Morphological and Molecular Characterization of Three Endolichenic Isolates of Xylaria (Xylariaceae), from Cladonia curta Ahti & Marcelli (Cladoniaceae)
Abstract
1. Introduction
2. Results
2.1. Isolation of Endolichenic Fungi
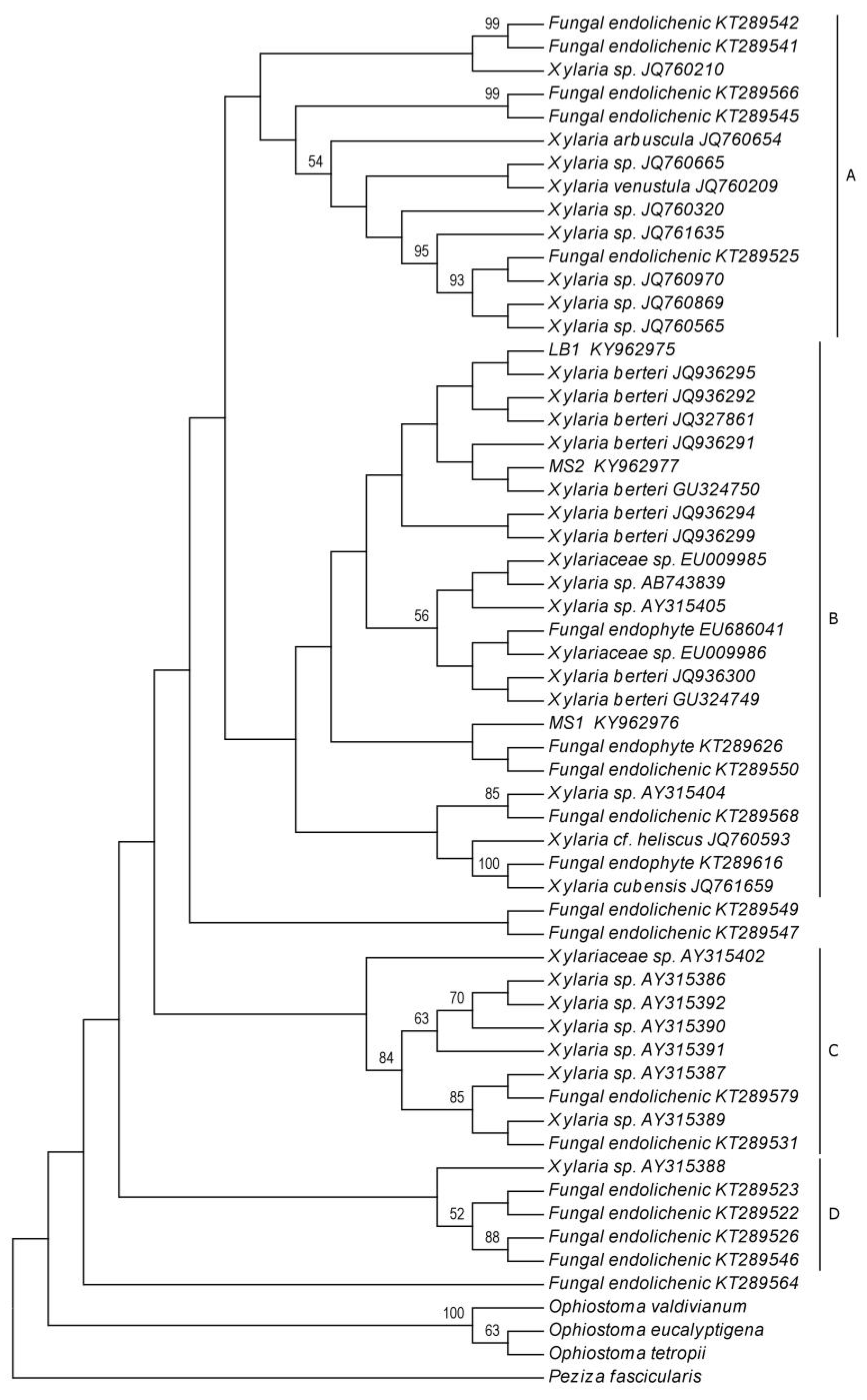
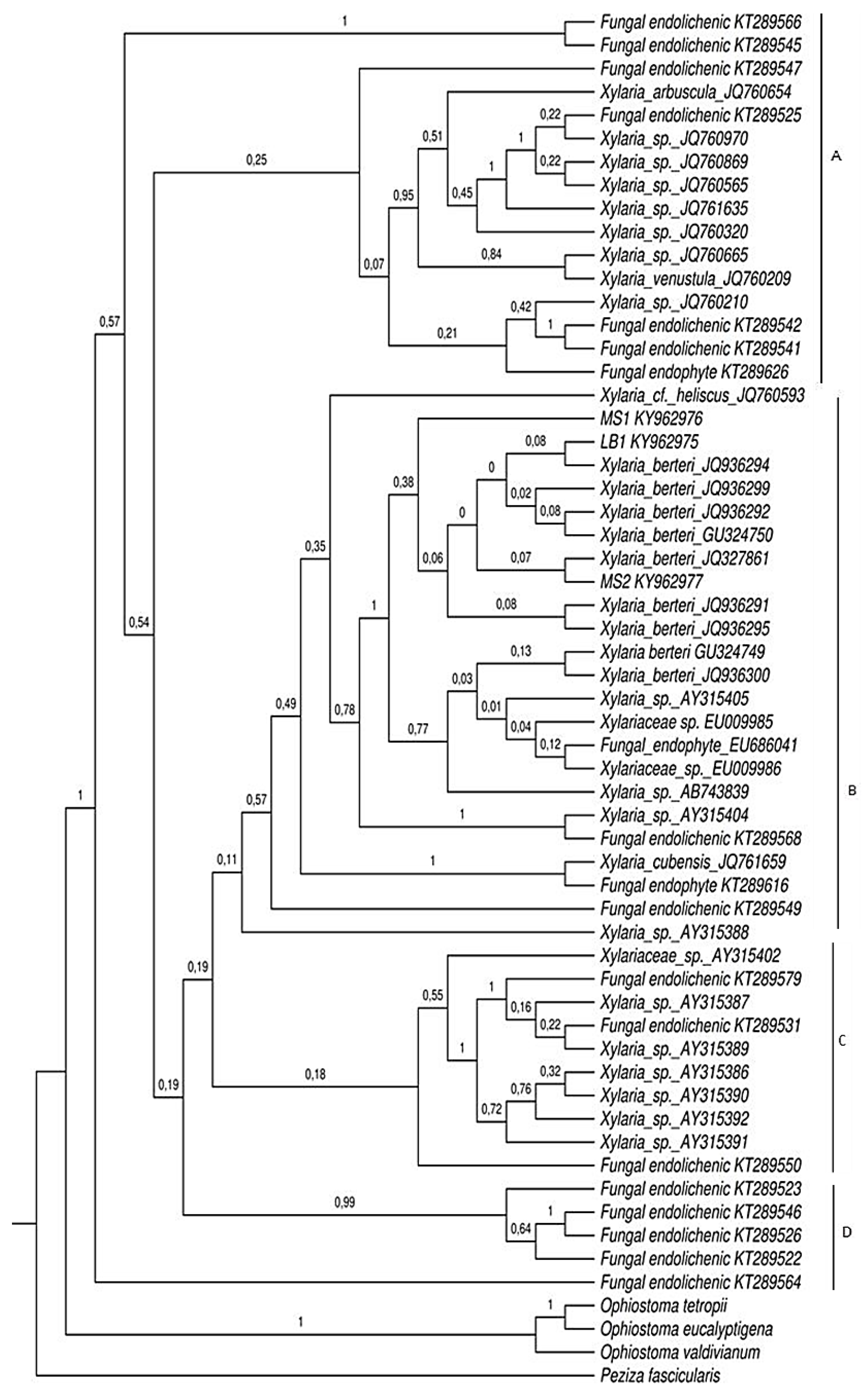
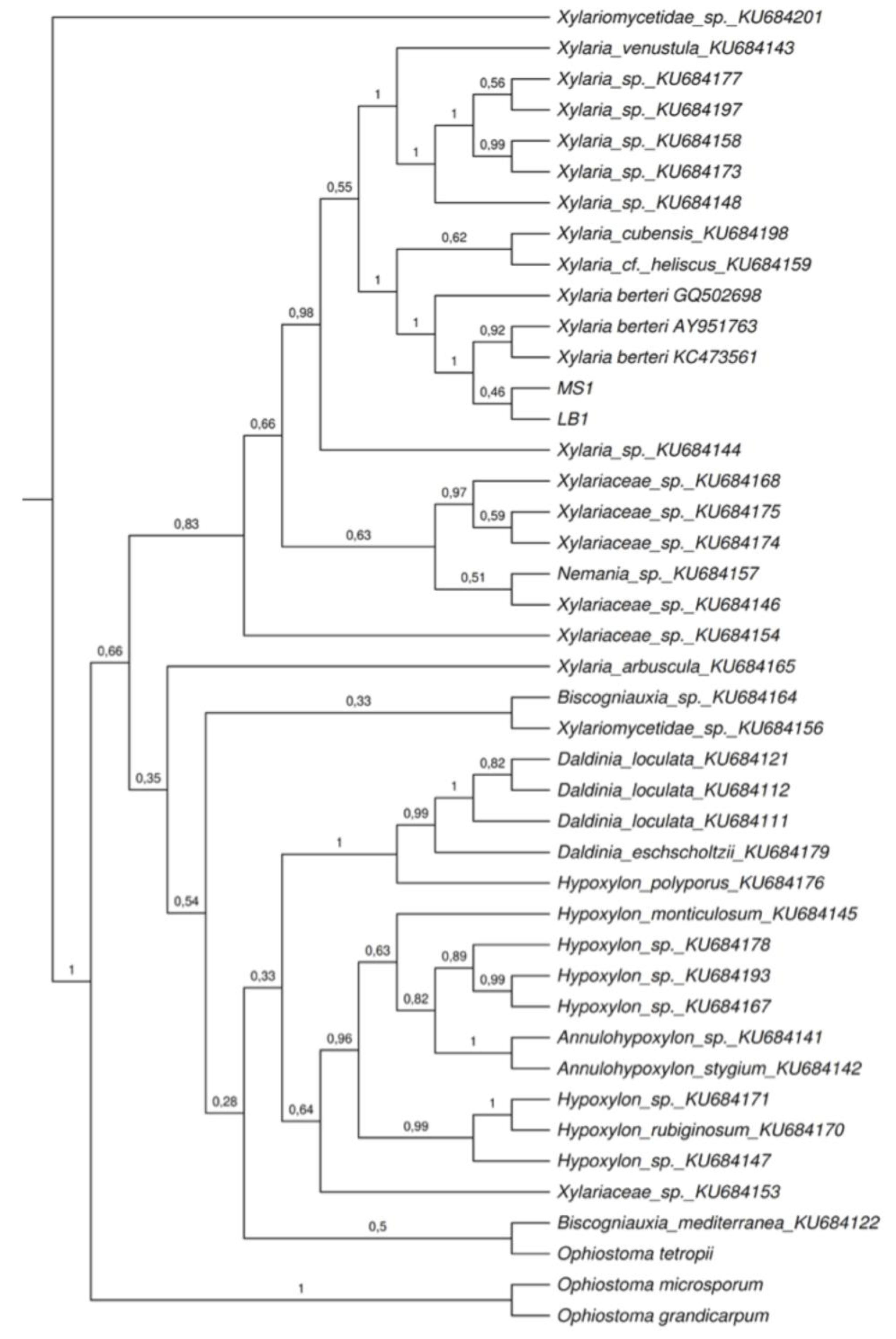
2.2. Phylogenetic Analyses
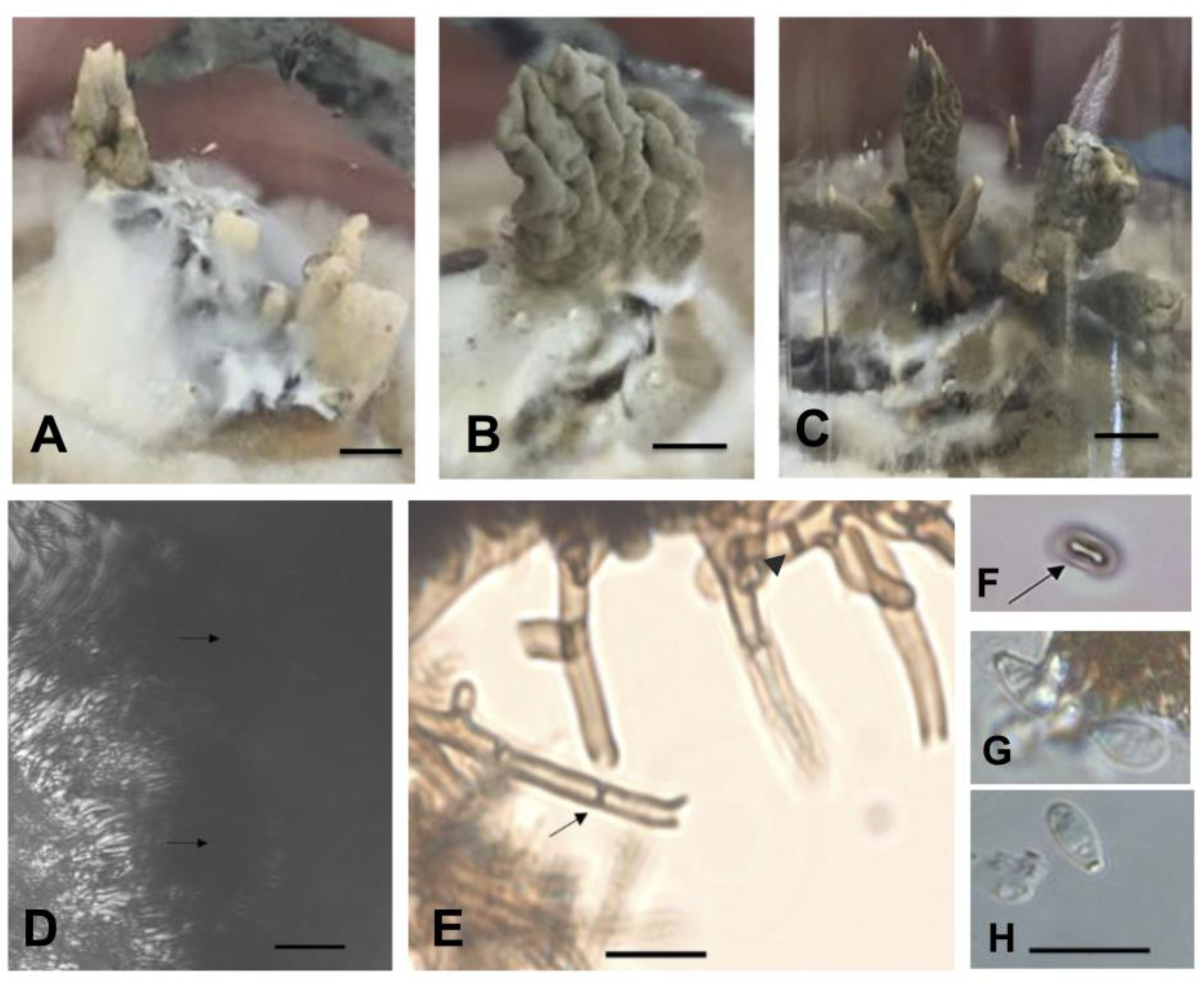
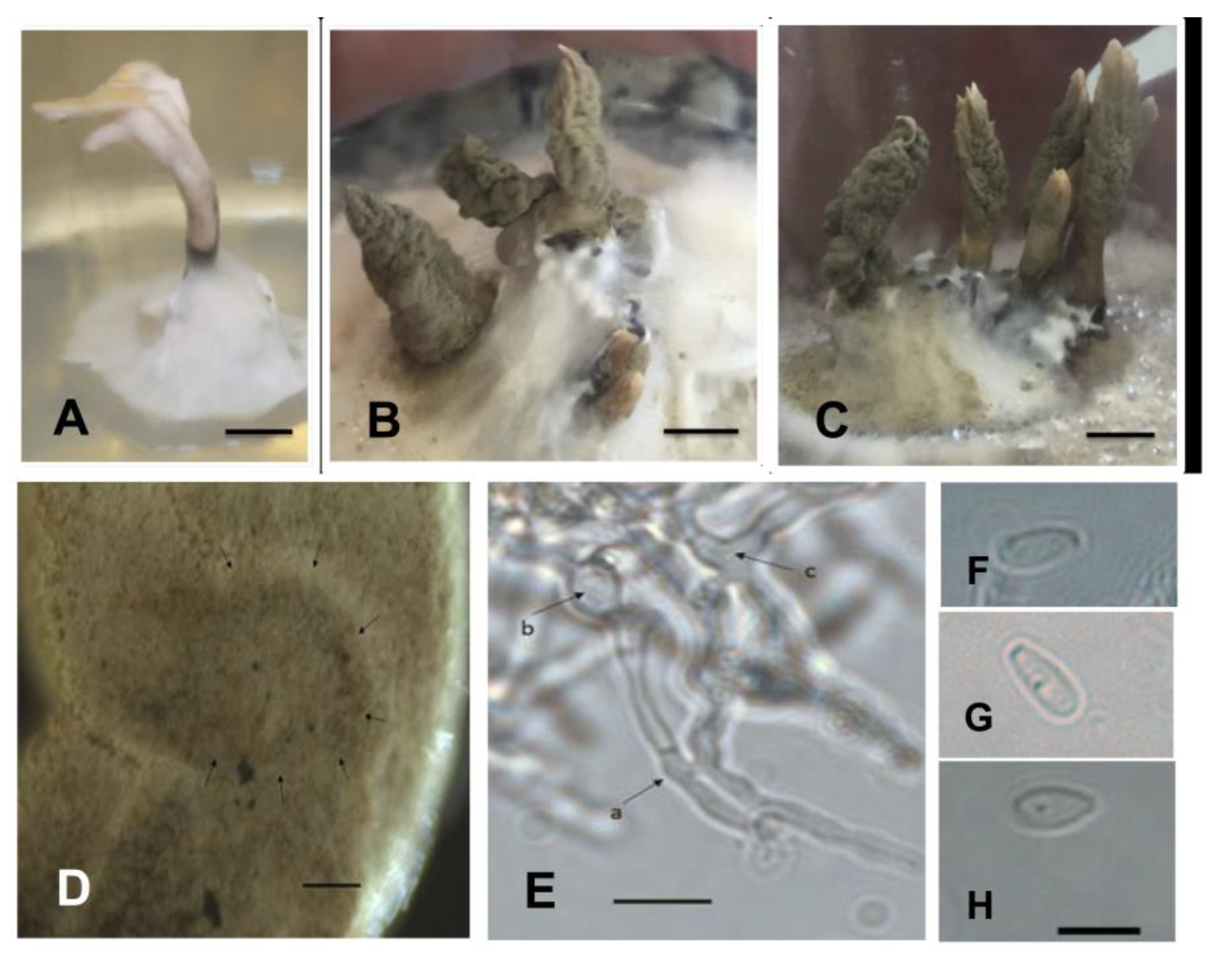
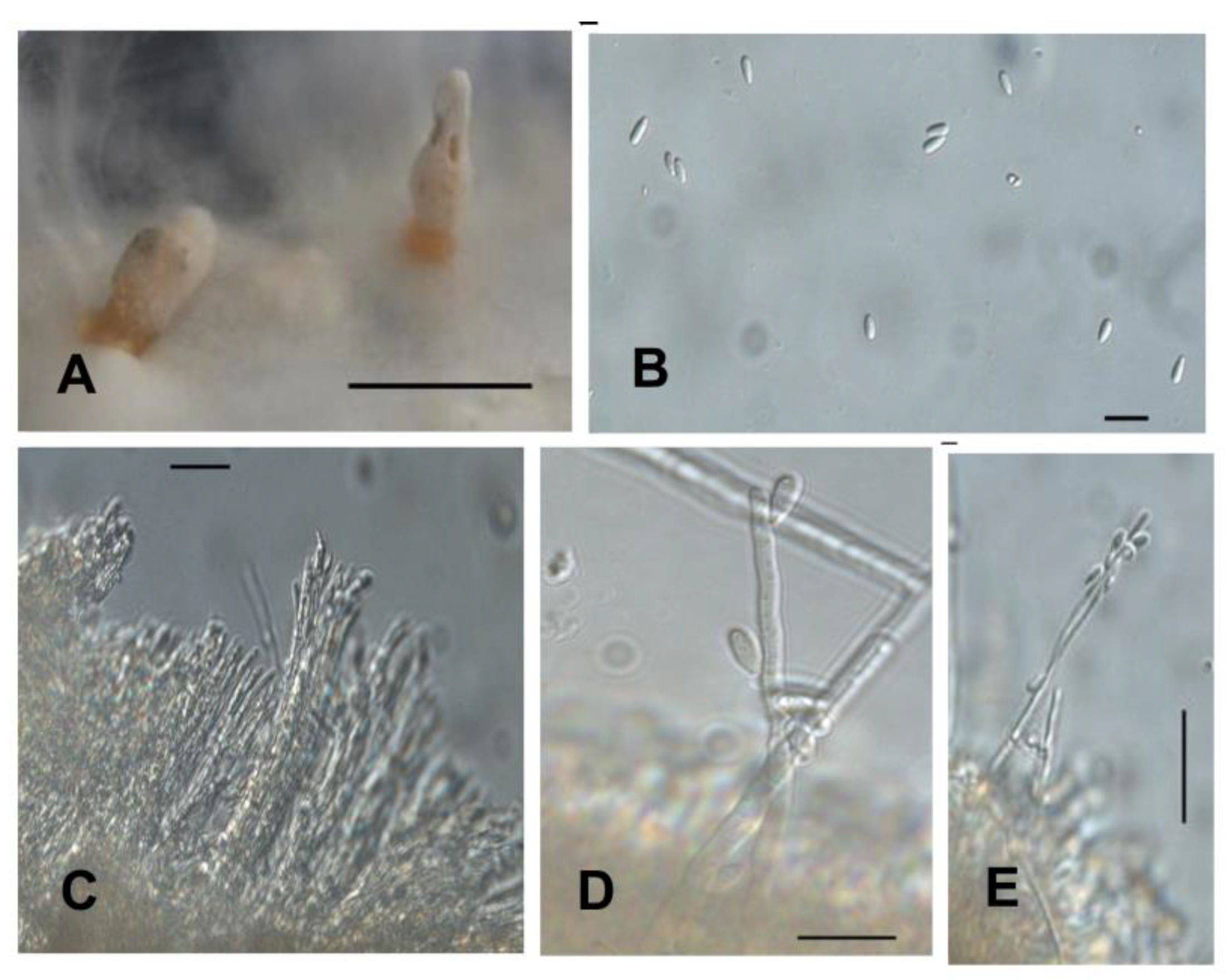
2.3. Phenotypic Characterization of Isolates in Culture Medium
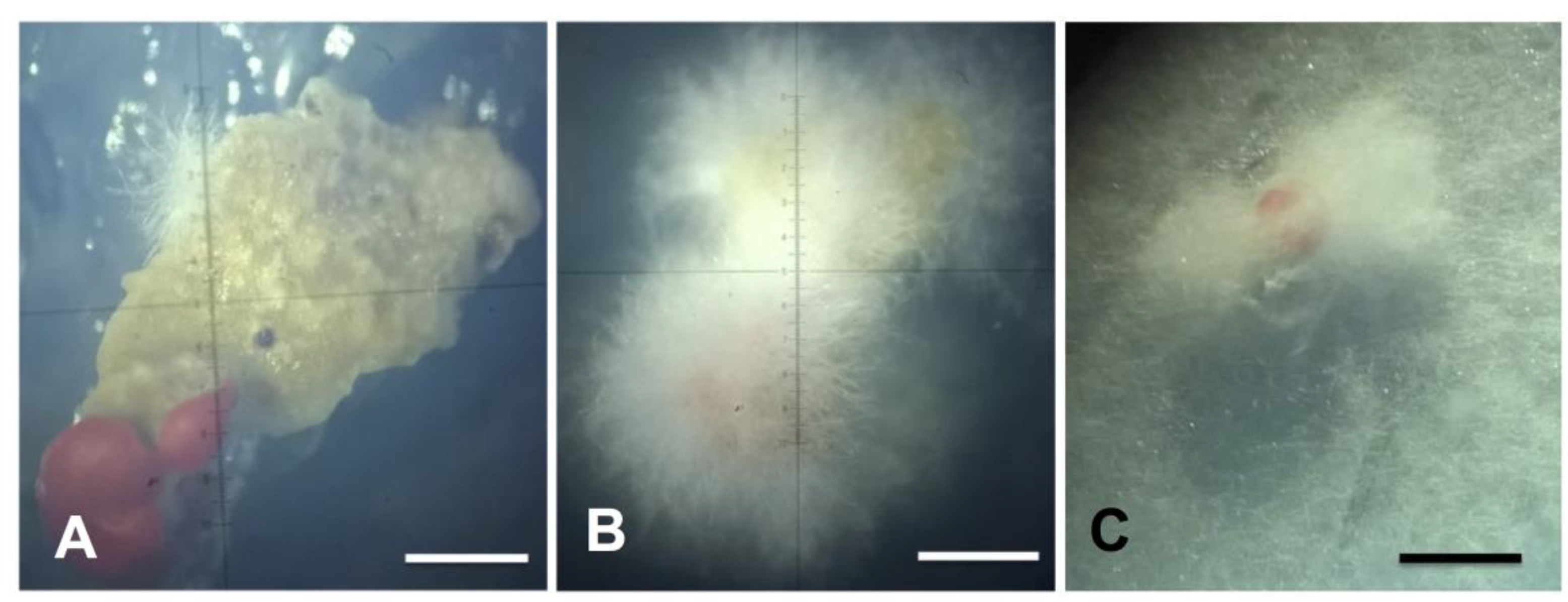
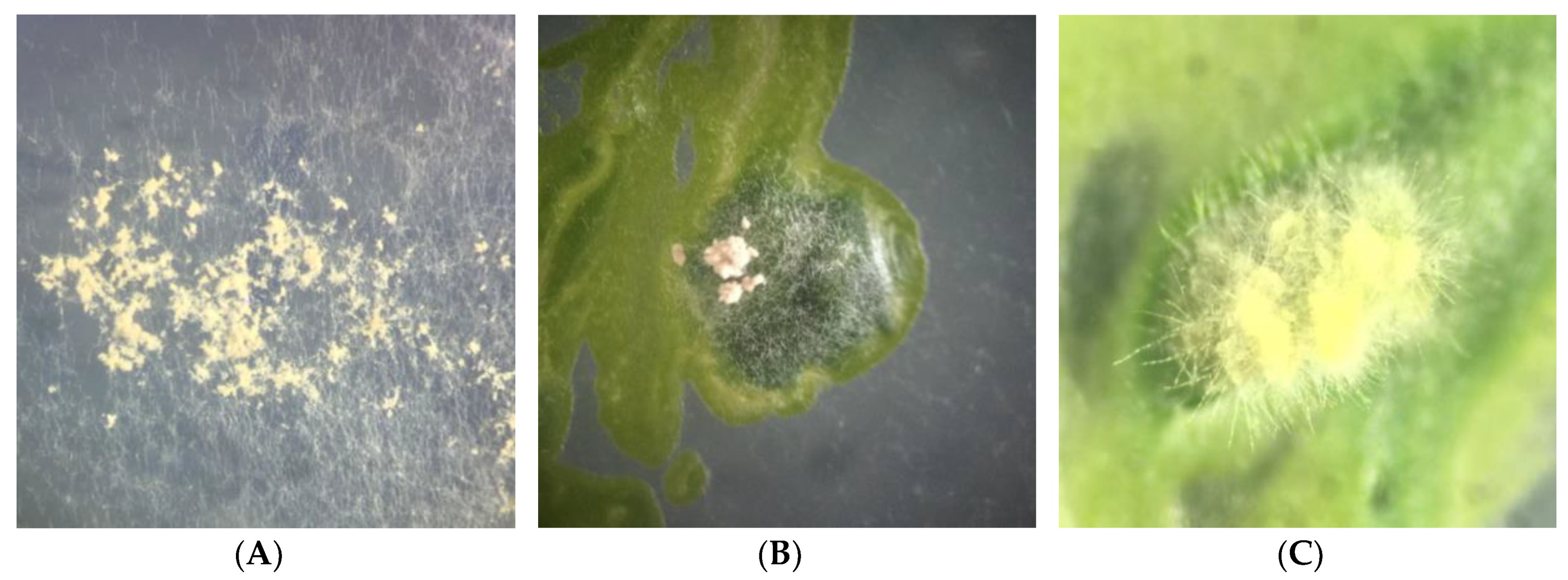
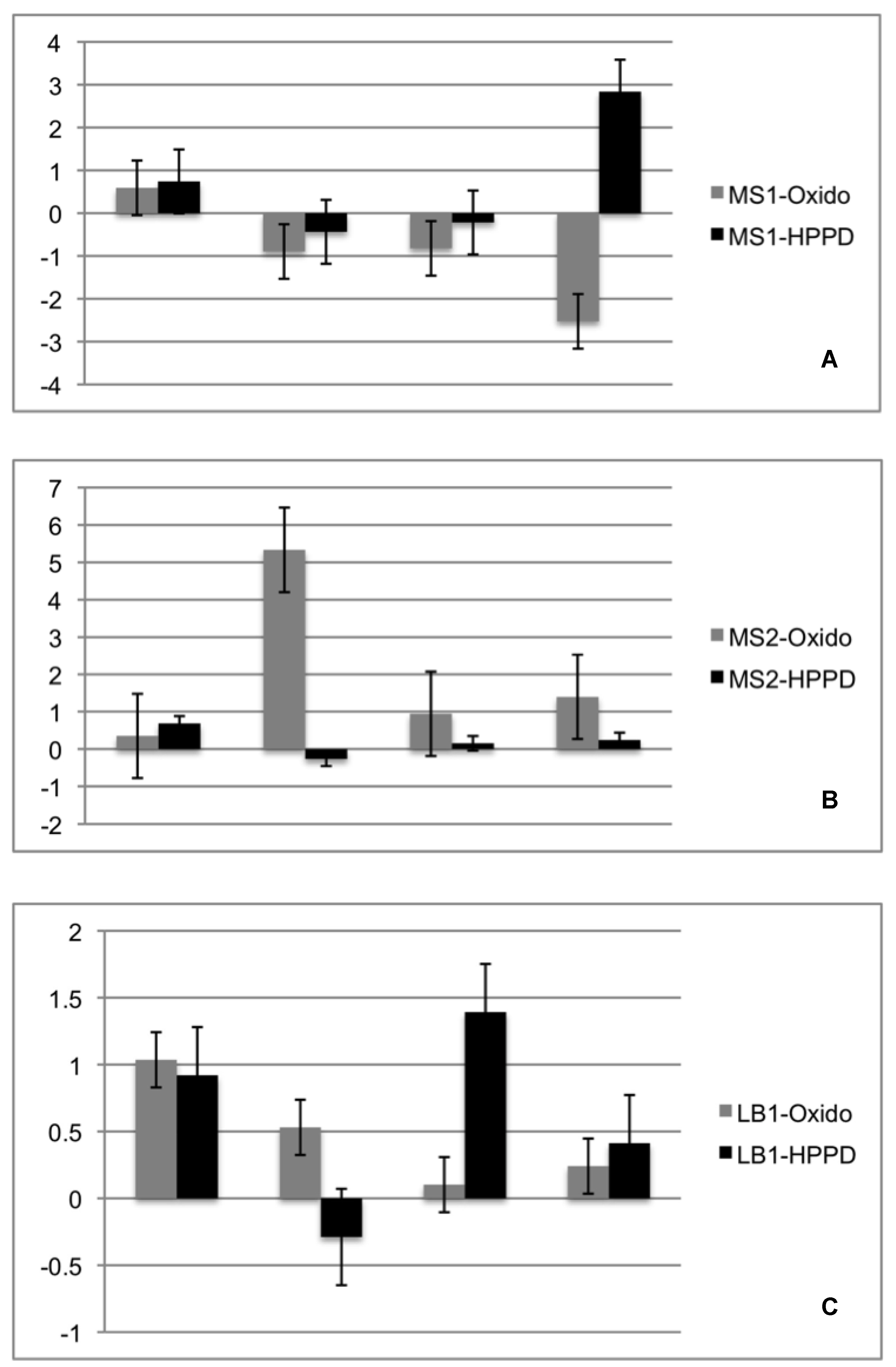
2.4. Re-Synthesis and qPCR Evaluation on the Endophytic Role in the Lichen Symbiosis
3. Discussion
4. Materials and Methods
4.1. Collection
4.2. Fungal Isolation and Culture
4.3. Phenotypic Characterization of Isolates in Culture Medium
4.4. DNA Extraction, Amplification, PCR Purification, and Sequencing
4.5. Phylogenetic Analyses
4.6. Re-Synthesis and qPCR Evaluation on the Endophytic Role in the Lichen Symbiosis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arnold, A.E.; Maynard, Z.; Gilbert, G.S.; Coley, P.D.; Kursar, T.A. Are Tropical Fungal Endophytes Hyperdiverse? Ecol. Lett. 2000, 3, 267–274. [Google Scholar] [CrossRef]
- Suryanarayanan, T.S.; Thirunavukkarasu, N.; Hariharan, G.N.; Balaji, P. Occurrence of Non-Obligate Microfungi inside Lichen Thalli. Sydowia-Horn- 2005, 57, 120. [Google Scholar]
- Suryanarayanan, T.S.; Govindarajulu, M.B.; Rajamani, T.; Tripathi, M.; Joshi, Y. Endolichenic fungi in lichens of Champawat district, Uttarakhand, Northern India. Mycol. Prog. 2017, 16, 205–211. [Google Scholar] [CrossRef]
- Tripathi, M.; Joshi, Y. Endolichenic fungi in Kumaun Himalaya: A case study. Recent Adv. Lichenol. 2015, 6, 111–120. [Google Scholar]
- U’Ren, J.M.; Lutzoni, F.; Miadlikowska, J.; Arnold, A.E. Community analysis reveals close affinities between endophytic and endolichenic fungi in mosses and lichens. Microb. Ecol. 2010, 60, 340–353. [Google Scholar] [CrossRef]
- Wu, W.; Dai, H.; Bao, L.; Ren, B.; Lu, J.; Luo, Y.; Guo, L.; Zhang, L.; Liu, H. Isolation and Structural Elucidation of Proline-Containing Cyclopentapeptides from an Endolichenic Xylaria Sp. J. Nat. Prod. 2011, 74, 1303–1308. [Google Scholar] [CrossRef]
- Kannangara, B.T.S.D.P.; Rajapaksha, R.S.C.G.; Paranagama, P.A. Nature and bioactivities of endolichenic fungi in Pseudocyphellaria sp., Parmotrema sp. and Usnea sp. at Hakgala montane forest in Sri Lanka. Lett. Appl. Microbiol. 2009, 48, 203–209. [Google Scholar] [CrossRef] [PubMed]
- U’Ren, J.M.; Lutzoni, F.; Miadlikowska, J.; Laetsch, A.D.; Arnold, A.E. Host and geographic structure of endophytic and endolichenic fungi at a continental scale. Am. J. Bot. 2012, 99, 898–914. [Google Scholar] [CrossRef]
- U’Ren, J.M.; Miadlikowska, J.; Zimmerman, N.B.; Lutzoni, F.; Stajich, J.E.; Arnold, A.E. Contributions of North American endophytes to the phylogeny, ecology, and taxonomy of Xylariaceae (Sordariomycetes, Ascomycota). Mol. Phylogenetics Evol. 2016, 98, 210–232. [Google Scholar] [CrossRef]
- Li, W.C.; Zhou, J.; Guo, S.Y.; Guo, L.D. Endophytic fungi associated with lichens in Baihua mountain of Beijing, China. Fungal Divers. 2007, 25, 69–80. [Google Scholar]
- Padhi, S.; Tayung, K. In Vitro Antimicrobial Potentials of Endolichenic Fungi Isolated from Thalli of Parmelia Lichen against Some Human Pathogens. Beni-Suef Univ. J. Basic Appl. Sci. 2015, 4, 299–306. [Google Scholar] [CrossRef]
- Tripathi, M.; Joshi, Y.; Gupta, R.C. Assessment of endolichenic fungal diversity in some forests of Kumaun Himalaya. Curr. Sci. 2014, 107, 745–748. [Google Scholar]
- Wang, Y.; Zheng, Y.; Wang, X.; Wei, X.; Wei, J. Lichen-associated fungal community in Hypogymnia hypotrypa (Parmeliaceae, Ascomycota) affected by geographic distribution and altitude. Front. Microbiol. 2016, 7, 1231. [Google Scholar] [CrossRef] [PubMed]
- Grube, M.; Cernava, T.; Soh, J.; Fuchs, S.; Aschenbrenner, I.; Lassek, C.; Wegner, U.; Becher, D.; Riedel, K.; Sensen, C.W.; et al. Exploring functional contexts of symbiotic sustain within lichen-associated bacteria by comparative omics. ISME J. 2015, 9, 412. [Google Scholar] [CrossRef] [PubMed]
- Armaleo, D.; Müller, O.; Lutzoni, F.; Andrésson, O.S.; Blanc, G.; Bode, H.B.; Collart, F.R.; Dal Grande, F.; Dietrich, F.; Grigoriev, I.V.; et al. The lichen symbiosis re-viewed through the genomes of Cladonia grayi and its algal partner Asterohloris glomerata. BMC Genom. 2019, 20, 605. [Google Scholar] [CrossRef] [PubMed]
- Guzow-Krzeminska, B.; Stocker-Wörgötter, E. In vitro culturing and resynthesis of the mycobionte Protoparmeliopssis muralis with albal bionts. Lichenologist 2013, 45, 65–76. [Google Scholar] [CrossRef]
- Joneson, S.; Armaleo, D.; Lutzoni, F. Fungal and algal gene expression in early developmental stages of lichen-symbiosis. Mycologia 2011, 1032, 291–306. [Google Scholar] [CrossRef] [PubMed]
- Chagnon, P.L.; U’Ren, J.M.; Miadlikowska, J.; Lutzoni, F.; Arnold, A.E. Interaction type influences ecological network structure more than local abiotic conditions: Evidence from endophytic and endolichenic fungi at a continental scale. Oecologia 2016, 180, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.M.; Lin, C.R.; Fang, M.J.; Rogers, J.D.; Lechat, J.F.C.; Ju, Y.M. Phylogenetic status of Xylaria subgenus Pseudoxylaria among taxa of the subfamily Xylarioideae (Xylariaceae) and phylogeny of the taxa involved in the subfamily. Mol. Phylogenetics Evol. 2010, 54, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.; Ju, Y.M. The Xylariaceae of the Hawaiian Islands. N. Am. Fungi 2012, 7, 1–5. [Google Scholar] [CrossRef]
- Stadler, M.; Kuhnert, E.; Peršoh, D.; Fournier, J. The Xylariaceae as model example for a unified nomenclature following the ‘‘One Fungus-One Name”. (1F1N) concept. Mycology 2013, 4, 5–21. [Google Scholar]
- Lee, J.S.; Ko, K.S.; Jung, H.S. Phylogenetic Analysis of Xylaria Based on Nuclear Ribosomal ITS1-5.8 S-ITS2 Sequences. FEMS Microbiol. Lett. 2000, 187, 89–93. [Google Scholar] [CrossRef]
- Rogers, J.D. Thoughts and musings on tropical Xylariaceae. Mycol. Res. 2000, 104, 1412–1420. [Google Scholar] [CrossRef]
- Davis, E.C.; Shaw, A.J. Biogeographic and phylogenetic patterns in diversity of liverwort-associated endophytes. Am. J. Bot. 2008, 95, 914–924. [Google Scholar] [CrossRef]
- Guo, L.D.; Hyde, K.D.; Liew, E.C.Y. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 2000, 147, 617–630. [Google Scholar] [CrossRef]
- Shibuya, H.; Kitamura, C.; Maehara, S.; Nagahata, M.; Winarno, H.; Simanjuntak, P.; Kim, H.S.; Wataya, Y.; Ohashi, K. Transformation of Cinchona alkaloids into 1-N-oxide derivatives by endophytic Xylaria sp. isolated from Cinchona pubescens. Chem. Pharm. Bull. 2003, 51, 71–74. [Google Scholar] [CrossRef]
- Bayman, P.; Angulo-Sandoval, P.; Báez-ortiz, Z.; Lodge, D.J. Distribution and dispersal of Xylaria endophytes in two tree species in Puerto Rico. Mycol. Res. 1998, 102, 944–948. [Google Scholar] [CrossRef]
- Vaz-Aline, B.M.; Fontenla, S.; Rocha, F.S.; Brandão, L.R.; Vieira, M.L.A.; Garcia, V.; Góes-Neto, A.; Rosa, C.A. Fungal endophyte β-diversity associated with Myrtaceae species in an Andean Patagonian Forest (Argentina) and an Atlantic Forest (Brazil). Fungal Ecol. 2014, 8, 28–36. [Google Scholar]
- Callan, B.E.; Rogers, J.D. A synoptic key to Xylaria species from continental United States and Canada based on cultural anamorphic features. Mycotaxon 1993, XLVI, 141–154. [Google Scholar]
- Rogers, J.D. Xylaria bulbosa, Xylaria curta, and Xylaria longipes in Continental United States. Mycologia 1983, 75, 457. [Google Scholar] [CrossRef]
- Petrini, O.; Petrini, L. Xylariaceous fungi as endophytes. Sydowia Ann. Mycol. Ser. II 1985, 38, 216–234. [Google Scholar]
- Gumboski, E.L.; Peña-Cañón, E.R.; De Albuquerque, M.; Putzke, J.; Pereira, A.B. Occurrence of Cladonia curta Ahti & Marcelli (Cladoniaceae, lichenized Ascomycota) in the Brazilian Pampa biome. R. bras. Bioci. 2018, 16, 69–71. [Google Scholar]
- Petrini, O.; Hake, U.; Dreyfuss, M.M. An Analysis of Fungal Communities Isolated from Fruticose Lichens. Mycologia 1990, 82, 444. [Google Scholar] [CrossRef]
- Ahti, T. Cladoniaceae. Flora Neotrop. 2000, 78, 1–362. [Google Scholar]
- Douanla-Meli, C.; Langer, E. Diversity and Molecular Phylogeny of Fungal Endophytes Associated with Diospyros crassiflora. Mycology 2012, 3, 175–187. [Google Scholar]
- Gazis, R.; Chaverri, P. Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea Brasiliensis) in Peru. Fungal Ecol. 2009, 3, 240–254. [Google Scholar] [CrossRef]
- Hsieh, H.M.; Ju, Y.M.; Rogers, J.D. Molecular phylogeny of Hypoxylon and closely related genera. Mycologia 2005, 97, 844–865. [Google Scholar] [CrossRef]
- De Souza Leite, T.; Cnossen-Fassoni, A.; Pereira, O.L.; Mizubuti, E.S.G.; Araújo, E.F.; Queiroz, M.V. Novel and highly diverse fungal endophytes in soybean revealed by the consortium of two different techniques. J. Microbiol. 2013, 51, 56–69. [Google Scholar] [CrossRef]
- Rojas-Jimenez, K.; Hernandez, M.; Blanco, J.; Vargas, L.D.; Acosta-Vargas, L.G.; Tamayo, G. Richness of Cultivable Endophytic Fungi along an Altitudinal Gradient in Wet Forests of Costa Rica. Fungal Ecol. 2016, 20, 124–131. [Google Scholar] [CrossRef]
- Smith, S.A.; Tank, D.C.; Boulanger, L.A.; Bascom-Slack, C.A.; Kaury Kingery, E.D.; Babbs, B. Bioactive Endophytes Warrant Intensified Exploration and Conservation. PLoS ONE 2008, 3, e3052. [Google Scholar] [CrossRef]
- Vega, F.E.; Simpkins, A.; Aime, M.C.; Posada, F.; Peterson, S.W.; Rehner, S.A.; Infante, F.; Castillo, A.; Arnold, A.E. Fungal endophyte diversity in coffee plants from Colombia, Hawai’i, Mexico and Puerto Rico. Fungal Ecol. 2010, 3, 122–138. [Google Scholar] [CrossRef]
- Fu, C.H.; Hsieh, H.M.; Chen, C.Y.; Chang, T.T.; Huang, Y.M.; Ju, Y.M. Ophiodiaporthe Cyatheae gen. et sp. nov., a diaporthalean pathogen causing a cevastating wilt disease of Cyathea Lepifera in Taiwan. Mycologia 2013, 105, 861–872. [Google Scholar] [CrossRef]
- Davis, E.C.; Franklin, J.B.; Shaw, A.J.; Vilgalys, R. Endophytic Xylaria (Xylariaceae) among Liverworts and Angiosperms: Phylogenetics, Distribution, and Symbiosis. Am. J. Bot. 2003, 90, 1661–1667. [Google Scholar] [CrossRef]
- Rogers, J.D.; Ju, Y.M. Key to Hawaiian Rosellinia taxa and additions to host-fungus index. N. Am. Fungi 2015, 10, 1–4. [Google Scholar]
- Arnold, A.E.; Miadlikowska, J.; Higgins, K.L.; Sarvate, S.D.; Gugger, P.; Way, A.; Hofstetter, V.; Kauff, F.; Lutzoni, F. A Phylogenetic Estimation of Trophic Transition Networks for Ascomycetous Fungi: Are Lichens Cradles of Symbiotrophic Fungal Diversification? Syst. Biol. 2009, 58, 283–297. [Google Scholar] [CrossRef]
- Ko Ko, T.W.; Stephenson, S.L.; Bahkali, A.H.; Hyde, K.D. From morphology to molecular biology: Can we use sequence data to identify fungal endophytes? Fungal Divers. 2011, 50, 113–120. [Google Scholar] [CrossRef]
- Thomas, D.C.; Vandegrift, R.; Ludden, A.; Carroll, G.C.; Bitty, A.R. Spatial Ecology of the Fungal Genus Xylaria in a Tropical Cloud Forest. Biotropica 2016, 48, 381–393. [Google Scholar] [CrossRef]
- Ju, J.M.; Hsieh, H.M.; Rogers, J.D.; Fournier, J.; Jaklitsch, W.M.; Courtecuisse, R. New and interesting penzigioid Xylaria species with small, soft stromata. Mycologia 2012, 104, 766–776. [Google Scholar] [CrossRef]
- Da Silva Cruz, K.; Cortez, V.G. Xylaria (Xylariaceae, Ascomycota) in the Parque Estadual de São Camilo. Acta Biológica Parana. 2015, 44, 129–144. [Google Scholar]
- Hladki, A.I.; Romero, A.I. A preliminary account of Xylaria in the Tucuman Province, Argentina, with a key to the known species from the Northern Provinces. Fungal Divers. 2010, 42, 79–96. [Google Scholar] [CrossRef]
- Medel, R.; Guzmán, G.; Castillo, R. Adiciones Al Conocimiento de Xylaria (Ascomycota, Xylariales) En México. Rev. Mex. Micol. 2010, 31, 9–18. [Google Scholar]
- Callan, B.E.; Rogers, J.D. Teleomorph-anamorph, connection and correlations in some Xylaria species. Mycotaxon 1990, 36, 343–369. [Google Scholar]
- Galvez-Valdivieso, G.; Pineda, M.; Aguilar, M. Functional characterization and expression analysis of p-hydroxyphenylpyruvate dioxygenase from the green alga Chlamydomonas reinhardtii (Chlorophyta). J. Phycol. 2011, 46, 297–308. [Google Scholar] [CrossRef]
- McDonald, T.R.; Gaya, E.; Lutzoni, F. Twenty-five cultures of lichenizing fungi available for experimental studies on symbiotic systems. Symbiosis 2013, 59, 165–171. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Persǒh, D.; Melcher, M.; Graf, K.; Fournier, J.; Stadler, M.; Rambold, G. Molecular and morphological evidence for the delimitation of Xylaria hypoxylon. Mycologia 2009, 101, 256–268. [Google Scholar] [CrossRef]
- Hartun de Capriles, C.; Mata, S.; Middelveen, M. Preservation of fungi in water (Castellani): 20 years. Mycopathologia 1989, 106, 73–80. [Google Scholar] [CrossRef]
- Maerz, A.; Paul, M. A Dictionary of Color, 1st ed.; McGraw-Hill Book Company, Inc.: New York, NY, USA, 1930; 207p. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Application; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Nilsson, R.H.; Kristiansson, E.; Ryberg, M.; Hallenberg, N.; Larsson, K.H. Intraspecific ITS variability in the kingdom Fungi as expressed in the International Sequence Databases and its implications for molecular species identification. Evol. Bioinform. Online 2008, 4, 193–201. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer v1.6. 2014. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 4 November 2016).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| ID Isolate (Size) | Region | Sequences of Genbank | ||||||
|---|---|---|---|---|---|---|---|---|
| Nearest Match | Accession | Query Cover | Max Identity | Substrate Types | Host Species | Reference | ||
| LB1 (565 bp) | ITS rDNA | Xylaria berteri | GU324749 | 99% | 98% | Bark | Dicotyledons | [19] |
| Xylaria berteri | JQ936299 | 98% | 99% | Leaves | Glycine max | [38] | ||
| Xylariaceae sp. | EU009985 | 98% | 98% | Leaves | Coffea arabica | [41] | ||
| Xylaria berteri | JQ327861 | 99% | 98% | Leaves | Myrceugenia ovata var. nanophylla | [28] | ||
| Xylaria berteri | JQ936295 | 99% | 98% | Leaves | Glycine max | [38] | ||
| Xylariaceae sp. | EU009986 | 98% | 98% | Leaves | Coffea arabica | [41] | ||
| Xylaria berteri | JQ936294 | 99% | 98% | Leaves | Glycine max | [38] | ||
| Xylaria berteri | JQ936291 | 98% | 98% | Leaves | Glycine max | [38] | ||
| Xylaria sp. | AB743839 | 99% | 98% | Stems | Cinchona pubescens | [26] | ||
| Xylaria berteri | GU324750 | 99% | 98% | Bark | Dicotyledons | [19] | ||
| MS1 (566 bp) | ITS rDNA | Xylaria berteri | GU324749 | 100% | 99% | Bark | Dicotyledons | [19] |
| Xylaria berteri | JQ327861 | 100% | 98% | Leaves | Myrceugenia ovata var. nanophylla | [28] | ||
| Xylariaceae sp. | EU009985 | 98% | 99% | Leaves | Coffea arabica | [41] | ||
| Xylaria berteri | JQ936295 | 100% | 98% | Leaves | Glycine max | [38] | ||
| Xylariaceae sp. | EU009986 | 98% | 98% | Leaves | Coffea arabica | [28] | ||
| Xylaria berteri | JQ936294 | 100% | 98% | Leaves | Glycine max | [38] | ||
| Xylaria berteri | JQ936291 | 99% | 98% | Leaves | Glycine max | [38] | ||
| Xylaria allantoidea | KR534643 | 98% | 98% | Leaves | Nertera granadensis | [39] | ||
| Xylaria berteri | GU324750 | 100% | 98% | Bark | Dicotyledons | [19] | ||
| MS2 (515 bp) | ITS rDNA | Fungal endophyte | EU686041 | 99% | 99% | leaves | Plagiochila sp. | [24] |
| Xylaria berteri | GU324749 | 100% | 99% | Bark | Dicotyledons | [19] | ||
| Xylaria berteri | JQ936300 | 99% | 99% | Leaves | Glycine max | [38] | ||
| Xylaria berteri | JQ936295 | 100% | 99% | Leaves | Glycine max | [38] | ||
| Xylaria berteri | JQ327861 | 100% | 99% | Leaves | Myrceugenia ovata var. nanophylla | [41] | ||
| Xylaria berteri | JQ936294 | 100% | 99% | Leaves | Glycine max | [38] | ||
| Xylaria berteri | JQ936291 | 100% | 99% | Leaves | Glycine max | [38] | ||
| Xylaria allantoidea | KR534643 | 100% | 98% | Leaves | Nertera granadensis | [39] | ||
| Xylaria berteri | JQ936297 | 100% | 98% | Leaves | Glycine max | [38] | ||
| Xylaria sp. | AB743839 | 100% | 99% | Stems | Cinchona pubescens | [26] | ||
| Xylaria berteri | JQ936292 | 100% | 99% | Leaves | Glycine max | [38] | ||
| Xylaria berteri | JQ936301 | 98% | 98% | Leaves | Glycine max | [38] | ||
| Xylaria berteri | JQ936298 | 98% | 98% | Leaves | Glycine max | [38] | ||
| Xylaria allantoidea | KR534722 | 100% | 98% | Leaves | Leandra longicoma | [39] | ||
| Xylaria sp. | JQ341065 | 100% | 98% | Leaves | Diospyros crassiflora | [34] | ||
| Xylaria allantoidea | FJ884194 | 99% | 98% | Leaves | Hevea brasiliensis | [35] | ||
| Xylaria berteri | GU324750 | 98% | 99% | Bark | Dicotyledons | [19] | ||
| Fungal endophyte | EU977315 | 99% | 98% | Twigs | Angiosperm | [40] | ||
| LB1 (404 bp) | β-tubulin | Xylaria berteri | AY951763 | 100% | 98% | On wood | Castanopsis carlesii var. sessilis | [37] |
| Xylaria berteri | KC473561 | 100% | 98% | Bark | Cyathea lepifera | [42] | ||
| MS1 (409 bp) | β-tubulin | Xylaria berteri | AY951763 | 100% | 97% | On wood | Castanopsis carlesii var. sessilis | [37] |
| Xylaria berteri | KC473561 | 100% | 97% | Bark | Cyathea lepifera | [42] | ||
| MS2 (242 bp) | β-tubulin | Xylaria berteri | FJ904911 | 90% | 77% 1-21 | Leaves | Grevillea robusta | Unpublished |
| Xylaria berteri | AY951763 | 90% | 77% 1-21 | On wood | Castanopsis carlesii var. sessilis | [37] | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cañón, E.R.P.; de Albuquerque, M.P.; Alves, R.P.; Pereira, A.B.; Victoria, F.d.C. Morphological and Molecular Characterization of Three Endolichenic Isolates of Xylaria (Xylariaceae), from Cladonia curta Ahti & Marcelli (Cladoniaceae). Plants 2019, 8, 399. https://doi.org/10.3390/plants8100399
Cañón ERP, de Albuquerque MP, Alves RP, Pereira AB, Victoria FdC. Morphological and Molecular Characterization of Three Endolichenic Isolates of Xylaria (Xylariaceae), from Cladonia curta Ahti & Marcelli (Cladoniaceae). Plants. 2019; 8(10):399. https://doi.org/10.3390/plants8100399
Chicago/Turabian StyleCañón, Ehidy Rocio Peña, Margeli Pereira de Albuquerque, Rodrigo Paidano Alves, Antonio Batista Pereira, and Filipe de Carvalho Victoria. 2019. "Morphological and Molecular Characterization of Three Endolichenic Isolates of Xylaria (Xylariaceae), from Cladonia curta Ahti & Marcelli (Cladoniaceae)" Plants 8, no. 10: 399. https://doi.org/10.3390/plants8100399
APA StyleCañón, E. R. P., de Albuquerque, M. P., Alves, R. P., Pereira, A. B., & Victoria, F. d. C. (2019). Morphological and Molecular Characterization of Three Endolichenic Isolates of Xylaria (Xylariaceae), from Cladonia curta Ahti & Marcelli (Cladoniaceae). Plants, 8(10), 399. https://doi.org/10.3390/plants8100399







