Drawing a Line: Grasses and Boundaries
Abstract
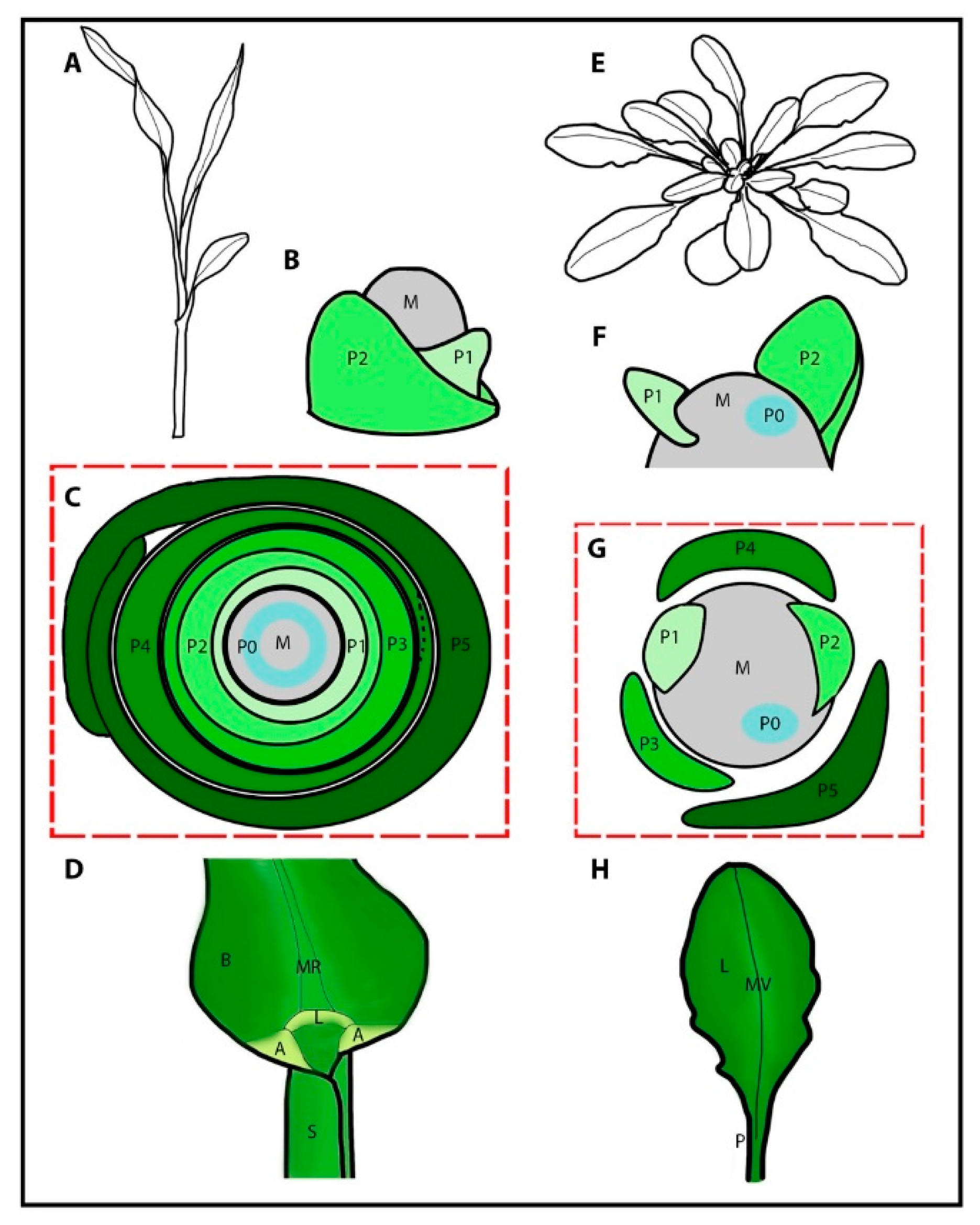
1. Organogenesis
2. Boundaries and Plant Development
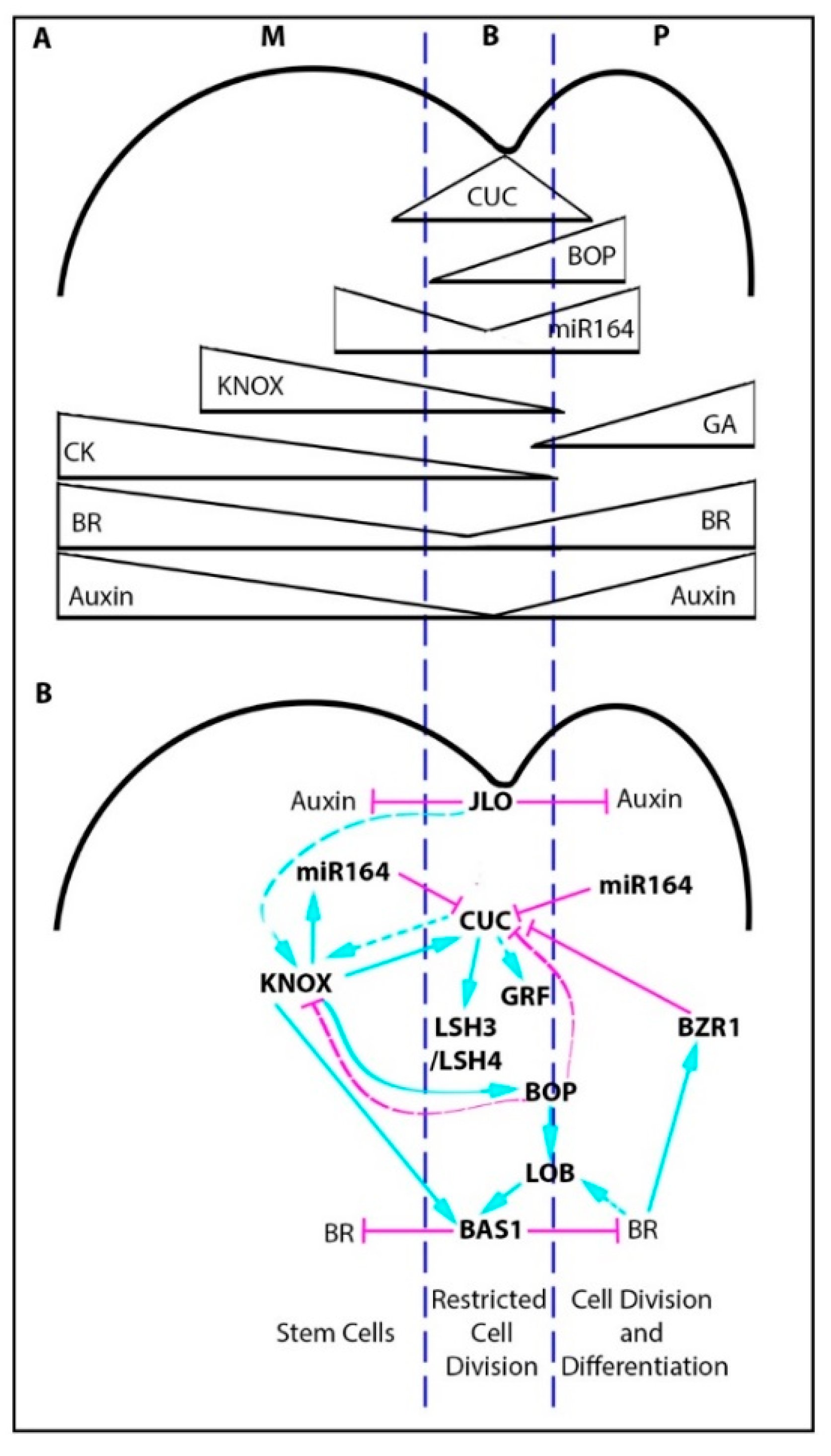
2.1. The Boundary Gene Regulatory Network
2.2. The Boundary Gene Regulatory Network and Leaf Margin Development
3. Vegetative Organogenesis in Grass Crops
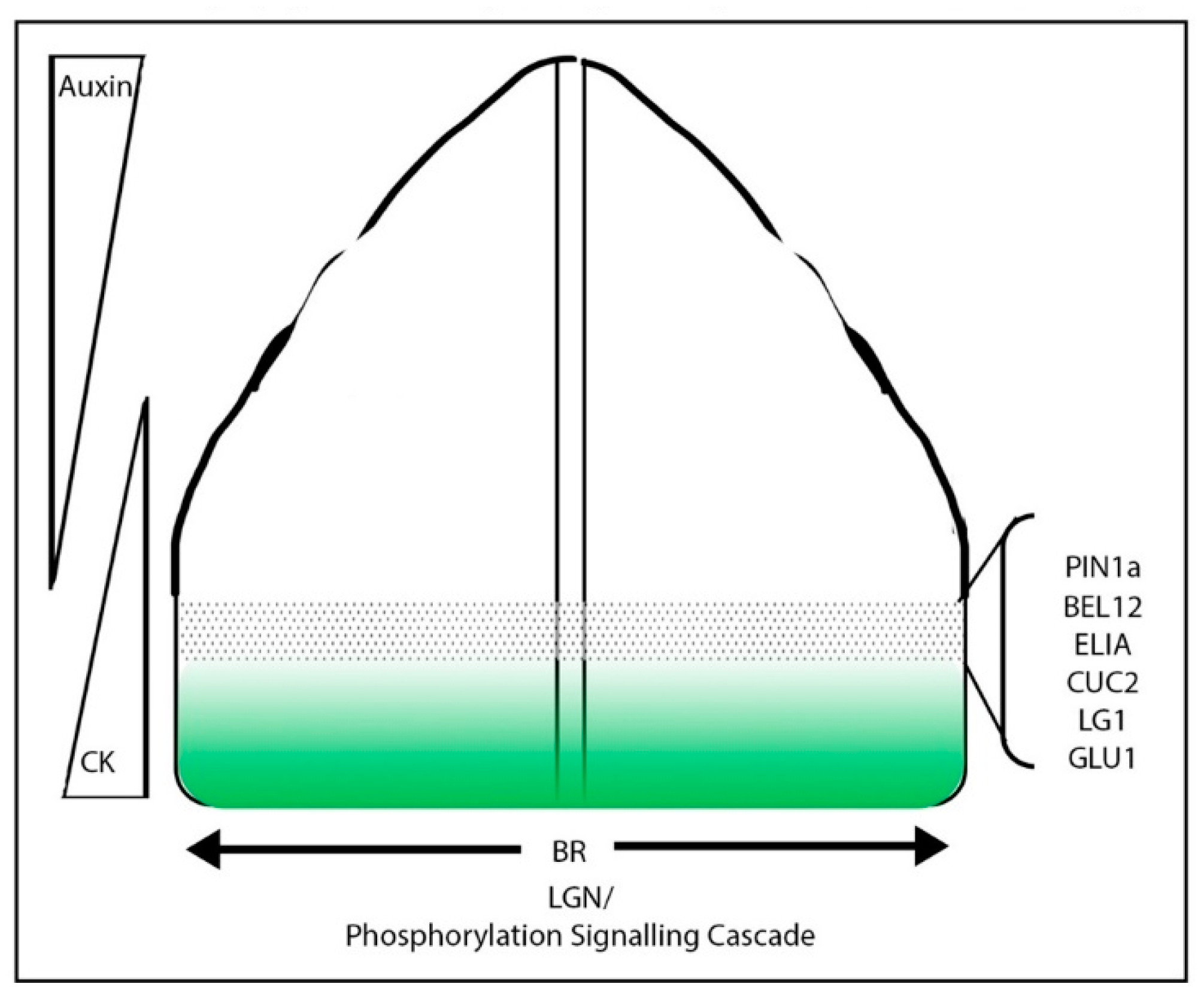
4. Boundary Specification in Grass Crops
4.1. Meristem/Organ Boundaries in the Grasses
4.2. Intra-whorl Boundaries (the Boundary Between the Overlapping Margins of the Sheath) in the Grasses
4.3. Within-Organ Boundaries (the Blade/Sheath Boundary and the Development of the Ligule and Auricle) in the Grasses
4.3.1. Liguleless Mutants and the Patterning of the Ligule
4.3.2. Ectopic Induction of New Blade/Sheath Boundaries
4.3.3. A Proposed Model of Blade/Sheath Boundary Specification
5. Pleiotropy and Boundaries
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jackson, D.; Veit, B.; Hake, S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 1994, 120, 405–413. [Google Scholar]
- Smith, L.G.; Greene, B.; Veit, B.; Hake, S. A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 1992, 116, 21–30. [Google Scholar] [PubMed]
- Long, J.A.; Moan, E.I.; Medford, J.I.; Barton, M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 1996, 379, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Heisler, M.G.; Hamant, O.; Krupinski, P.; Uyttewaal, M.; Ohno, C.; Jönsson, H.; Traas, J.; Meyerowitz, E.M. Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol. 2010, 8, e1000516. [Google Scholar] [CrossRef] [PubMed]
- Abley, K.; De Reuille, P.B.; Strutt, D.; Bangham, A.; Prusinkiewicz, P.; Marée, A.F.M.; Grieneisen, V.A.; Coen, E. An intracellular partitioning-based framework for tissue cell polarity in plants and animals. Development 2013, 140, 2061–2074. [Google Scholar] [CrossRef]
- Bayer, E.M.; Smith, R.S.; Mandel, T.; Nakayama, N.; Sauer, M.; Prusinkiewicz, P.; Kuhlemeier, C. Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 2009, 23, 373–384. [Google Scholar] [CrossRef]
- Bhatia, N.; Bozorg, B.; Larsson, A.; Ohno, C.; Jönsson, H.; Heisler, M.G. Auxin acts through MONOPTEROS to regulate plant cell polarity and pattern phyllotaxis. Curr. Biol. 2016, 26, 3202–3208. [Google Scholar] [CrossRef]
- Jönsson, H.; Heisler, M.G.; Shapiro, B.E.; Meyerowitz, E.M.; Mjolsness, E. An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA 2006, 103, 1633–1638. [Google Scholar] [CrossRef]
- Smith, R.S.; Guyomarc’h, S.; Mandel, T.; Reinhardt, D.; Kuhlemeier, C.; Prusinkiewicz, P. A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA 2006, 103, 1301–1306. [Google Scholar] [CrossRef]
- Stoma, S.; Lucas, M.; Chopard, J.; Schaedel, M.; Traas, J.; Godin, C. Flux-based transport enhancement as a plausible unifying mechanism for auxin transport in meristem development. PLoS Comput. Biol. 2008, 4, e1000207. [Google Scholar] [CrossRef]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of Auxin Transport and Gene Expression during Primordium Development Revealed by Live Imaging of the Arabidopsis Inflorescence Meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, D.; Mandel, T.; Kuhlemeier, C. Auxin Regulates the Initiation and Radial Position of Plant Lateral Organs. Plant Cell 2000, 12, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, D.; Pesce, E.-R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Traas, J.; Friml, J.; Kuhlemeier, C. Regulation of phyllotaxis by polar auxin transport. Nature 2003, 426, 255. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Hibara, K.; Karim, M.R.; Takada, S.; Taoka, K.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 Regulates Postembryonic Shoot Meristem and Organ Boundary Formation. Plant Cell 2006, 18, 2946–2957. [Google Scholar] [CrossRef] [PubMed]
- Hamant, O.; Heisler, M.G.; Jönsson, H.; Krupinski, P.; Uyttewaal, M.; Bokov, P.; Corson, F.; Sahlin, P.; Boudaoud, A.; Meyerowitz, E.M. Developmental patterning by mechanical signals in Arabidopsis. Science 2008, 322, 1650–1655. [Google Scholar] [CrossRef] [PubMed]
- Abad, U.; Sassi, M.; Traas, J. Flower development: From morphodynamics to morphomechanics. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2017, 372. [Google Scholar] [CrossRef] [PubMed]
- Green, P.B. Mechanism for Plant Cellular Morphogenesis. Science 1962, 138, 1404–1405. [Google Scholar] [CrossRef]
- Hamant, O.; Traas, J. The mechanics behind plant development. New Phytol. 2010, 185, 369–385. [Google Scholar] [CrossRef]
- Coen, E.; Rebocho, A.B. Resolving conflicts: Modeling genetic control of plant morphogenesis. Dev. Cell 2016, 38, 579–583. [Google Scholar] [CrossRef] [PubMed]
- Rebocho, A.B.; Kennaway, J.R.; Bangham, J.A.; Coen, E. Formation and Shaping of the Antirrhinum Flower through Modulation of the CUP Boundary Gene. Curr. Biol. 2017, 27, 2610–2622. [Google Scholar] [CrossRef] [PubMed]
- Juarez, M.T.; Twigg, R.W.; Timmermans, M.C.P. Specification of adaxial cell fate during maize leaf development. Development 2004, 131, 4533–4544. [Google Scholar] [CrossRef] [PubMed]
- McConnell, J.R.; Emery, J.; Eshed, Y.; Bao, N.; Bowman, J.; Barton, M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001, 411, 709. [Google Scholar] [CrossRef] [PubMed]
- Waites, R.; Hudson, A. Phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 1995, 121, 2143–2154. [Google Scholar]
- Kumar, S.; Mishra, R.K.; Kumar, A.; Srivastava, S.; Chaudhary, S. Regulation of stipule development by COCHLEATA and STIPULE-REDUCED genes in pea Pisum sativum. Planta 2009, 230, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Harlan, H.V. The origin of hooded barley. J. Hered. 1931, 22, 265–272. [Google Scholar] [CrossRef]
- Müller, K.J.; Romano, N.; Gerstner, O.; Garcia-Marotot, F.; Pozzi, C.; Salamini, F.; Rohde, W. The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 1995, 374, 727. [Google Scholar] [CrossRef]
- Stebbins, G.L.; Yagil, E. The morphogenetic effects of the hooded gene in barley I: The course of development in hooded and awned genotypes. Genetics 1966, 54, 727. [Google Scholar]
- Williams-Carrier, R.E.; Lie, Y.S.; Hake, S.; Lemaux, P.G. Ectopic expression of the maize kn1 gene phenocopies the Hooded mutant of barley. Development 1997, 124, 3737–3745. [Google Scholar]
- Richardson, A.E.; Rebocho, A.B.; Coen, E.S. Ectopic KNOX expression affects plant development by altering tissue cell polarity and identity. Plant Cell 2016. [Google Scholar] [CrossRef]
- Chuck, G.; Lincoln, C.; Hake, S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 1996, 8, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Shuai, B.; Reynaga-Peña, C.G.; Springer, P.S. The Lateral Organ Boundaries Gene Defines a Novel, Plant-Specific Gene Family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Jeon, J.O.; Lee, M.M.; Kim, J.H. Genetic interaction between GROWTH-REGULATING FACTOR and CUP-SHAPED COTYLEDON in organ separation. Plant Signal. Behav. 2015, 10, e988071. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bell, E.M.; Lin, W.; Husbands, A.Y.; Yu, L.; Jaganatha, V.; Jablonska, B.; Mangeon, A.; Neff, M.M.; Girke, T.; Springer, P.S. Arabidopsis LATERAL ORGAN BOUNDARIES negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA 2012, 109, 21146–21151. [Google Scholar] [CrossRef] [PubMed]
- Vroemen, C.W.; Mordhorst, A.P.; Albrecht, C.; Kwaaitaal, M.A.C.J.; de Vries, S.C. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 2003, 15, 1563–1577. [Google Scholar] [CrossRef]
- Takeda, S.; Hanano, K.; Kariya, A.; Shimizu, S.; Zhao, L.; Matsui, M.; Tasaka, M.; Aida, M. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 2011, 66, 1066–1077. [Google Scholar] [CrossRef]
- Tian, C.; Zhang, X.; He, J.; Yu, H.; Wang, Y.; Shi, B.; Han, Y.; Wang, G.; Feng, X.; Zhang, C.; et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 2014, 10, 755. [Google Scholar] [CrossRef]
- Borghi, L.; Bureau, M.; Simon, R. Arabidopsis JAGGED LATERAL ORGANS is Expressed in Boundaries and Coordinates KNOX and PIN Activity. Plant Cell 2007, 19, 1795–1808. [Google Scholar] [CrossRef]
- Spinelli, S.V.; Martin, A.P.; Viola, I.L.; Gonzalez, D.H.; Palatnik, J.F. A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol. 2011. [Google Scholar] [CrossRef]
- Ha, C.M.; Jun, J.H.; Fletcher, J.C. Control of Arabidopsis Leaf Morphogenesis Through Regulation of the YABBY and KNOX Families of Transcription Factors. Genetics 2010, 186, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Ichihashi, Y.; Aguilar-Martínez, J.A.; Farhi, M.; Chitwood, D.H.; Kumar, R.; Millon, L.V.; Peng, J.; Maloof, J.N.; Sinha, N.R. Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc. Natl. Acad. Sci. USA 2014. [Google Scholar] [CrossRef]
- Norberg, M.; Holmlund, M.; Nilsson, O. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 2005, 132, 2203–2213. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, N.; Yilmaz, A.; Mejia-Guerra, M.K.; Morohashi, K.; O’connor, D.; Grotewold, E.; Hake, S. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 2012, 26, 1685–1690. [Google Scholar] [CrossRef] [PubMed]
- Žádníková, P.; Simon, R. How boundaries control plant development. Curr. Opin. Plant Biol. 2014, 17, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Scofield, S.; Murison, A.; Jones, A.; Fozard, J.; Aida, M.; Band, L.R.; Bennett, M.; Murray, J.A.H. Coordination of meristem and boundary functions by transcription factors in the SHOOT MERISTEMLESS regulatory network. Development 2018. [Google Scholar] [CrossRef] [PubMed]
- Clouse, S.D.; Langford, M.; McMorris, T.C. A Brassinosteroid-Insensitive Mutant in Arabidopsis thaliana Exhibits Multiple Defects in Growth and Development. Plant Physiol. 1996, 111, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.M.; Jun, J.H.; Nam, H.G.; Fletcher, J.C. BLADE-ON-PETIOLE1 and 2 Control Arabidopsis Lateral Organ Fate through Regulation of LOB Domain and Adaxial-Abaxial Polarity Genes. Plant Cell 2007, 19, 1809–1825. [Google Scholar] [CrossRef]
- Hepworth, S.R.; Zhang, Y.; McKim, S.; Li, X.; Haughn, G.W. BLADE-ON-PETIOLE–dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 2005, 17, 1434–1448. [Google Scholar] [CrossRef]
- Bar, M.; Ori, N. Compound leaf development in model plant species. Curr. Opin. Plant Biol. 2015, 23, 61–69. [Google Scholar] [CrossRef]
- Blein, T.; Pulido, A.; Vialette-Guiraud, A.; Nikovics, K.; Morin, H.; Hay, A.; Johansen, I.E.; Tsiantis, M.; Laufs, P. A Conserved Molecular Framework for Compound Leaf Development. Science 2008, 322, 1835–1839. [Google Scholar] [CrossRef] [PubMed]
- Barkoulas, M.; Hay, A.; Kougioumoutzi, E.; Tsiantis, M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 2008, 40, 1136. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Tsiantis, M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 2006, 38, 942. [Google Scholar] [CrossRef] [PubMed]
- Bilsborough, G.D.; Runions, A.; Barkoulas, M.; Jenkins, H.W.; Hasson, A.; Galinha, C.; Laufs, P.; Hay, A.; Prusinkiewicz, P.; Tsiantis, M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 2011, 108, 3424–3429. [Google Scholar] [CrossRef] [PubMed]
- Nikovics, K.; Blein, T.; Peaucelle, A.; Ishida, T.; Morin, H.; Aida, M.; Laufs, P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 2006, 18, 2929–2945. [Google Scholar] [CrossRef] [PubMed]
- Hareven, D.; Gutfinger, T.; Parnis, A.; Eshed, Y.; Lifschitz, E. The Making of a Compound Leaf: Genetic Manipulation of Leaf Architecture in Tomato. Cell 1996, 84, 735–744. [Google Scholar] [CrossRef]
- Gourlay, C.W.; Hofer, J.M.I.; Ellis, T.H.N. Pea Compound Leaf Architecture Is Regulated by Interactions among the Genes UNIFOLIATA, COCHLEATA, AFIL, and TENDRIL-LESS. Plant Cell 2000, 12, 1279–1294. [Google Scholar] [CrossRef] [PubMed]
- Vlad, D.; Kierzkowski, D.; Rast, M.I.; Vuolo, F.; Ioio, R.D.; Galinha, C.; Gan, X.; Hajheidari, M.; Hay, A.; Smith, R.S. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 2014, 343, 780–783. [Google Scholar] [CrossRef]
- Chaw, S.-M.; Chang, C.-C.; Chen, H.-L.; Li, W.-H. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 2004, 58, 424–441. [Google Scholar]
- Sharman, B.C. Developmental anatomy of the shoot of Zea mays L. Ann. Bot. 1942, 6, 245–282. [Google Scholar] [CrossRef]
- Poethig, R.S.; Szymkowiak, E.J. Clonal analysis of leaf development in maize. Available online: http://agris.fao.org/agris-search/search.do?recordID=IT9561182 (accessed on 24 December 2018).
- Scanlon, M.J. The Polar Auxin Transport Inhibitor N-1-Naphthylphthalamic Acid Disrupts Leaf Initiation, KNOX Protein Regulation, and Formation of Leaf Margins in Maize. Plant Physiol. 2003, 133, 597–605. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, D.L.; Elton, S.; Ticchiarelli, F.; Hsia, M.M.; Vogel, J.P.; Leyser, O. Cross-species functional diversity within the PIN auxin efflux protein family. Elife 2017, 6, e31804. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, D.L.; Runions, A.; Sluis, A.; Bragg, J.; Vogel, J.P.; Prusinkiewicz, P.; Hake, S. A Division in PIN-Mediated Auxin Patterning during Organ Initiation in Grasses. PLOS Comput. Biol. 2014, 10, e1003447. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.C.; Koenig, D.; Chitwood, D.H.; Sinha, N.R. A sister of PIN1 gene in tomato (Solanum lycopersicum) defines leaf and flower organ initiation patterns by maintaining epidermal auxin flux. Dev. Biol. 2016, 419, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Han, L.; Wang, Z.-Y. Potential but limited redundant roles of MtPIN4, MtPIN5 and MtPIN10/SLM1 in the development of Medicago truncatula. Plant Signal. Behav. 2011, 6, 1834–1836. [Google Scholar] [CrossRef] [PubMed]
- Pepper, G.E.; Pearce, R.B.; Mock, J.J. Leaf Orientation and Yield of Maize1. Crop Sci. 1977, 17, 883–886. [Google Scholar] [CrossRef]
- Araus, J.L.; Reynolds, M.P.; Acevedo, E. Leaf Posture, Grain Yield, Growth, Leaf Structure, and Carbon Isotope Discrimination in Wheat. Crop Sci. 1993, 33, 1273–1279. [Google Scholar] [CrossRef]
- Nan, S.S.; Ootsuki, Y.; Adachi, S.; Yamamoto, T.; Ueda, T.; Tanabata, T.; Motobayashi, T.; Ookawa, T.; Hirasawa, T. A near-isogenic rice line carrying a QTL for larger leaf inclination angle yields heavier biomass and grain. Field Crop. Res. 2018, 219, 131–138. [Google Scholar] [CrossRef]
- Pendleton, J.W.; Smith, G.E.; Winter, S.R.; Johnston, T.J. Field Investigations of the Relationships of Leaf (Angle in Corn (Zea mays L.) to Grain Yield and Apparent Photosynthesis1. Agron. J. 1968, 60, 422–424. [Google Scholar] [CrossRef]
- Duvick, D.N. The contribution of breeding to yield advances in maize (Zea mays L.). Adv. Agron. 2005, 86, 83–145. [Google Scholar]
- Mantilla-Perez, M.B.; Salas Fernandez, M.G. Differential manipulation of leaf angle throughout the canopy: Current status and prospects. J. Exp. Bot. 2017, 68, 5699–5717. [Google Scholar] [CrossRef] [PubMed]
- Lambert, R.J.; Johnson, R.R. Leaf Angle, Tassel Morphology, and the Performance of Maize Hybrids 1. Crop Sci. 1978, 18, 499–502. [Google Scholar] [CrossRef]
- Duncan, W.G. Leaf Angles, Leaf Area, and Canopy Photosynthesis 1. Crop Sci. 1971, 11, 482–485. [Google Scholar] [CrossRef]
- Tian, F.; Bradbury, P.J.; Brown, P.J.; Hung, H.; Sun, Q.; Flint-Garcia, S.; Rocheford, T.R.; McMullen, M.D.; Holland, J.B.; Buckler, E.S. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011, 43, 159. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, M.L.; Passas, H.J.; Smith, L.G. Clonal Analysis of Epidermal Patterning during Maize Leaf Development. Dev. Biol. 1999, 216, 646–658. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sylvester, A.W.; Cande, W.Z.; Freeling, M. Division and differentiation during normal and liguleless-1 maize leaf development. Development 1990, 110, 985–1000. [Google Scholar] [PubMed]
- Weir, I.; Lu, J.; Cook, H.; Causier, B.; Schwarz-Sommer, Z.; Davies, B. CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development 2004, 131, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Ha, C.M.; Kim, G.-T.; Kim, B.C.; Jun, J.H.; Soh, M.S.; Ueno, Y.; Machida, Y.; Tsukaya, H.; Nam, H.G. The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 2003, 130, 161–172. [Google Scholar] [CrossRef]
- Zimmermann, R.; Werr, W. Pattern Formation in the Monocot Embryo as Revealed by NAM and CUC3 Orthologues from Zea mays L. Plant Mol. Biol. 2005, 58, 669–685. [Google Scholar] [CrossRef]
- Adam, H.; Marguerettaz, M.; Qadri, R.; Adroher, B.; Richaud, F.; Collin, M.; Thuillet, A.-C.; Vigouroux, Y.; Laufs, P.; Tregear, J.W.; et al. Divergent Expression Patterns of miR164 and CUP-SHAPED COTYLEDON Genes in Palms and Other Monocots: Implication for the Evolution of Meristem Function in Angiosperms. Mol. Biol. Evol. 2011, 28, 1439–1454. [Google Scholar] [CrossRef] [PubMed]
- Vialette-Guiraud, A.C.M.; Adam, H.; Finet, C.; Jasinski, S.; Jouannic, S.; Scutt, C.P. Insights from ANA-grade angiosperms into the early evolution of CUP-SHAPED COTYLEDON genes. Ann. Bot. 2011, 107, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Li, W.; Unger-Wallace, E.; Yang, J.; Vollbrecht, E.; Chuck, G. Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc. Natl. Acad. Sci. USA 2017, 114, E8656–E8664. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhou, X.; Zheng, Y.; Zhang, W.; Zhu, J.-K. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Sieber, P.; Wellmer, F.; Gheyselinck, J.; Riechmann, J.L.; Meyerowitz, E.M. Redundancy and specialization among plant microRNAs: Role of the MIR164 family in developmental robustness. Development 2007, 134, 1051–1060. [Google Scholar] [CrossRef]
- Li, J.; Guo, G.; Guo, W.; Guo, G.; Tong, D.; Ni, Z.; Sun, Q.; Yao, Y. miRNA164-directed cleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.). BMC Plant Biol. 2012, 12, 220. [Google Scholar] [CrossRef]
- Xiong, Y.; Liu, T.; Tian, C.; Sun, S.; Li, J.; Chen, M. Transcription Factors in Rice: A Genome-wide Comparative Analysis between Monocots and Eudicots. Plant Mol. Biol. 2005, 59, 191–203. [Google Scholar] [CrossRef]
- Tsuda, K.; Akiba, T.; Kimura, F.; Ishibashi, M.; Moriya, C.; Nakagawa, K.; Kurata, N.; Ito, Y. ONION2 fatty acid elongase is required for shoot development in rice. Plant Cell Physiol. 2013, 54, 209–217. [Google Scholar] [CrossRef]
- Tsuda, K.; Ito, Y.; Yamaki, S.; Miyao, A.; Hirochika, H.; Kurata, N. Isolation and mapping of three rice mutants that showed ectopic expression of KNOX genes in leaves. Plant Sci. 2009, 177, 131–135. [Google Scholar] [CrossRef]
- Takasugi, T.; Ito, Y. Altered expression of auxin-related genes in the fatty acid elongase mutant oni1 of rice. Plant Signal Behav. 2011, 6. [Google Scholar] [CrossRef]
- Akiba, T.; Hibara, K.I.; Kimura, F.; Tsuda, K.; Shibata, K.; Ishibashi, M.; Moriya, C.; Nakagawa, K.; Kurata, N.; Itoh, J.I.; et al. Organ fusion and defective shoot development in oni3 mutants of rice. Plant Cell Physiol. 2014, 55, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Sinha, N.; Lynch, M. Fused organs in the adherent1 mutation in maize show altered epidermal walls with no perturbations in tissue identities. Planta 1998, 206, 184–195. [Google Scholar] [CrossRef]
- La Rocca, N.; Manzotti, P.S.; Cavaiuolo, M.; Barbante, A.; Dalla Vecchia, F.; Gabotti, D.; Gendrot, G.; Horner, D.S.; Krstajic, J.; Persico, M.; et al. The maize fused leaves1 (fdl1) gene controls organ separation in the embryo and seedling shoot and promotes coleoptile opening. J. Exp. Bot. 2015, 66, 5753–5767. [Google Scholar] [CrossRef] [PubMed]
- Tavakol, E.; Okagaki, R.; Verderio, G.; Shariati, V.; Hussien, A.; Bilgic, H.; Scanlon, M.J.; Todt, N.R.; Close, T.J.; Druka, A. The barley Uniculme4 gene encodes a BLADE-ON-PETIOLE-like protein that controls tillering and leaf patterning. Plant Physiol. 2015, 168, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Mangeon, A.; Lin, W.; Springer, P.S. Functional divergence in the Arabidopsis LOB-domain gene family. Plant Signal. Behav. 2012, 7, 1544–1547. [Google Scholar] [CrossRef] [PubMed]
- Bortiri, E.; Chuck, G.; Vollbrecht, E.; Rocheford, T.; Martienssen, R.; Hake, S. Ramosa2 Encodes a LATERAL ORGAN BOUNDARY Domain Protein That Determines the Fate of Stem Cells in Branch Meristems of Maize. Plant Cell 2006, 18, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Johnston, R.; Leiboff, S.; Scanlon, M.J. Ontogeny of the sheathing leaf base in maize (Zea mays). New Phytol. 2014, 306–315. [Google Scholar] [CrossRef]
- Scanlon, M.J.; Chen, K.D.; McKnight, C.C. The narrow sheath Duplicate Genes: Sectors of Dual Aneuploidy Reveal Ancestrally Conserved Gene Functions During Maize Leaf Development. Genetics 2000, 155, 1379–1389. [Google Scholar]
- Moon, J.; Candela, H.; Hake, S. The Liguleless narrow mutation affects proximal-distal signaling and leaf growth. Development 2013, 140, 405–412. [Google Scholar] [CrossRef]
- Johnston, R.; Wang, M.; Sun, Q.; Sylvester, A.W.; Hake, S.; Scanlon, M.J. Transcriptomic Analyses Indicate That Maize Ligule Development Recapitulates Gene Expression Patterns That Occur during Lateral Organ Initiation. Plant Cell Online 2014. [Google Scholar] [CrossRef]
- Moreno, M.A.; Harper, L.C.; Krueger, R.W.; Dellaporta, S.L.; Freeling, M. liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 1997, 11, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, J.J.; Kim, S.L.; Yim, J.; An, G. Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol. Biol. 2007, 65, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Rossini, L.; Vecchietti, A.; Nicoloso, L.; Stein, N.; Franzago, S.; Salamini, F.; Pozzi, C. Candidate genes for barley mutants involved in plant architecture: An in silico approach. Theor. Appl. Genet. 2006, 112, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Harper, L.; Freeling, M. Interactions of liguleless1 and liguleless2 function during ligule induction in maize. Genetics 1996, 144, 1871–1882. [Google Scholar] [PubMed]
- Walsh, J.; Waters, C.A.; Freeling, M. The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev. 1998, 12, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Hake, S. The dominant mutant Wavy auricle in blade1 disrupts patterning in a lateral domain of the maize leaf. Plant Physiol. 2004, 135, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Schütze, K.; Harter, K.; Chaban, C. Post-translational regulation of plant bZIP factors. Trends Plant Sci. 2008, 13, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Ding, Z.; Li, P. Maize network analysis revealed gene modules involved in development, nutrients utilization, metabolism, and stress response. BMC Plant Biol. 2017, 17, 131. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Morinaka, Y.; Ohnishi, T.; Sunohara, H.; Fujioka, S.; Ueguchi-Tanaka, M.; Mizutani, M.; Sakata, K.; Takatsuto, S.; Yoshida, S. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 2006, 24, 105. [Google Scholar] [CrossRef]
- Hong, Z.; Ueguchi-Tanaka, M.; Umemura, K.; Uozu, S.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 2003, 15, 2900–2910. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Ueguchi-Tanaka, M.; Shimizu-Sato, S.; Inukai, Y.; Fujioka, S.; Shimada, Y.; Takatsuto, S.; Agetsuma, M.; Yoshida, S.; Watanabe, Y. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002, 32, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, C.; Ihara, Y.; Wu, X.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 2000, 12, 1591–1605. [Google Scholar] [CrossRef] [PubMed]
- Kir, G.; Ye, H.; Nelissen, H.; Neelakandan, A.K.; Kusnandar, A.S.; Luo, A.; Inzé, D.; Sylvester, A.W.; Yin, Y.; Becraft, P.W. RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol. 2015, 169, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, L.; Wang, M.; Xu, Y.; Luo, W.; Liu, Y.; Xu, Z.; Li, J.; Chong, K. Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 2009, 7, 791–806. [Google Scholar] [CrossRef]
- Makarevitch, I.; Thompson, A.; Muehlbauer, G.J.; Springer, N.M. Brd1 Gene in Maize Encodes a Brassinosteroid C-6 Oxidase. PLoS ONE 2012, 7, e30798. [Google Scholar] [CrossRef]
- Okagaki, R.J.; Haaning, A.; Bilgic, H.; Heinen, S.; Druka, A.; Bayer, M.; Waugh, R.; Muehlbauer, G.J. ELIGULUM-A regulates lateral branch and leaf development in barley. Plant Physiol. 2018. [Google Scholar] [CrossRef]
- Vollbrecht, E.; Veit, B.; Sinha, N.; Hake, S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 1991, 350, 241. [Google Scholar] [CrossRef]
- Ramirez, J.; Bolduc, N.; Lisch, D.; Hake, S. Distal expression of knotted1 in maize leaves leads to reestablishment of proximal/distal patterning and leaf dissection. Plant Physiol. 2009, 151, 1878–1888. [Google Scholar] [CrossRef]
- Lewis, M.W.; Bolduc, N.; Hake, K.; Htike, Y.; Hay, A.; Candela, H.; Hake, S. Gene regulatory interactions at lateral organ boundaries in maize. Development 2014, 141, 4590–4597. [Google Scholar] [CrossRef]
- Foster, T.; Yamaguchi, J.; Wong, B.C.; Veit, B.; Hake, S. Gnarley1 is a dominant mutation in the knox4 homeobox gene affecting cell shape and identity. Plant Cell 1999, 11, 1239–1252. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.; Veit, B.; Hake, S. Mosaic analysis of the dominant mutant, Gnarley1-R, reveals distinct lateral and transverse signaling pathways during maize leaf development. Development 1999, 126, 305–313. [Google Scholar] [PubMed]
- Muehlbauer, G.J.; Fowler, J.E.; Girard, L.; Tyers, R.; Harper, L.; Freeling, M. Ectopic Expression of the Maize Homeobox Gene Liguleless3 Alters Cell Fates in the Leaf 1. Plant Physiol. 1999, 119, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Fowler, J.E.; Muehlbauer, G.J.; Freeling, M. Mosaic analysis of the liguleless3 mutant phenotype in maize by coordinate suppression of mutator-insertion alleles. Genetics 1996, 143, 489–503. [Google Scholar] [PubMed]
- Muehlbauer, G.J.; Fowler, J.E.; Freeling, M. Sectors expressing the homeobox gene liguleless3 implicate a time-dependent mechanism for cell fate acquisition along the proximal-distal axis of the maize leaf. Development 1997, 124, 5097–5106. [Google Scholar] [PubMed]
- Bertrand-Garcia, R.; Freeling, M. Hairy-Sheath Frayed # 1-0: A Systemic, Heterochronic Mutant of Maize that Specifies Slow Developmental Stage Transitions. Am. J. Bot. 1991, 78, 747–765. [Google Scholar] [CrossRef]
- Saberman, J.; Bertrand-Garcia, R. Hairy-sheath-frayed 1-O is a Non-Cell-Autonomous Mutation That Regulates Developmental Stage Transitions in Maize. J. Hered. 1997, 88, 549–553. [Google Scholar] [CrossRef][Green Version]
- Tsuda, K.; Abraham-Juarez, M.J.; Maeno, A.; Dong, Z.; Aromdee, D.; Meeley, R.; Shiroishi, T.; Nonomura, K.; Hake, S. KNOTTED1 cofactors, BLH12 and BLH14, regulate internode patterning and vein anastomosis in maize. Plant Cell 2017. [Google Scholar] [CrossRef]
- Osmont, K.S.; Sadeghian, N.; Freeling, M. Mosaic analysis of extended auricle1 (eta1) suggests that a two-way signaling pathway is involved in positioning the blade/sheath boundary in Zea mays. Dev. Biol. 2006, 295, 1–12. [Google Scholar] [CrossRef][Green Version]
- Osmont, K.S.; Jesaitis, L.A.; Freeling, M. The extended auricle1 (eta1) Gene Is Essential for the Genetic Network Controlling Postinitiation Maize Leaf Development. Genetics 2003, 165, 1507–1519. [Google Scholar]
- Tsiantis, M.; Brown, M.I.N.; Skibinski, G.; Langdale, J.A. Disruption of Auxin Transport Is Associated with Aberrant Leaf Development in Maize. Plant Physiol. 1999, 121, 1163–1168. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.W.; Bolduc, N.; Hake, K.; Htike, Y.; Hay, A.; Candela, H. Recruitment of regulatory interactions from the inflorescence to the leaf in the dominant Wavy auricle in blade mutant. Development 2014, 141, 4590–4597. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.; Freeling, M. The liguleless2 gene of maize functions during the transition from the vegetative to the reproductive shoot apex. Plant J. 1999, 19, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Busch, B.L.; Schmitz, G.; Rossmann, S.; Piron, F.; Ding, J.; Bendahmane, A.; Theres, K. Shoot branching and leaf dissection in tomato are regulated by homologous gene modules. Plant Cell 2011. [Google Scholar] [CrossRef] [PubMed]
- Raman, S.; Greb, T.; Peaucelle, A.; Blein, T.; Laufs, P.; Theres, K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008, 55, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-K.; Geisler, M.; Springer, P.S. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 2009, 136, 2423–2432. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Mislata, A.; Bencivenga, S.; Bush, M.; Schiessl, K.; Boden, S.; Sablowski, R. DELLA genes restrict inflorescence meristem function independently of plant height. Nat. Plants 2017, 3, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Schürholz, A.-K.; López-Salmerón, V.; Li, Z.; Forner, J.; Wenzl, C.; Gaillochet, C.; Augustin, S.; Barro, A.V.; Fuchs, M.; Gebert, M.; et al. A Comprehensive Toolkit for Inducible, Cell Type-Specific Gene Expression in Arabidopsis. Plant Physiol. 2018, 178, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Reddy, G.V.; Heisler, M.G.; Ehrhardt, D.W.; Meyerowitz, E.M. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development 2004, 131, 4225–4237. [Google Scholar] [CrossRef] [PubMed]
- Soyk, S.; Lemmon, Z.H.; Oved, M.; Fisher, J.; Liberatore, K.L.; Park, S.J.; Goren, A.; Jiang, K.; Ramos, A.; van der Knaap, E.; et al. Bypassing Negative Epistasis on Yield in Tomato Imposed by a Domestication Gene. Cell 2017, 169, 1142–1155. [Google Scholar] [CrossRef]
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480. [Google Scholar] [CrossRef] [PubMed]

| Arabidopsis | Maize | Barley | Description |
|---|---|---|---|
| SHOOTMERISTEMLESS (STM) | KNOTTED1 (KN1) | BARLEY KNOTTED 3 (BKn3) | KNOX transcription factor |
| CUP-SHAPED COTYLEDON 1,2,3 (CUC1, 2, 3) | NO APICAL MERISTEM 1 and 2 (NAM 1,2), CUC3 | NAC domain transcription factor | |
| BLADE ON PETIOLE 1 (BOP) | TASSELS REPLACE UPPER EARS 1 (TRU1) and TRU1-like | UNICULME4 (CUL4) | Ankyrin repeat domain protein |
| miR164 a/b/c | miR164 a/b/c/d/e/f/g/h | microRNA | |
| PIN-FORMED 1 (PIN1) | PINFORMED 1a and 1b (PIN1a, PIN1b) | Auxin transporter | |
| not present in Arabidopsis | SISTER OF PIN1 (SoPIN1) | Auxin transporter |
| Gene Name | Species | Description |
|---|---|---|
| KNOTTED 1 (KN1) | Maize | KNOX Transcription Factor, meristem identity |
| NO APICAL MERISTEM 1 and 2 (NAM 1,2), CUC3 | Maize | NAC domain, transcription factor, expressed in boundary domains |
| TASSELS REPLACE UPPER EARS 1 (TRU1) and TRU1-like | Maize | Ankyrin repeat domain protein expressed in the sheath and in axillary meristems. |
| PINFORMED 1a and 1b (PIN1a, PIN1b) | Maize | Auxin transporter |
| SISTER OF PIN1 (SoPIN1) | Maize | Auxin transporter |
| RAMOSA 2 (RA2) | Maize | Lateral organ boundary domain transcription factor, involved in axillary meristem development. |
| SPARSE INFLORESENCE 1 (SPI1) | Maize | YUCCA gene, auxin biosynthesis. |
| NARROWSHEATH 1 and 2 | Maize | WOX genes, involved in leaf development |
| LIGULELESS1 (LG1) | Maize | Squamosa Binding Protein transcription factor, involved in ligule development. |
| LIGULELESS2 (LG2) | Maize | BZIP/DOG domain transcription factor, involved in ligule development. |
| LIGULELESS NARROW (LGN) | Maize | Serine-threonine kinase, involved in ligule development. |
| LIGULELESS3 (LG3) | Maize | KNOX transcription factor, ectopic expression of LG3 induces ectopic blade/sheath boundaries. |
| LIGULELESS4 (LG4) | Maize | KNOX transcription factor, ectopic expression of LG4 induces ectopic blade/sheath boundaries. |
| GNARLEY4 (GN4) | Maize | KNOX transcription factor, ectopic expression of LG4 induces ectopic blade/sheath boundaries |
| WAVY AURICLES IN BLADE 1 (WAB1) | Maize | TCP transcription factor, ectopic expression of WAB1 induces ectopic blade/sheath boundaries |
| BEL1-like homeodomain 12 and 14 (BEL12/14) | Maize | BEL1-like homeodomain transcription factors, expressed in the developing ligule |
| BRASSINOSTEROID INSENSITIVE 1 (BRI1) | Maize | Brassinosteroid receptor, involved in auricle development and leaf angle |
| BRASSINOSTEROID-DEFICIENT DWARF1 (BRD1) | Maize | Brassinosteroid C6-oxidase, involved in brassino-steroid synthesis, expressed in the base of leaves. Involved in ligule and auricle development. |
| BETA-D-GULCOSIDASE 1 (GLU1) | Maize | Expressed in developing ligules |
| UNICULME4 (CUL4) | Barley | Ankyrin repeat domain protein, expressed in the sheath and involved in ligule development |
| ELIGULUM A (ELIA) | Barley | RNase H domain protein, involved in ligule development |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richardson, A.E.; Hake, S. Drawing a Line: Grasses and Boundaries. Plants 2019, 8, 4. https://doi.org/10.3390/plants8010004
Richardson AE, Hake S. Drawing a Line: Grasses and Boundaries. Plants. 2019; 8(1):4. https://doi.org/10.3390/plants8010004
Chicago/Turabian StyleRichardson, Annis E, and Sarah Hake. 2019. "Drawing a Line: Grasses and Boundaries" Plants 8, no. 1: 4. https://doi.org/10.3390/plants8010004
APA StyleRichardson, A. E., & Hake, S. (2019). Drawing a Line: Grasses and Boundaries. Plants, 8(1), 4. https://doi.org/10.3390/plants8010004




