leaf senescence in Gossypium hirsutum. J. Exp. Bot. 2015, 66, 4669–4682. [Google Scholar] [CrossRef] [PubMed]
- Elasad, M.; Ondati, E.; Wei, H.; Wang, H.; Su, J.; Fan, S.; Pang, C.; Yu, S. Functional analysis of nine cotton genes related to leaf senescence in Gossypium hirsutum L. Physiol. Mol. Biol. Plants 2018, 24, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Dou, L.; Guo, Y.; Wang, H.; Li, L.; Wang, C.; Ma, L.; Wei, H.; Yu, S. The WRKY transcription factor GhWRKY27 coordinates the senescence regulatory pathway in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 2019, 19, 116. [Google Scholar] [CrossRef] [PubMed]
- Christ, B.; Süssenbacher, I.; Moser, S.; Bichsel, N.; Egert, A.; Müller, T.; Kräutler, B.; Hörtensteiner, S. Cytochrome P450 CYP89A9 Is Involved in the Formation of Major Chlorophyll Catabolites during Leaf Senescence in Arabidopsis. Plant Cell 2013, 25, 1868–1880. [Google Scholar] [CrossRef]
- Diago, M.P.; Correa, C.; Millan, B.; Barreiro, P.; Valero, C.; Tardáguila, J. Grapevine Yield and Leaf Area Estimation Using Supervised Classification Methodology on RGB Images Taken under Field Conditions. Sensors 2012, 12, 16988–17006. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. French-Italian Public Consortium for Grapevine Genome Characterization The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463. [Google Scholar] [CrossRef]
- Le Hénanff, G.; Profizi, C.; Courteaux, B.; Rabenoelina, F.; Gérard, C.; Clément, C.; Baillieul, F.; Cordelier, S.; Dhondt-Cordelier, S. Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J. Exp. Bot. 2013, 64, 4877–4893. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, G.; Yan, C.; Liu, L.; Zhang, Q.; Han, Z.; Li, B. DRL1, Encoding A NAC Transcription Factor, Is Involved in Leaf Senescence in Grapevine. Int. J. Mol. Sci. 2019, 20, 2678. [Google Scholar] [CrossRef]
- Katsumoto, Y.; Fukuchi-Mizutani, M.; Fukui, Y.; Brugliera, F.; Holton, T.A.; Karan, M.; Nakamura, N.; Yonekura-Sakakibara, K.; Togami, J.; Pigeaire, A.; et al. Engineering of the Rose Flavonoid Biosynthetic Pathway Successfully Generated Blue-Hued Flowers Accumulating Delphinidin. Plant Cell Physiol. 2007, 48, 1589–1600. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Haruta, K.S.; Pitaksutheepong, C.; Abe, Y.; Kakizaki, Y.; Yamamoto, K.; Shimada, N.; Yamamura, S.; Nishihara, M. Identification and Characterization of R2R3-MYB and bHLH Transcription Factors Regulating Anthocyanin Biosynthesis in Gentian Flowers. Plant Cell Physiol. 2008, 49, 1818–1829. [Google Scholar] [CrossRef]
- Shibuya, K.; Shimizu, K.; Niki, T.; Ichimura, K. Identification of a NAC transcription factor, EPHEMERAL1, that controls petal senescence in Japanese morning glory. Plant J. 2014, 79, 1044–1051. [Google Scholar] [CrossRef]
- De La Torre, M.C.P.; Fernandez, P.; Greppi, J.A.; Coviella, M.A.; Fernández, M.N.; Astigueta, F.; Mata, D.A.; Trupkin, S.A. Transformation of Mecardonia (Plantaginaceae) with wild-type Agrobacterium rhizogenes efficiently improves compact growth, branching and flower related ornamental traits. Sci. Hortic. 2018, 234, 300–311. [Google Scholar] [CrossRef]
- Trupkin, S.A.; Astigueta, F.H.; Baigorria, A.H.; García, M.N.; Delfosse, V.C.; González, S.A.; de la Torre, M.C.; Moschen, S.; Lía, V.V.; Fernández, P.; et al. Identification and expression analysis of NAC transcription factors potentially involved in leaf and petal senescence in Petunia hybrida. Plant Sci. 2019, 287, 10195. [Google Scholar] [CrossRef]
- Agarwal, P.; Kapoor, S.; Tyagi, A.K. Transcription factors regulating the progression of monocot and dicot seed development. BioEssays 2011, 33, 189–202. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. Curr. Neuropharmacol. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Tranbarger, T.J.; Fooyontphanich, K.; Roongsattham, P.; Pizot, M.; Collin, M.; Jantasuriyarat, C.; Suraninpong, P.; Tragoonrung, S.; Dussert, S.; Verdeil, J.L.; et al. Transcriptome Analysis of Cell Wall and NAC Domain Transcription Factor Genes during Elaeis guineensis Fruit Ripening: Evidence for Widespread Conservation within Monocot and Eudicot Lineages. Front. Plant Sci. 2017, 8, 603. [Google Scholar] [CrossRef]
- Pereira-Santana, A.; Alcaraz, L.D.; Castaño, E.; Sánchez-Calderón, L.; Sánchez-Teyer, F.; Rodriguez-Zapata, L. Comparative Genomics of NAC Transcriptional Factors in Angiosperms: Implications for the Adaptation and Diversification of Flowering Plants. PLoS ONE 2015, 10, e0141866. [Google Scholar] [CrossRef]
- Jin, X.; Ren, J.; Nevo, E.; Yin, X.; Sun, D.; Peng, J. Divergent Evolutionary Patterns of NAC Transcription Factors Are Associated with Diversification and Gene Duplications in Angiosperm. Front. Plant Sci. 2017, 8, 1156. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Rozhon, W.; Poppenberger, B. The role of hormones in the aging of plants—A mini-review. Gerontology 2013, 60, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, M.; Chambrier, P.; Rodrigues Bento, S.; Morel, P. Petunia, Your Next Supermodel? Front. Plant Sci. 2016, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
| Species | Code ID/Name | Orthologue in A. thaliana | Expression in Leaf Senescence | Function | Reference |
|---|---|---|---|---|---|
| Arabidopsis thaliana | AT1G01720/ATAF1 | - | Increase | Promote leaf senescence | [44] |
| AT1G52890/ANAC019 | - | Increase | Promote leaf senescence | [73] | |
| AT3G15500/ANAC055 | - | Increase | Promote leaf senescence | [73] | |
| AT4G27410/ANAC072 | - | Increase | Promote leaf senescence | [73] | |
| AT1G34180/ANAC016 | - | Increase | Promote leaf senescence | [43] | |
| AT1G69490/NAP | - | Increase | Promote leaf senescence | [31] | |
| AT5G13180/VNI2 | - | Increase | Delay leaf senescence | [46] | |
| AT5G39610/ORE1 | - | Increase | Promote leaf senescence | [17] | |
| AT2G38440/ORS1 | - | Increase | Promote leaf senescence | [30] | |
| AT2G43000/JUB1 | - | Increase | Delay leaf senescence | [45] | |
| Oryza sativa | Os04g0460600/OsNAC002 | ORE1 | Increase | Promote leaf senescence | [119] |
| Os01g0884300/OsNAC006 | ANAC072 | Increase | unclear | [123] | |
| Os03g21060/OsNAP | NAP | Increase | Promote leaf senescence | [113] | |
| Os11g0184900/OsNAC005 | ATAF1 | Increase | unclear | [113] | |
| Os03g0815100/OsNAC009 | NAP | Increase | unclear | [122] | |
| Os11g03300/OsNAC010 | NAP | Increase | unclear | [122] | |
| Os06g0675600/ONAC011/OsY37 | ANAC022 | Increase | Promote leaf senescence | [124] | |
| Os08g0433500/ONAC106 | ANAC100 | Decrease | Delay leaf senescence | [125] | |
| Triticum aestivum | TaNAM-A1 (GPC-A1) | ANAC025 | Increase | Promote leaf senescence | [136] |
| TaNAM-D1 (GPC-D1) | ANAC025 | Increase | Promote leaf senescence | [136] | |
| TaNAM-B1 (GPC-B1) | ANAC025 | Increase | Promote leaf senescence | [97] | |
| TaNAC-S | ANAC001 | Decrease | Delay leaf senescence | [101] | |
| TraesCS5A02G143200 | NAP | Increase | unclear | [111] | |
| TraesCS5B02G142100 | NAP | Increase | unclear | [111] | |
| TraesCS1A02G2466300 | ANAC082 | Increase | unclear | [111] | |
| TraesCS1B02G77300 | ANAC082 | Increase | unclear | [111] | |
| TraesCS5A02G127200 | ANAC090 | Increase | unclear | [111] | |
| Hordeum vulgare | HvNAC026 | ANAC104 | Increase | unclear | [138] |
| HvNAC023 | NAP | Increase | unclear | [138] | |
| HvNAC027 | ANAC025 | Increase | unclear | [138] | |
| HvNAC029 | ANAC025 | Increase | unclear | [138] | |
| HvNAC030 | ANAC018 | Increase | unclear | [138] | |
| HvNAC005 | NAP | Increase | Promote leaf senescence | [139] | |
| HvNAC001 | ANAC022 | Increase | unclear | [137] | |
| HvNAC013 | ANAC100 | Increase | unclear | [137] | |
| HvWRKY12 | AtWRKY018 | Increase | unclear | [140] | |
| Zea mays | GRMZM2G104400 | VNI2 | Increase | unclear | [155] |
| GRMZM2G475014 | ORE1 | Increase | unclear | [155] | |
| GRMZM2G146380 | ANAC046 | Increase | unclear | [155] | |
| GRMZM2G163251 | JUB1 | Increase | unclear | [155] | |
| GRMZM2G109627 | ANAC047 | Increase | unclear | [155] | |
| GRMZM2G011598 | ANAC025 | Increase | unclear | [155] | |
| GRMZM2G114850/ZmNAC007 | ANAC022 | Increase | Promote leaf senescence | [156] | |
| Sorghum bicolor | Sb01g036590 | ORE1 | Increase | unclear | [159] |
| Sb01g006410 | JUB1 | Increase | unclear | [159] | |
| Glycine max | Gm0266x00007/GmNAC1 | NAP | Increase | Promote leaf senescence | [166] |
| Gm0025x00889/GmNAC5 | ANAC079 | Increase | Promote leaf senescence | [166] | |
| Glyma.08G360200/GmNAC065 | VNI2 | Increase | Promote leaf senescence | [164] | |
| Glyma.12G149100/GmNAC085 | ANAC072 | Decrease | Promote leaf senescence | [164] | |
| Glyma.12G022700/GmNAC81 | ANAC036 | Increase | Promote leaf senescence | [170] | |
| Glyma.05G195000/GmNAC30 | ATAF | Increase | unclear | [164] | |
| Helianhtus annuus | HanXRQChr13g0397761/HaNAC001 | ANAC100 | Increase | unclear | [21] |
| HanXRQChr13g0407321/HaNAC002 | ANAC072 | Decrease | unclear | [21] | |
| HanXRQChr11g0327521/HaNAC004 | ANAC047 | Increase | unclear | [21] | |
| HanXRQChr03g0079641/HaNAC005 | ANAC019 | Increase | unclear | [21] | |
| HanXRQChr08g0210751/HaNAC003 | NAP | Increase | unclear | [21] | |
| Han003584/HaHB-4 | AtHB7 | Increase | Delay leaf senescence | [180] | |
| Gossypium hirsutum | GhNAC12 | ORS1 | Increase | Promote leaf senescence | [187] |
| GhNAP | NAP | Increase | Promote leaf senescence | [188] | |
| CotAD_37422 | GRF2 | Increase | unclear | [190] | |
| KF669775/GhWRKY27 | AtWRKY041 | Increase | Promote leaf senescence | [191] | |
| Vitis vinifera | VvNAC1 | NAP | Increase | unclear | [195] |
| XP-002281816/VvDRL1 | ANAC036 | Decrease | Delay leaf senescence | [196] | |
| Petunia hybrida | Comp559557_c0_seq1/PhNAC024 | NAP | Increase | unclear | [201] |
| Comp22005_c0_seq3/PhNAC017 | ORE1 | Increase | unclear | [201] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bengoa Luoni, S.; Astigueta, F.H.; Nicosia, S.; Moschen, S.; Fernandez, P.; Heinz, R. Transcription Factors Associated with Leaf Senescence in Crops. Plants 2019, 8, 411. https://doi.org/10.3390/plants8100411
Bengoa Luoni S, Astigueta FH, Nicosia S, Moschen S, Fernandez P, Heinz R. Transcription Factors Associated with Leaf Senescence in Crops. Plants. 2019; 8(10):411. https://doi.org/10.3390/plants8100411
Chicago/Turabian StyleBengoa Luoni, Sofia, Francisco H. Astigueta, Salvador Nicosia, Sebastian Moschen, Paula Fernandez, and Ruth Heinz. 2019. "Transcription Factors Associated with Leaf Senescence in Crops" Plants 8, no. 10: 411. https://doi.org/10.3390/plants8100411
APA StyleBengoa Luoni, S., Astigueta, F. H., Nicosia, S., Moschen, S., Fernandez, P., & Heinz, R. (2019). Transcription Factors Associated with Leaf Senescence in Crops. Plants, 8(10), 411. https://doi.org/10.3390/plants8100411






