Molecular Analyses of the Distribution and Function of Diazotrophic Rhizobia and Methanotrophs in the Tissues and Rhizosphere of Non-Leguminous Plants
Abstract
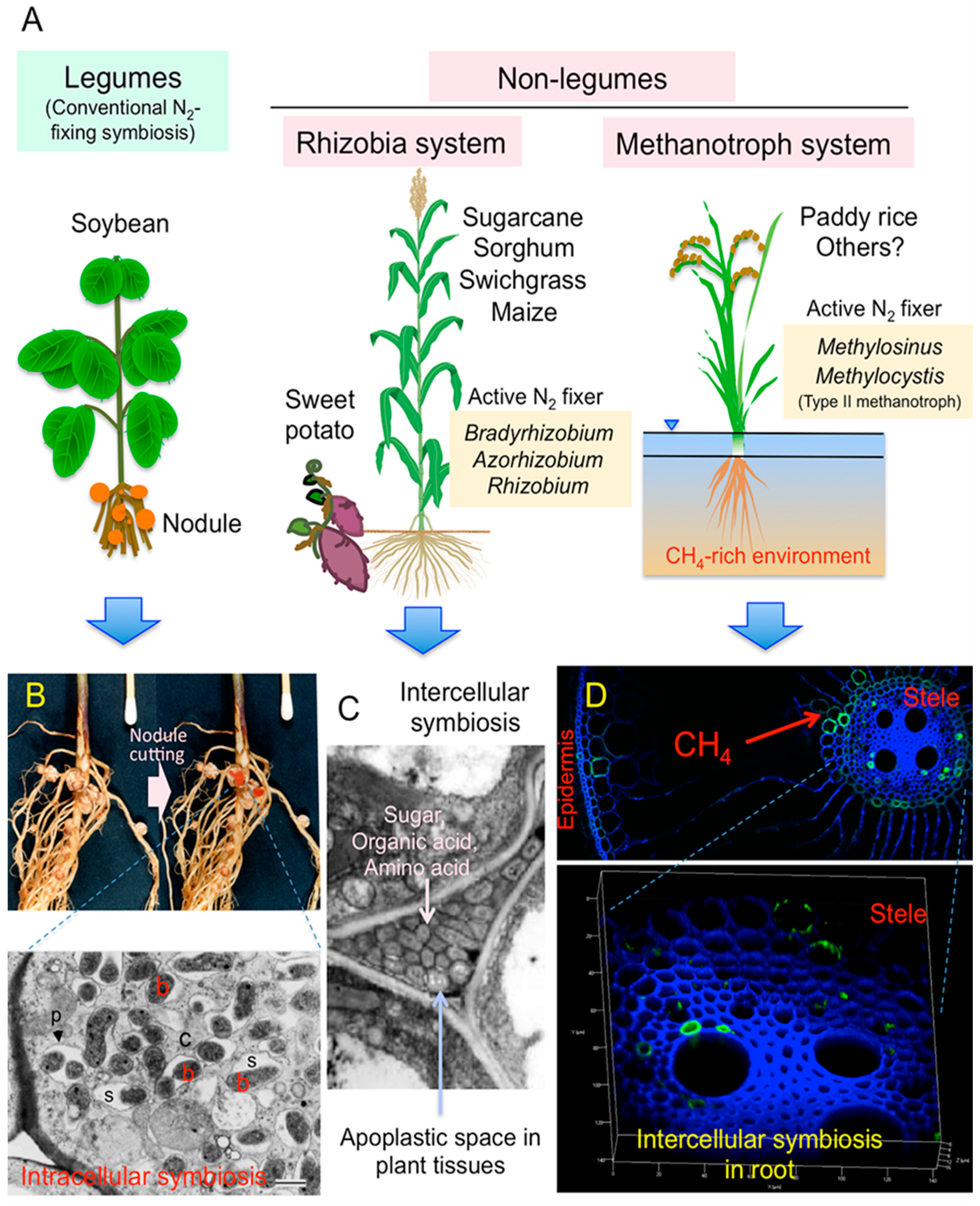
1. Introduction
2. Case Studies to Search the Actively N2-Fixing Diazotrophs in Non-Legumes
2.1. Detection of Expression of nifH Genes in Young Sugarcane Stems
2.2. Detection of Diazotrophic Methanotrophs in Rice Roots by Metaproteomics
3. Molecular Analyses of Diazotrophs in Non-Legumes
3.1. Endophytic Diazotrophs in Maize Plants
3.2. Endophytic Diazotrophs in Sorghum
3.3. Endophytic Diazotrophs in Switchgrass
3.4. Endophytic Diazotrophs in Sugarcane Plants
3.5. Endophytic Diazotrophs in Sweet Potato Plants
3.6. Diazotrophs Associated with Paddy Rice
4. Distribution and Ecophysiological Characteristics of Rhizobia in Non-Leguminous Plants
5. Ecosystem Functioning of Diazotrophic Methanotrophs
6. Research Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Galloway, J.N.; Dentener, F.J.; Capone, D.G.; Boyer, E.W.; Howarth, R.W.; Seizinger, S.P.; Asner, G.P.; Cleveland, C.C.; Green, P.A.; Holland, E.A.; et al. Nitrogen cycles: Past, present, and future. Biogeochemistry 2004, 70, 153–226. [Google Scholar] [CrossRef]
- Yoneyama, T.; Terakado-Toonooka, J.; Minamisawa, K. Exploration of bacterial N2-fixation systems in association with soil-grown sugarcane, sweet potato, and paddy rice: A review and synthesis. Soil Sci. Plant Nutr. 2017, 63, 578–590. [Google Scholar] [CrossRef]
- Elbeltagy, A.; Nishioka, K.; Sato, T.; Suzuki, H.; Ye, B.; Hamada, T.; Isawa, T.; Mitsui, H.; Minamisawa, K. Endophytic colonization and in planta nitogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl. Environ. Microbiol. 2001, 67, 5285–5293. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Okubo, T.; Kubota, K.; Kasahara, Y.; Tsurumaru, H.; Anda, M.; Ikeda, S.; Minamisawa, K. Metaproteomic identification of diazotrophic methanotrophs and their localization in root tissues of field-grown rice plants. Appl. Environ. Microbiol. 2014, 80, 5043–5052. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.C.; Cleveland, C.C.; Townsend, A.R. Functional ecology of free-living nitrogen fixation: A contemporary perspective. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 489–512. [Google Scholar] [CrossRef]
- James, E.K. Nitrogen fixation in endophytic and associative symbiosis. Field Crops Res. 2000, 65, 197–209. [Google Scholar] [CrossRef]
- Yoneyama, T.; Muraoka, T.; Kim, T.H.; Dacanay, E.V.; Nakanishi, Y. The natural 15N abundance of sugarcane and neighboring plants in Brazil, the Philippines and Miyako (Japan). Plant Soil 1997, 189, 239–244. [Google Scholar] [CrossRef]
- Yoneyama, T.; Terakado, J.; Masuda, T. Natural abundance of 15N in sweet potato, pumpkin, sorghum and castor bean: Possible input of N2-derived nitrogen in sweet potato. Biol. Fertil. Soils 1997, 26, 152–154. [Google Scholar] [CrossRef]
- Ladha, J.K.; Tirol-Padre, A.; Reddy, C.K.; Cassman, K.G.; Verma, S.; Powlson, D.S.; van Kessel, C.; Richter, D.B.; Chakraborty, D.; Pathak, H. Global nitrogen budgets in cereals: A 50-year assessment for maize, rice, and wheat production systems. Sci. Rep. 2016, 6, 19355. [Google Scholar] [CrossRef]
- Thaweenut, N.; Hachisuka, Y.; Ando, S.; Yanagisawa, S.; Yoneyama, T. Two seasons’ study on nifH gene expression and nitrogen fixation by diazotrophic endophytes in sugarcane (Saccharum spp. hybrids): Expression of nifH genes similar to those of rhizobia. Plant Soil 2011, 338, 435–449. [Google Scholar] [CrossRef]
- Ando, S.; Goto, M.; Meunchang, S.; Thongra-ar, P.; Fujiwara, T.; Hayashi, H.; Yoneyama, T. Detection of nifH sequences in sugarcane (Saccharum officinarum L.) and pineapple (Ananas comosus [L] Merr.). Soil Sci. Plant Nutr. 2005, 51, 303–308. [Google Scholar] [CrossRef]
- Gaby, J.C.; Rishishway, L.; Valderrama-Aguirre, L.C.; Green, S.J.; Valderrama-Aguirre, A.; Jordan, I.K.; Kostka, J.E. Diazotroph community characterization via a high-throughput nifH amplicon sequencing and analysis pipeline. Appl. Environ. Microbiol. 2018, 84, e1512–e1517. [Google Scholar]
- Ikeda, S.; Sasaki, K.; Okubo, T.; Yamashita, A.; Terasawa, K.; Bao, Z.; Liu, D.; Watanabe, T.; Murase, J.; Asakawa, S.; et al. Low nitrogen fertilization adapts rice root microbiome to low nutrient environment by changing biogeochemical functions. Microbes Environ. 2014, 29, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, Y.; Morimoto, H.; Kuwano, M.; Kadoya, R. Genome-wide analytical approaches using semi-quantitative expression proteomics for aromatic hydrocarbon metabolism in Pseudomonas pudida F1. J. Microbiol. Methods 2012, 91, 434–442. [Google Scholar] [CrossRef]
- Estrada, P.; Mavingui, P.; Cournyer, B.; Fontaine, F.; Balandreau, J.; Caballero-Mellado, J. A N2-fixing endophytic Burrkholderia sp. associated with maize plants cultivated in Mexico. Can. J. Microbiol. 2002, 48, 285–294. [Google Scholar] [CrossRef]
- Perin, L.; Martínez-Aguilar, L.; Castro-González, R.; Estrada-de los Santos, P.; Cabellos-Avelar, T.; Guedes, H.V.; Reis, V.M.; Caballero-Mellado, J. Diazotrophic Burkholderia species associated with field-grown maize and sugarcane. Appl. Environ. Microbiol. 2006, 72, 3103–3110. [Google Scholar] [CrossRef]
- Roesch, L.F.W.; Camargo, F.A.O.; Bento, F.M.; Triplett, E.W. Biodiversity of diazotrophic bacteria within the soil, root and stem of field-grown maize. Plant Soil 2008, 302, 91–104. [Google Scholar] [CrossRef]
- Van Deynze, A.; Zamora, P.; Delaux, P.-M.; Heitmann, C.; Jayaraman, D.; Rajasekar, S.; Graham, D.; Maeda, J.; Gibson, D.; Schwartz, K.D.; et al. Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLOS Biol. 2018, 16, e2006352. [Google Scholar] [CrossRef]
- Coelho, M.R.R.; de Vos, M.; Carneiro, N.P.; Marriel, I.E.; Paiva, E.; Seldin, L. Diversity of nifH gene pools in the rhizosphere of two cultivars of sorghum (Sorghum bicolor) treated with contrasting levels of nitrogen fertilizer. Fems Microbiol. Lett. 2008, 279, 15–22. [Google Scholar] [CrossRef]
- Hara, S.; Morikawa, T.; Wasai, S.; Kasahara, Y.; Koshiba, T.; Yamazaki, K.; Fujiwara, T.; Tokunaga, T.; Minamisawa, K. Identification of nitrogen-fixing Bradyrhizobium associated with roots of field-grown sorghum by metagenome and proteome analyses. Front. Microbiol. 2019, 10, 407. [Google Scholar] [CrossRef]
- Bahulikar, R.A.; Torres-Jerez, I.; Worley, E.; Craven, K.; Udvardi, M.K. Diversity of nitrogen-fixing bacteria associated with switchgrass in the native tallgrass prairie of Northern Oklahoma. Appl. Environ. Microbiol. 2014, 80, 5636–5643. [Google Scholar] [CrossRef] [PubMed]
- Lowman, S.; Kim-Dura, S.; Mei, C.; Nowak, J. Strategies for enhancement of switchgrass (Panicum virgatum L.) performance under limited nitrogen supply based on utilization of N-fixing bacterial endophytes. Plant Soil 2016, 405, 47–63. [Google Scholar] [CrossRef]
- Roley, S.S.; Duncan, D.S.; Liang, D.; Garoutte, A.; Jackson, R.D.; Tiedje, J.M.; Robertson, G.P. Associative nitrogen fixation (ANF) in switchgrass (Panicum virgatum) across a nitrogen input gradient. PLoS ONE 2018, 13, e0197320. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, V.A.; Dobereiner, J. A new acid tolerant nitrogen-fixing bacterium associated with sugarcane. Plant Soil 1988, 108, 23–31. [Google Scholar] [CrossRef]
- Gillis, M.; Kersters, K.; Hoste, B.; Janssens, D.; Kroppenstedt, D.; Stephen, M.P.; Teixeira, K.R.S.; Döbereiner, J.; de Ley, J. Acetobacter diazotrophicus sp. nov., a nitrogen fixing acetic acid bacterium associated with sugarcane. Int. J. Syst. Bacteriol. 1989, 39, 361–364. [Google Scholar] [CrossRef]
- Pimentel, J.P.; Olivares, F.; Pitard, R.M.; Urquiaga, S.; Akiba, F.; Döbereiner, J. Dinitrogen fixation and infection of grass leaves by Psudomonas rubrisubalbicans and Herbaspirillum seropedicae. Plant Soil 1991, 137, 61–65. [Google Scholar] [CrossRef]
- Olivares, F.L.; Baldani, V.L.D.; Reis, V.M.; Baldani, J.I.; Döbereiner, J. Occurrence of the endophytic diazotrophs Herbaspirillum spp. in roots, stems, and leaves, predominantly of Gramineae. Biol. Fertil. Soils 1996, 21, 197–200. [Google Scholar] [CrossRef]
- Sevilla, M.; Meletzus, D.; Teixeira, K.; Lee, S.; Nutakki, A.; Baldani, I.; Kennedy, C. Analysis of nif and regulatory genes in Acetobacter diazotrophicus. Soil Biol. Biochem. 1997, 29, 871–874. [Google Scholar] [CrossRef]
- Sevilla, M.; Oliveira, A.; Baldani, I.; Kennedy, C. Contributions of the bacterial endophyte Acetobacter diazotrophicus to sugarcane nutrition: A preliminary study. Symbiosis 1998, 25, 181–191. [Google Scholar]
- Li, R.P.; MacRae, I.C. Specific identification and enumeration of Acetobacter diazotrophicus in sugarcane. Soil Biol. Biochem. 1991, 24, 413–419. [Google Scholar] [CrossRef]
- Fuentes-Ramirez, L.E.; Jimenez-Salgado, T.; Abarca-Ocampo, I.R.; Caballero-Mellado, J. Acetobacter diazotrophicus, an indoleacetic acid producing bacterium isolated from sugarcane cultivars of México. Plant Soil 1993, 154, 145–150. [Google Scholar] [CrossRef]
- Asis, C.A.; Kubota, M.; Ohta, H.; Arima, Y.; Chebotar, V.K.; Tsuchiya, K.; Akao, S. Isolation and partial characterization of endophytic diazotrophs associated with Japanese sugarcane cultivar. Soil Sci. Plant Nutr. 2000, 46, 759–765. [Google Scholar] [CrossRef]
- Bastián, F.; Cohen, A.; Piccoli, P.; Luna, V.; Baraldi, R.; Bottini, R. Production of indole-3-acetic acid and gibberellins A1 and A3 by Acetobacter diazotrophicus and Herbaspirillum seropedicae in chemically-defined culture media. Plant Growth Regul. 1998, 24, 7–11. [Google Scholar] [CrossRef]
- Cavalcante, J.J.V.; Vargas, C.; Nogueira, E.M.; Vinagre, F.; Schwarcz, K.; Baldani, J.I.; Ferreira, P.C.G.; Hemerly, A.S. Members of the ethylene signaling pathway are regulated in sugarcane during the association with nitrogen-fixing endophytic bacteria. J. Exp. Bot. 2007, 58, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Castro-González, R.; Martínez-Aguilar, L.; Ramirez-Trujíllo, A.; Santos, P.E.; Caballero-Mellado, J. High diversity of culturable Burkholderia species associated with sugarcane. Plant Soil 2011, 345, 155–169. [Google Scholar] [CrossRef]
- Okon, Y.; Labandera-Gonzalez, C.A. Agronomic applications of Azospirillum: An evaluation of 20 years worldwide field inoculation. Soil Biol. Biochem. 1994, 26, 1591–1601. [Google Scholar] [CrossRef]
- Dobbelaere, S.; Vanderleyden, J.; Okon, Y. Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit. Rev. Plant Sci. 2003, 22, 107–149. [Google Scholar] [CrossRef]
- Muñoz-Rojas, J.; Caballero-Mellado, J. Population dynamics of Gluconacetobacter diazotrophicus in sugarcane cultivars and its effect on plant growth. Microb. Ecol. 2003, 46, 454–464. [Google Scholar] [CrossRef]
- Beneduzi, A.; Moreira, F.; Costa, P.B.; Vargas, L.K.; Lisboa, B.B.; Favreto, R.; Baldani, J.I.; Passaglia, L.M.P. Diversity and plant growth promoting evaluation abilities of bacteria isolated from sugarcane cultivated in the South of Brazil. Appl. Soil Ecol. 2013, 63, 94–104. [Google Scholar] [CrossRef]
- Carvalho, T.L.G.; Balsemao-Pires, E.; Saraiva, R.M.; Ferreira, P.C.G.; Hemerly, A.S. Nitrogen signaling in plant interactions with associative and endophytic dizotrophic bacteria. J. Exp. Bot. 2014, 65, 5631–5642. [Google Scholar] [CrossRef]
- Fischer, D.; Pfitzner, B.; Schmid, M.; Simõnes-Araújo, J.L.; Reis, V.M.; Pereira, W.; Ormeño-Orrillo, E.; Hai, B.; Hofmann, A.; Schloter, M.; et al. Molecular characterization of the diazotrophic bacterial community in uninoculated and inoculated field-grown sugarcane (Saccharum sp.). Plant Soil 2012, 356, 83–99. [Google Scholar] [CrossRef]
- Rouws, L.F.M.; Leite, J.; de Matos, G.F.; Zilli, J.E.; Coelho, M.R.R.; Xavier, G.R.; Fisher, D.; Hartmann, A.; Reis, V.M.; Baldani, J.I. Endophytic Bradyrhizobium spp. isolates from sugarcane obtained through different culture strategies. Environ. Microbiol. Rep. 2014, 6, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Matos, G.F.; Zilli, J.E.; Araújo, J.L.S.; Parma, M.M.; Melo, I.S.; Radl, V.; Baldani, J.V.; Rouws, L.F.M. Bradyrhizobium sacchari sp. nov., a legume nodulating bacterium isolated from sugarcane roots. Arch. Microbiol. 2017, 199, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Biggs, I.M.; Stewart, G.R.; Wilson, J.R.; Critchley, C. 15N natural abundance studies in Australian commercial sugarcane. Plant Soil 2002, 238, 21–30. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Paungfoo-Lonhienne, C.; Dennis, P.G.; Robinson, N.; Ragan, M.A.; Schmidt, S.; Hugenholtz, P. The core root microbiome of sugarcane cultivated under varying nitrogen fertilizer application. Environ. Microbiol. 2016, 18, 1338–1351. [Google Scholar] [CrossRef] [PubMed]
- Paungfoo-Lonhienne, C.; Lonhienne, T.G.A.; Yeoh, Y.K.; Webb, R.I.; Laskhmanan, P.; Chan, C.X.; Lim, P.-E.; Ragan, M.A.; Schmidt, S.; Hugenholtz, P. A new species of Burkholderia isolated from sugarcane roots promotes plant growth. Microb. Biotechnol. 2014, 7, 142–154. [Google Scholar] [CrossRef]
- Reiter, B.; Bürgmann, H.; Burg, K.; Sessitsch, A. Endophytic nifH gene diversity in African sweet potato. Can. J. Microbiol. 2003, 49, 549–555. [Google Scholar] [CrossRef][Green Version]
- Terakado-Tonooka, J.; Ohwaki, Y.; Yamakawa, H.; Tanaka, F.; Yoneyama, T.; Fujihara, S. Expressed nifH genes of endophytic bacteria detected in field-grown sweet potatoes (Ipomoea batatas L.). Microbes Environ. 2008, 23, 89–93. [Google Scholar] [CrossRef]
- Terakado-Tonooka, J.; Fuhihara, S.; Ohwaki, Y. Possible contribution of Bradyrhizobium on nitrogen fixation in sweet potatoes. Plant Soil 2013, 367, 639–650. [Google Scholar] [CrossRef]
- Nozué, A.; Miché, L.; Klonowska, A.; Laguerre, G.; de Lajudie, P.; Moulin, L. Multilocus sequence analysis of bradyrhizobia isolated from Aeschynomene species in Senegal. Syst. Appl. Microbiol. 2009, 32, 400–412. [Google Scholar] [CrossRef]
- Okubo, T.; Piromyou, P.; Tittabutr, P.; Teaumroong, N.; Minamisawa, T. Origin and evolution of nitrogen fixation genes on symbiosis islands and plasmid in Bradyrhizobum. Microbes Environ. 2016, 31, 260–267. [Google Scholar] [CrossRef]
- Hirano, K.; Hayatsu, M.; Nioh, I.; Nakai, H. Comparison of nitrogen-fixing bacterial flora of rice rhizosphere in the fields treated long-term with agrochemicals and non-agrochemicals. Microbes Environ. 2001, 16, 155–160. [Google Scholar] [CrossRef]
- Chaintreuil, C.; Giraud, E.; Prin, Y.; Lorquin, J.; Bâ, A.; Gillis, M.; Lajudie, P.; Dreyfus, B. Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl. Environ. Microbiol. 2000, 66, 5437–5447. [Google Scholar] [CrossRef] [PubMed]
- Piromyou, P.; Greetatorn, T.; Teamtisong, K.; Okubo, T.; Shinoda, R.; Nuntakij, A.; Tittabutr, P.; Boonkerd, N.; Minamisawa, K.; Teaumroong, N. Preferential association of endophytic bradyrhizobia with different rice cultivars and its implications for rice endophyte evolution. Appl. Environ. Microbiol. 2015, 81, 3049–3061. [Google Scholar] [CrossRef]
- Elbeltagy, A.; Nishioka, K.; Suzuki, H.; Sato, T.; Sato, Y.; Morisaki, H.; Mitsui, H.; Minamisawa, K. Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil Sci. Plant Nutr. 2000, 46, 617–629. [Google Scholar] [CrossRef]
- Videira, S.S.; Araujo, J.L.S.; Rodrigues, L.S.; Baldani, V.L.D.; Baldani, J.I. Occurrence and diversity of nitrogen-fixing Sphingomonas bacteria associated with rice plants grown in Brazil. FEMS Microbiol. Lett. 2009, 293, 11–19. [Google Scholar] [CrossRef]
- Chi, F.; Shen, S.-H.; Cheng, H.-P.; Jing, Y.-X.; Yanni, Y.G.; Dazzo, F.B. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl. Environ. Microbiol. 2005, 71, 7271–7278. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Suga, Y.; Yahiro, N.; Matsuguchi, T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J. Bacteriol. 1995, 177, 1414–1417. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Hardoim, P.; Döring, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomics analysis. Mol. Plant Microbe Interact. 2012, 25, 28–36. [Google Scholar] [CrossRef]
- Mårtensson, L.; Díez, B.; Wartainen, I.; Zheng, W.; El-Shehawy, R.; Rasmussen, U. Diazotrophic diversity, nifH gene expression and nitrogenase activity in a rice paddy field in Fujian, China. Plant Soil 2009, 325, 207–218. [Google Scholar] [CrossRef]
- Knief, C.; Delmotte, N.; Chaffron, S.; Stark, M.; Innerebner, G.; Wassmann, R.; von Mering, C.; Vorholt, J.A. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012, 6, 1378–1390. [Google Scholar] [CrossRef] [PubMed]
- Nouchi, I.; Mariko, S.; Aoki, K. Mechanism of methane transport from the rhizosphere to the atmosphere through rice plants. Plant Physiol. 1990, 94, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Jeong, S.E.; Kim, P.J.; Madsen, E.L.; Jeon, C.O. High resolution depth distribution of Bacteria, Archaea, methanotrops, and methanogens in the bulk and rhizosphere soils of a flooded rice paddy. Front. Microbiol. 2015, 6, 639. [Google Scholar] [CrossRef] [PubMed]
- Auman, A.J.; Speake, C.C.; Lidstrom, M.E. nifH sequences and nitrogen fixation in type I and type II methanotrophs. Appl. Environ. Microbiol. 2001, 67, 4009–4016. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.S.; Bender, G.L.; Shine, J.; Rolfe, B.G.; Gresshoff, P.M. In vitro expression of nitrogenase activity in Parasponia-Rhizobium strain ANU289. Arch. Microbiol. 1989, 134, 12–16. [Google Scholar] [CrossRef]
- Moulin, L.; Munlve, A.; Dreyfus, B.; Bolvin-Masson, C. Nodulation of legumes by members of the β-subclass of proteobacteria. Nature 2001, 411, 948–950. [Google Scholar] [CrossRef]
- Chen, W.-M.; Moulin, L.; Bontemps, C.; Vandamme, P.; Béna, G.; Boivin-Masson, C. Legume symbiotic nitrogen fixation by β-proteobacteria is widespread in nature. J. Bacteriol. 2003, 185, 7266–7272. [Google Scholar] [CrossRef]
- Compant, S.; Nowa, J.; Coenye, T.; Clement, C.; Barka, E.A. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 2008, 32, 607–626. [Google Scholar] [CrossRef]
- Sy, A.; Giraud, E.; Jourand, P.; Garcia, N.; Willems, A.; de Lajudie, P.; Neyra, M.; Gillis, M.; Boivin-Masson, C.; Dreyfus, B. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J. Bacteriol. 2001, 183, 214–220. [Google Scholar] [CrossRef]
- Jourand, P.; Giraud, E.; Béna, G.; Sy, A.; Willems, A.; Gillis, M.; Dreyfus, B.; de Lajudie, P. Methylobacterium nodulans sp. nov., for a group of aerobic, facultatively methylotrophic, legume root-nodule-forming and nitrogen-fixing bacteria. Int. J. Syst. Evol. Microbiol. 2004, 54, 2269–2273. [Google Scholar] [CrossRef] [PubMed]
- Renier, A.; de Faria, S.M.; Jourand, P.; Giraud, E.; Dreyfus, B.; Rapior, S.; Prin, Y. Nodulation of Crotalaria podocarpa DC. by Methylobacterium nodulans displays very unusual features. J. Exp. Bot. 2011, 62, 3693–3697. [Google Scholar] [CrossRef] [PubMed]
- Sprent, J.I.; Ardley, J.; James, E.K. Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 2017, 215, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef] [PubMed]
- Giraud, E.; Moulin, L.; Vallenet, D.; Barbe, V.; Cytryn, E.; Avarre, J.C.; Jaubert, M.; Simon, D.; Cartieaux, F.; Prin, Y.; et al. Legumes symbioses: Absence of nod genes in photosynthetic bradyrhizobia. Science 2007, 316, 1307–1312. [Google Scholar] [CrossRef]
- Bonaldi, K.; Gourion, B.; Fardoux, J.; Hannibal, L.; Cartieaux, F.; Boursot, M.; Vallennet, D.; Chaintreuil, C.; Prin, Y.; Nouwen, N.; et al. Large-scale transposon mutagenesis of photosynthetic Bradyrhizobium sp. strain ORS278 reveals new genetic loci putatively important for nod-independent symbiosis with Aeschynomene indica. Mol. Plant-Microbe Interact. 2010, 23, 760–770. [Google Scholar] [CrossRef]
- Chaintreuil, C.; Arrighi, J.-F.; Giraud, E.; Miché, L.; Moulin, L.; Dreyfus, B.; Munive-Hernández, J.-A.; Villegas-Hernandez, M.C.; Béna, G. Evolution of symbiosis in the legume genus Aeschynomene. New Phytol. 2013, 200, 1247–1259. [Google Scholar] [CrossRef]
- Okubo, T.; Fukushima, S.; Itakura, M.; Oshima, K.; Longtonglang, A.; Teaumroong, N.; Mitsui, H.; Hattori, M.; Hattori, R.; Hattori, T.; et al. Genome analysis suggests that the soil oligotrophic bacterium Agromonas oligotrophica (Bradyrhizobium oligotrophicum) is a nitrogen-fixing symbiont of Aeschynomene indica. Appl. Environ. Microbiol. 2013, 79, 2542–2551. [Google Scholar] [CrossRef]
- Dreyfus, B.L.; Elmerich, C.; Dommergues, Y.R. Free-living Rhizobium strain able to grow on N2 as the sole nitrogen sources. Appl. Environ. Microbiol. 1983, 45, 711–713. [Google Scholar]
- Gebhardt, C.; Turner, G.L.; Gibson, A.H.; Dreyfus, B.L.; Bergersen, F.J. Nitrogen-fixing growth in continuous culture of a strain of Rhizobium sp. isolated from stem nodules on Sesbania rostrate. J. Gen. Microbiol. 1984, 130, 843–848. [Google Scholar]
- Alazard, D. Nitrogen fixation in pure culture by rhizobia isolated from stem nodules of tropical Aeschynomene species. FEMS Microbiol. Lett. 1990, 68, 177–182. [Google Scholar] [CrossRef]
- Elliott, G.N.; Chen, W.-M.; Chou, J.-H.; Wang, H.-C.; Sheu, S.-Y.; Perin, L.; Reis, V.M.; Simon, M.F.; Bontemps, C.; Sutherland, J.M.; et al. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 2007, 173, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Welbaum, G.E.; Meinzer, F.C. Compartmentation of solutes and water in developing sugarcane stalk tissue. Plant Physiol. 1990, 93, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, A.; Botha, F.C. Carbon partitioning during sucrose accumulation in sugarcane intermodal tissue. Plant Physiol. 1997, 115, 1651–1659. [Google Scholar] [CrossRef] [PubMed]
- Asis, C.A.; Shimizu, T.; Khan, M.K.; Akao, S. Organic acid and sugar contents in sugarcane stem apoplast solution and their role as carbon source for endophytic diazotrophs. Soil Sci. Plant Nutr. 2003, 49, 915–920. [Google Scholar] [CrossRef]
- Pagan, J.D.; Child, J.J.; Scowcroft, W.R.; Gibson, A.H. Nitrogen fixation by Rhizobium cultured on a defined medium. Nature 1975, 256, 406–407. [Google Scholar] [CrossRef]
- Kurz, W.G.W.; LaRue, T.A. Nitrogenase activity in rhizobia in absence of plant host. Nature 1975, 256, 407–409. [Google Scholar] [CrossRef]
- McComb, J.A.; Elliot, J.; Dilworth, M.J. Acetylene reduction by Rhizobium in pure culture. Nature 1975, 256, 409–410. [Google Scholar] [CrossRef]
- Stam, H.; van Verseveld, H.W.; Stouthamer, A.H. Derepression of nitrogenase in chemostat cultures of the fast growing Rhizobium leguminosarum. Arch. Microbiol. 1983, 135, 199–204. [Google Scholar] [CrossRef]
- Urban, J.E.; Davis, L.C.; Brown, S.J. Rhizobium trifolii 0403 is capable of growth in the absence of combined nitrogen. Appl. Environ. Microbiol. 1986, 52, 1060–1067. [Google Scholar]
- Sindhu, S.S.; Dararwal, K.R. Ex planta nitrogenase induction and uptake hydrogenase in Rhizobium sp. (cowpea miscellany). Soil Biol. Biochem. 1986, 18, 291–295. [Google Scholar] [CrossRef]
- Patriarca, E.J.; Tate, R.; Iaccarino, M. Key role of bacterial NH4+ metabolism in Rhizobium-plant symbiosis. Microbiol. Mol. Biol. Rev. 2002, 66, 203–222. [Google Scholar] [CrossRef]
- Tjepkema, J.; Evans, H.J. Nitrogen fixation by free-living Rhizobium in a defined liquid medium. Biochem. Biophys. Res. Commun. 1975, 65, 625–628. [Google Scholar] [CrossRef]
- O’Gara, F.; Shanmugam, K.T. Regulation of nitrogen fixation by rhizobia. Export of fixed N2 as NH4+. Biochim. Biophys. Acta 1976, 437, 313–321. [Google Scholar] [CrossRef]
- Tubb, R.S. Regulation of nitrogen fixation in Rhizobium sp. Appl. Environ. Microbiol. 1976, 32, 483–488. [Google Scholar] [PubMed]
- Bergersen, F.J.; Turner, G.L. Activity of nitrogenase and glutamine synthetase in relation to availability of oxygen in continuous cultures of a strain of cowpea Rhizobium sp. supplied with excess ammonium. Biochim. Biophys. Acta 1978, 538, 406–416. [Google Scholar] [CrossRef]
- Dong, Z.; Canny, M.J.; McCully, M.E.; Roboredo, M.R.; Cabadilla, C.F.; Ortega, E.; Rodés, R. A nitrogen-fixing endophyte of sugarcane stems. A new role for the apoplast. Plant Physiol. 1994, 105, 1139–1147. [Google Scholar] [CrossRef]
- Nogueira, E.M.; Olivares, F.L.; Japiassu, J.C.; Vilar, C.; Vinagre, F.; Baldani, J.I.; Hemerly, A.S. Characterization of glutamine synthetase genes in sugarcane genotypes with different rates of biological nitrogen fixation. Plant Sci. 2005, 169, 819–832. [Google Scholar] [CrossRef]
- Yanni, Y.G.; Rizk, R.Y.; Corich, V.; Squartini, A.; Ninke, K.; Philip-Hollingsworth, S.; Orgambide, G.; de Bruijin, F. Natural endophytic association between Rhizobium leguminosarum bv. trifolii and rice roots and assessment of its potential to promote rice growth. Plant Soil 1997, 194, 99–114. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Sathya, A.; Vijayabharathi, R.; Varshney, R.K.; Gowda, C.L.L.; Krishnamurthy, L. Plant growth promoting rhizobia: Challenges and opportunities. 3 Biotech. 2015, 5, 355–377. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Ricke, P.; Liesack, W. NifH and NifD phylogenies: An evolutionary basis for understanding nitrogen fixation capabilities of methanotrophic bacteria. Microbiology 2004, 150, 1301–1313. [Google Scholar] [CrossRef]
- Tamas, I.; Smirnova, A.V.; He, Z.; Dunfield, P.F. The (d)evolution of methanotrophy in the Beijerinckiaceae–a comparative genomics analysis. ISME J. 2014, 8, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.B.; Coty, V.F.; Stanley, J.P. Atmospheric nitrogen fixation by methane-oxidizing bacteria. J. Bacteriol. 1964, 88, 468–472. [Google Scholar] [PubMed]
- Shinoda, R.; Bao, Z.; Minamisawa, K. CH4 oxidation-dependent 15N2 fixation in rice roots in a low-nitrogen paddy field and in Methylosinus sp. strain 3S-1 isolated from the roots. Soil Biol. Biochem. 2019, 132, 40–46. [Google Scholar] [CrossRef]
- Hanson, R.S.; Hanson, T.E. Methanotropic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar]
- Bodelier, P.L.E.; Laanbroek, H.J. Nitrogen as a regulatory factor of methane oxidation in soils and sediments. Fems Microbiol. Ecol. 2004, 47, 265–277. [Google Scholar] [CrossRef]
- Minamisawa, K.; Imaizumi-Anraku, H.; Bao, Z.; Shinoda, R.; Okubo, T.; Ikeda, S. Are symbiotic methanotrophs key microbes for N acquisition in paddy rice root? Microbes Environ. 2016, 31, 4–10. [Google Scholar] [CrossRef]
- Kip, N.; Ouyang, W.; Winden, J.; Raghoebarsing, A.; Niftrik, L.; Pol, A.; Pan, Y.; Bodrossy, L.; Donselaar, E.G.; Reicharrt, G.-J.; et al. Detection, isolation, and characterization of acidophilic methanotrophs from Sphagnum mosses. Appl. Environ. Microbiol. 2011, 77, 5643–5654. [Google Scholar] [CrossRef]
- Vile, M.A.; Wieder, R.K.; Živković, T.; Scott, K.D.; Vitt, D.H.; Hartsock, J.A.; Iosue, C.L.; Quinn, J.C.; Petix, M.; Fillingim, H.M.; et al. N2-fixation by methanotrophs sustains carbon and nitrogen accumulation in pristine peatlands. Biogeochemistry 2014, 121, 317–328. [Google Scholar] [CrossRef]
- Larmola, T.; Leppänen, S.M.; Tuittla, E.-S.; Aarva, M.; Merilä, P.; Fritze, H.; Tiirola, M. Methanotrophy induces nitrogen fixation during peatland development. Proc. Natl. Acad. Sci. USA 2014, 111, 734–739. [Google Scholar] [CrossRef]
- Warren, M.J.; Lin, X.; Gaby, J.C.; Kretz, C.B.; Kolton, M.; Morton, P.L.; Pett-Ridge, W.J.; Westo, D.J.; Schadt, C.W.; Kostka, J.E.; et al. Molybdenum-based diazotrophy in a Sphagnum peatland in Northern Minnesota. Appl. Environ. Microbiol. 2017, 83, e01174-17. [Google Scholar] [CrossRef]
- You, M.; Nishiguchi, T.; Saito, A.; Isawa, T.; Mitsui, H.; Minamisawa, K. Expression of the nifH gene of a Herbaspirillum endophyte in wild rice species: Daily rhythm during the light-dark cycle. Appl. Environ. Microbiol. 2005, 71, 8183–8190. [Google Scholar] [CrossRef] [PubMed]
- Hara, S.; Matsuda, M.; Minamisawa, K. Growth stage-dependent bacterial communities in soybean plant tissues: Methylorubrum transiently dominated in the flowering stage of soybean shoot. Microbes Environ. 2019, in press. [Google Scholar] [CrossRef] [PubMed]
- Wasai, S.; Minamisawa, K. Plant-associated microbes: From rhizobia to plant microbiomes. Microbes Environ. 2018, 33, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Narihiro, T.; Kamagata, Y. Genomics and metagenomics in microbial ecology: Recent advances and challenges. Microbes Environ. 2017, 32, 1–4. [Google Scholar] [CrossRef]
- Garrido-Oter, R.; Nakano, R.T.; Dombrowski, N.; Ma, K.-W.; The AgBiome Team; McHardy, A.C.; Schulze-Lefert, P. Modular traits of the Rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 2018, 24, 155–167. [Google Scholar] [CrossRef]
- Levy, A.; Salas Gonzalez, I.; Mittelviefhaus, M.; Clingenpeel, S.; Herrera Paredes, S.; Miao, J.; Wang, K.; Devescovi, G.; Stillman, K.; Monteiro, F.; et al. Genomic features of bacterial adaptation to plants. Nat. Genet. 2017, 50, 138–150. [Google Scholar] [CrossRef]
- Toju, H.; Peay, K.G.; Yamamichi, M.; Narisawa, K.; Hiruma, K.; Naito, K.; Fukuda, S.; Ushio, M.; Nakaoka, S.; Onoda, Y.; et al. Core microbiomes for sustainable agroecosystems. Nat. Plants 2018, 4, 247–257. [Google Scholar] [CrossRef]
| Site and Sample of Investigation | Detection of nifH Genes | Close Genus Abundance and Bacteria | |
|---|---|---|---|
| Stem harvested in six regions in Rio Grande do Sul, Brazil [17] | Sequencing of nifH DNA clones | 27% 20% 12.6% 7.4% 5.4% The others restricted to the stem | Ideonella Azospirillum Klebsiella Herbaspirillum Raoultella Methylosinus, Rhizobium |
| Root harvested in six regions in Rio Grande do Sul, Brazil [17] | Sequencing of nifH DNA clones | 30% 23.3% 11.9% The other restricted to the root | Bradyrhizobium Azospirillum Klebsiella Dechloromonas |
| Rhizosphere soil collected in six regions in Rio Grande do Sul, Brazil [17] | Sequencing of nifH DNA clones | 20.8% 11% 5% 0.2% The others restricted to the rhizosphere soil | Bradyrhizobium Ideonella Azospirillum Klebsiella Methylocystis, Beijerinckia, Geobacter, Rhodovulum, Methylobacterium, Gluconacetobacter, Methylocella, Delftia |
| Site and Sample of Investigation | Detection of nifHDK Genes and Their Proteins | Close Genus (Similarity >91%) Abundance and Bacteria | |
|---|---|---|---|
| Roots harvested from two sorghum lines (KM1, KM2) at late growth stage in a Fukushima field, Japan [20] | Metagenome for nifHDK genes | 68% (KM1), 88% (KM2) 1–3% (KM1, KM2) | Bradyrhizobium spp. (including B. sp. S23321 and B. oligotrophicum S58T) Azorhizobium sp. |
| Proteome for NifHDK proteins | 71% (KM1), 69% (KM2) | Bradyrhizobium spp. (including B. sp. S23321 and B. sp. S58T) | |
| Rhizosphere of two cultivars (IPA 1011, IS 5322-C) with low (LF) and high fertilizer (HF) in Cerrado soil, Brazil [19] | Sequencing of nifH DNA clones | In IPA-LF 43% 21% 7% 18% In IPA-HF 29% 10% 16% 7% 8% 5% In IS-LF 42% 10% 10% 13% In IS-HF 39% 23% 11% 10% 13% | Bradyrhizobium sp. B. sp. AF484629 Rhizobium etli Azohydromonas australica Bradyrhizobium sp. B. sp. AF484629 Rhizobium etli Azohydromonas australica Ideonella sp. Burkholderia vietnamiensis Bradyrhizobium sp. B. sp. AF484629 Azohydromonas australica Ideonella sp. Bradyrhizobium sp. B. sp. AF484629 Sinorhizobium sp. Azohydromonas australica Ideonella sp. |
| Site and Sample of Investigation | Detection of nifH Genes | Close Genus Abundance and Bacteria | |
|---|---|---|---|
| Shoots from the tallgrass prairie of northern Oklahoma, USA [21] | Sequencing of nifH DNA clones | 7% 19% 6% 15% 11% 12% 6% 5% 4% 4% 4% | Bradyrhizobium sp. BTAi1 B. sp. MAFF210318 Burkholderia spp. Sphingomonas azotifigens Rhizobium helanshanense Desulfuromonas spp. Azospirillum lipoferum Klebsiella sp. Anaeromyxobacter spp. Geobacter spp. Syntrophobacter fumaroxidans |
| Roots from the tallgrass prairie of northern Oklahoma, USA [21] | Sequencing of nifH DNA clones | 9% 12% 18% 17% 21% 13% 14% 6% 1.5% | Bradyrhizobium sp. BTAi1 B. sp. MAFF210318 B. japonicum Burkholderia spp. Sphingomonas azotifigens Anaeromyxobacter spp. Geobacter spp. Methylocystis sp. Methylobacterium nodulans |
| RT-PCR amplification of nifH RNA | 10% 36% 13% 7% 9% 15% | Burkholderia spp. Rhizobium helanshanense Desulfuromonas spp. Geobacter spp. Azoarcus sp. BH72 Methylobacterium nodulans | |
| Site and Sample of Investigation | Detection of nifH Genes | Close Genus Abundance and Bacteria | |
|---|---|---|---|
| Stems of 8-month-old sugarcane cv. KF92-93, cv. NCo310 and cv. NiF8 grown in Miyako Island, Japan [11] | Sequencing of nifH DNA clones | 100% (KF), 88% (NCo) 73% (NiF) 27% (NiF) | Bradyrhizobium spp. Klebsiella spp. Serratia spp. |
| Stems of 50- and 100-day-old sugarcane (cv. NiF8) grown on a commercial soil under high temperature [10] | Sequencing of nifH DNA clones | 22% (50), 19% (100) 17% (50), 16% (100) 19% (50), 19% (100) 15% (50), 13% (100) 22% (50), 13% (100) | Bradyrhizobium sp. BTAi1 B. sp. IRBG230 B. sp. MAFF210318 Azorhizobium caulinodans Rhizobium daejonense |
| RT-PCR amplification of nifH RNA | 87% (100) 5% (50), 4% (100) 52% (50) | B sp. IRBG230 B. sp. MAFF210318 Azorhizobium caulinodans | |
| Roots of 50- and 100-day-old sugarcane (cv. NiF8) grown on a commercial soil under high temperature [10] | Sequencing of nifH DNA clones | 30% (50), 32% (100) 25% (50), 25% (100) 12% (50), 13% (100) 7% (50), 9% (100) 23% (50), 22% (100) | B. sp. MAFF210318 B. sp. IRBG230, Azorhizobium caulinodans Rhizobium daejonense Beijerinckia derxii |
| RT-PCR amplification of nifH RNA | 19% (50) 100% (100) 5% (50) 17% (50) 23% (50) | B. sp. MAFF210318 B. sp. IRBG230, Azorhizobium caulinodans Sinorhizobium fredii Beijerinckia derxii | |
| Roots of 59- and 100-day-old sugarcane (cv. NiF8) grown on Ishigaki soil under low temperature [10] | Sequencing of nifH DNA clones | 14% (59), 19% (100) 20% (59), 19% (100) 39% (59), 37% (100) 6% (59), 7% (100) 14% (100) | B. sp. MAFF210318 B. sp. IRBG230, Rhizobium daejonense Methylocystis rosea Methylobacterium sp. |
| RT-PCR amplification of nifH RNA | 50% (59), 100%(100) 50% (59) | B. sp. BTAi1 Burkholderia ferrariae | |
| Roots of 59- and 100-day-old sugarcane (cv. NiF8) grown on Tanegashima soil under low temperature [10] | Sequencing of nifH DNA clones | 46% (59), 24% (100) 18% (59), 29% (100) 4% (59), 15% (100) 12% (100) 4% (59) 6% (59) | B. sp. MAFF210318 B. sp. IRBG230, B. sp. IRBG228, Methylobacterium nodulans Methylocella silvestris Azonexus caeni |
| RT-PCR amplification of nifH RNA | 100% (100) | B. sp. MAFF210318 | |
| Site and Sample of Investigation | Detection of nifH Genes | Close Genus Abundance and Bacteria | |
|---|---|---|---|
| Leaf sheath of 6-month-old sugarcane (cv. RB 867515) grown in EMBRAPA without fertilizer and inoculation [41] | Determination of 16S rRNA cDNA sequences | 81% 19% | α-Proteobacteria (mostly Gluconacetobacter) β-Proteobacteria (Burkholderia spp., Herbaspirillum spp.) |
| RT-PCR amplification of nifH RNA | 10% 6% 84% | Rhizobium spp. Paraburkholderia tropica Idenella/Herbaspirillum-like bacteria | |
| Root of 6-month-old sugarcane (cv. RB 867515) grown in EMBRAPA without fertilizer and inoculation [41] | Determination of 16S rRNA cDNA sequences | 42% 3% 11% 17% 2% 25% | α-Proteobacteria (Rhizobium spp., Bradyrhizobium spp.) β-Proteobacteria δ-Proteobacteria Actinobacteria Acidobacteria, Planctomycetes |
| RT-PCR amplification of nifH RNA | 8% 20% 36% 24% 12% | Azospirillum brasilense Bradyrhizobium spp. Methylocapsa spp. Paraburkholderia tropica Idenella/Herbaspirillum-like | |
| White shoot roots of 5-month-old sugarcane (cv. RB867515) grown on EMBRAPA field [42] | Trap-plant (siratro) isolates Determination of 16S rRNA and cDNA sequence | 96% 4% | Bradyrhizobium spp. Rhizobium sp. (no nodC) |
| Trap-plant (cowpea) isolates Determination of 16S rRNA and cDNA sequence | 23% 3% | Bradyrhizobium spp. Rhizobium spp. | |
| Direct plate isolates | 6/9 1/9 1/9 1/9 | Bradyrhizobium spp. (4 no nodC) Rhizobium sp. (no nodC) Methylobacterium Herbaspirillum | |
| Site and Sampling of Investigation | Detection of nifH Genes | Close Genus (Similarity >91%) Abundance and Bacteria | |
|---|---|---|---|
| Stem of African sweet potato grown in Uganda and Kenya [47] | Sequencing of nifH DNA clones | 17% (Kenya) 71% (Kenya) 83% (Uganda) | Bradyrhizobium sp. ANU 289 Azoarcus sp. BH72 Clostridium pasteurianum |
| Stem harvested in Oct. 2002, Aug. 2004 and Oct. 2004 from cv. Beniazuma grown in Andozol, Japan [48] | Sequencing of nifH DNA clones | 31% (O2) 18% (O4) 18% (O4) 100% (A4), 63% (O4) | Herbaspirillum seropedicae B. sp. MAFF210318 B. sp. IRBG230 Azohydromonas australica |
| PCR amplification of nifH RNA | 77% (O2) 22% (O2) 40% (A4) 60% (A4) | Bacillus sp. BT97 B. sp. IRBG228 B. sp. MAFF210318 B. sp. IRBG230 | |
| Stem harvested in Oct. 2005 and Aug. 2006 from cv. Ayamurasaki grown on a gray lowland soil, Japan [48] | Sequencing of nifH DNA clones | 100% (O5) 27% (A6) 18% (A6) 18% (A6) 36% (A6) | B. sp. IRBG230 Pelomonas saccharophila Azohydromonas australica Paraburkholderia unamae Tolypothrix sp. PCC7601 |
| PCR amplification of nifH RNA | 100% (A6) | Pelomonas saccharophila | |
| Tuber of African sweet potato grown in Uganda and Kenya [48] | Sequencing of nifH DNA clones | 28% (Kenya) 71% (Uganda), 14% (Kenya) 71% (Kenya) 29% (Uganda), 28% (Kenya) | Bradyrhizobium japonicum Sinorhizobium meliloti Azoarcus sp. BH72 Paenibacillus odorifer |
| Tuber harvested in Oct. 2002, and Oct. 2004 from cv. Beniazuma grown on an Andozol, Japan [48] | Sequencing of nifH DNA clones | 46% (O2) 23% (O2), 30% (O4) 70% (O4) | B. sp. MAFF210318 Bradyrhizobium japonicum Rhizobium leguminosarum |
| PCR amplification of nifH RNA | 15% (O2) 85% (O2) | Bradyrhizobium japonicum Bacillus sp. BT97 | |
| Tuber harvested in Oct. 2005, Aug. 2006, and Oct. 2006 from cv. Ayamurasaki grown on a gray lowland soil, Japan [48] | Sequencing of nifH DNA clones | 46% (O5) 25% (A6), 28% (O6) 14% (O6) 10% (O5), 33% (A6), 21% (O6) 13% (O5), 17% (A6), 14% (O6) 18% (O5) | Azorhizobium caulinodans B. sp. IRBG230 Sinorhizobium sp. Pelomonas saccharophila Azohydromonas australica Paraburkholderia vietnamiensis |
| PCR amplification of nifH RNA | 100% (A6) | B. sp. IRBG230 | |
| Site and Sample of Investigation | Detection of nifHDK Genes and Their Proteins | Closest Genus Abundance and Bacteria | |
|---|---|---|---|
| Roots harvested from paddy rice grown on Kyushu University field [58] | Sequencing of nifH DNA clones | γ-Proteobacteria (Klebsiella pneumoniae, Azotobacter) δ-Proteobacteria (Desulfovibrio gigas) | |
| Roots harvested from paddy rice (cv. IR55423-01) grown in IRRI field, the Philippines at flowering [59] | Metagenome for nifH DNA | 3/5 1/5 1/5 | Bradyrhizobium sp. BTAi1 Xanthobacter autotrophicus Dickeya dadantii |
| RT-PCR amplification of nifH RNA | Geobacter spp. | ||
| Roots harvested from cv. Nipponbare rice grown on Tohoku University field at flowering stage [4] | Metaproteome for NifHDK | 29.7% 21.8% 9.3% | Methylocystaceae (Methylosinus sp., Methylocystis sp.) Bradyrhizobiaceae (Bradyrhizobium, Rhodopseudomonas) Burkholderiaceae |
| Rhizosphere from paddy rice field of Fujian province, China [60] | RT-PCR amplification of nifH RNA | 4 clones 3 clones 4 clones | α-Proteobacteria (Rhizobium, Methylocystis) β-Proteobacteria (Azoarcus sp., Azospira oryzae, Azotobacter sp.) γ-Proteobacteria (Methylococcus) δ-Proteobacteria (Geobacter) Firmicutes (Helicobacter) |
| Rhizosphere collected at IRRI fields, the Philippines 59 to 76 days after rice transplanting [61] | Metagenome 16S rRNA | α-Proteobacteria (Rhizobium, Methylobacterium) Actinobacteria (Microbacterium) | |
| Sequencing of nifH DNA clones | Rhizobium, Methylococcus, Dechloromonas, Anaeromyxobacter, Syntrophobacter, some methanogenic archaea | ||
| Metaproteome | 33% | α-Proteobacteria (Bradyrhizobium, Rhodopseudomonas, Azospirillum, Methylobacterium, Magnetospirillum, Methylosinus) β-Proteobacteria (Dechloromonas, Acidovorax, Herbaspirillum) δ-Proteobacteria (Anaeromyxobacter, Geobacter, Desulfovibrio) | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoneyama, T.; Terakado-Tonooka, J.; Bao, Z.; Minamisawa, K. Molecular Analyses of the Distribution and Function of Diazotrophic Rhizobia and Methanotrophs in the Tissues and Rhizosphere of Non-Leguminous Plants. Plants 2019, 8, 408. https://doi.org/10.3390/plants8100408
Yoneyama T, Terakado-Tonooka J, Bao Z, Minamisawa K. Molecular Analyses of the Distribution and Function of Diazotrophic Rhizobia and Methanotrophs in the Tissues and Rhizosphere of Non-Leguminous Plants. Plants. 2019; 8(10):408. https://doi.org/10.3390/plants8100408
Chicago/Turabian StyleYoneyama, Tadakatsu, Junko Terakado-Tonooka, Zhihua Bao, and Kiwamu Minamisawa. 2019. "Molecular Analyses of the Distribution and Function of Diazotrophic Rhizobia and Methanotrophs in the Tissues and Rhizosphere of Non-Leguminous Plants" Plants 8, no. 10: 408. https://doi.org/10.3390/plants8100408
APA StyleYoneyama, T., Terakado-Tonooka, J., Bao, Z., & Minamisawa, K. (2019). Molecular Analyses of the Distribution and Function of Diazotrophic Rhizobia and Methanotrophs in the Tissues and Rhizosphere of Non-Leguminous Plants. Plants, 8(10), 408. https://doi.org/10.3390/plants8100408




