Translating Flowering Time from Arabidopsis thaliana to Brassicaceae and Asteraceae Crop Species
Abstract
1. Introduction
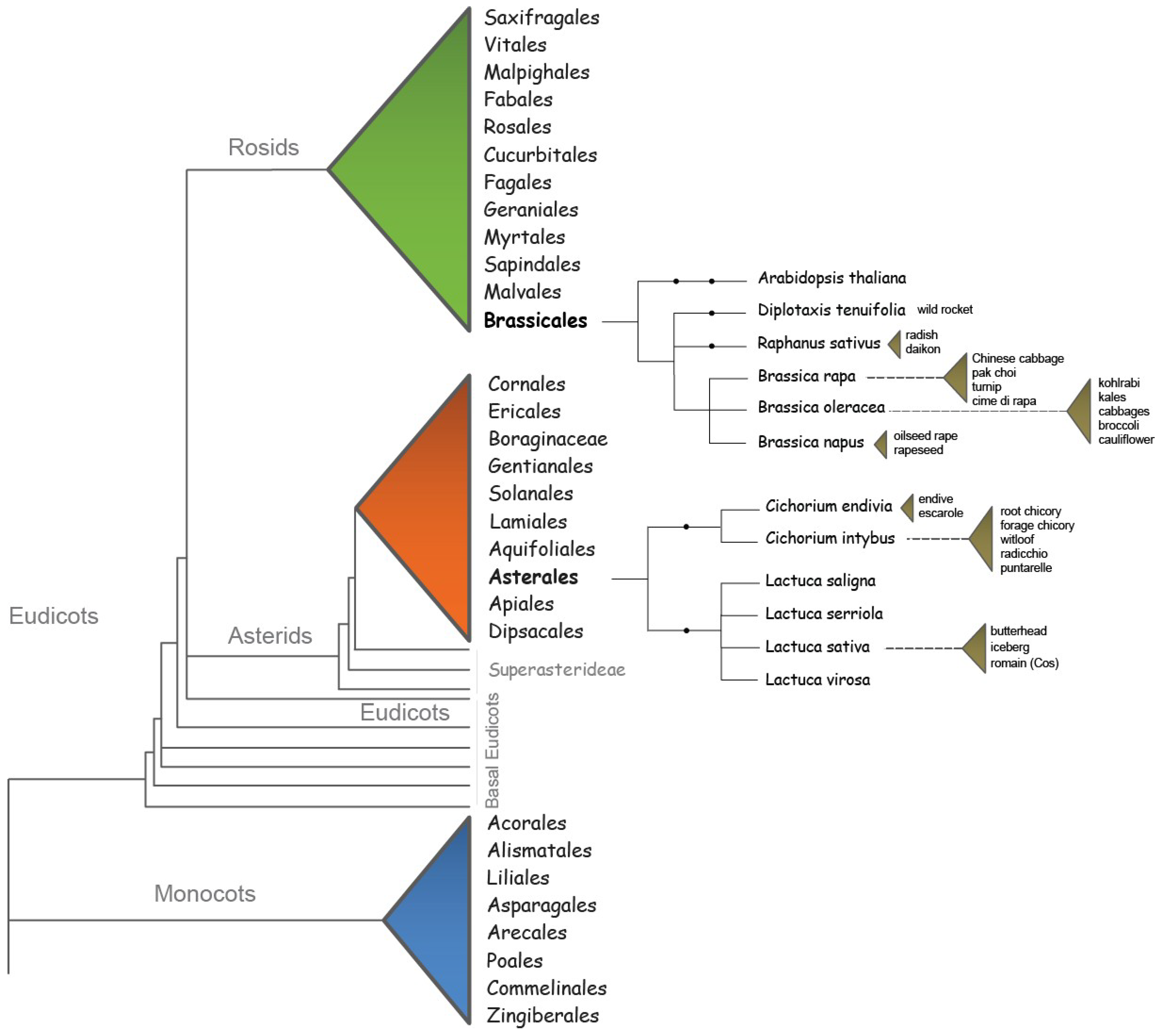
2. Flowering Requirements of Brassicaceae and Asteraceae Species
3. Breeding Goals
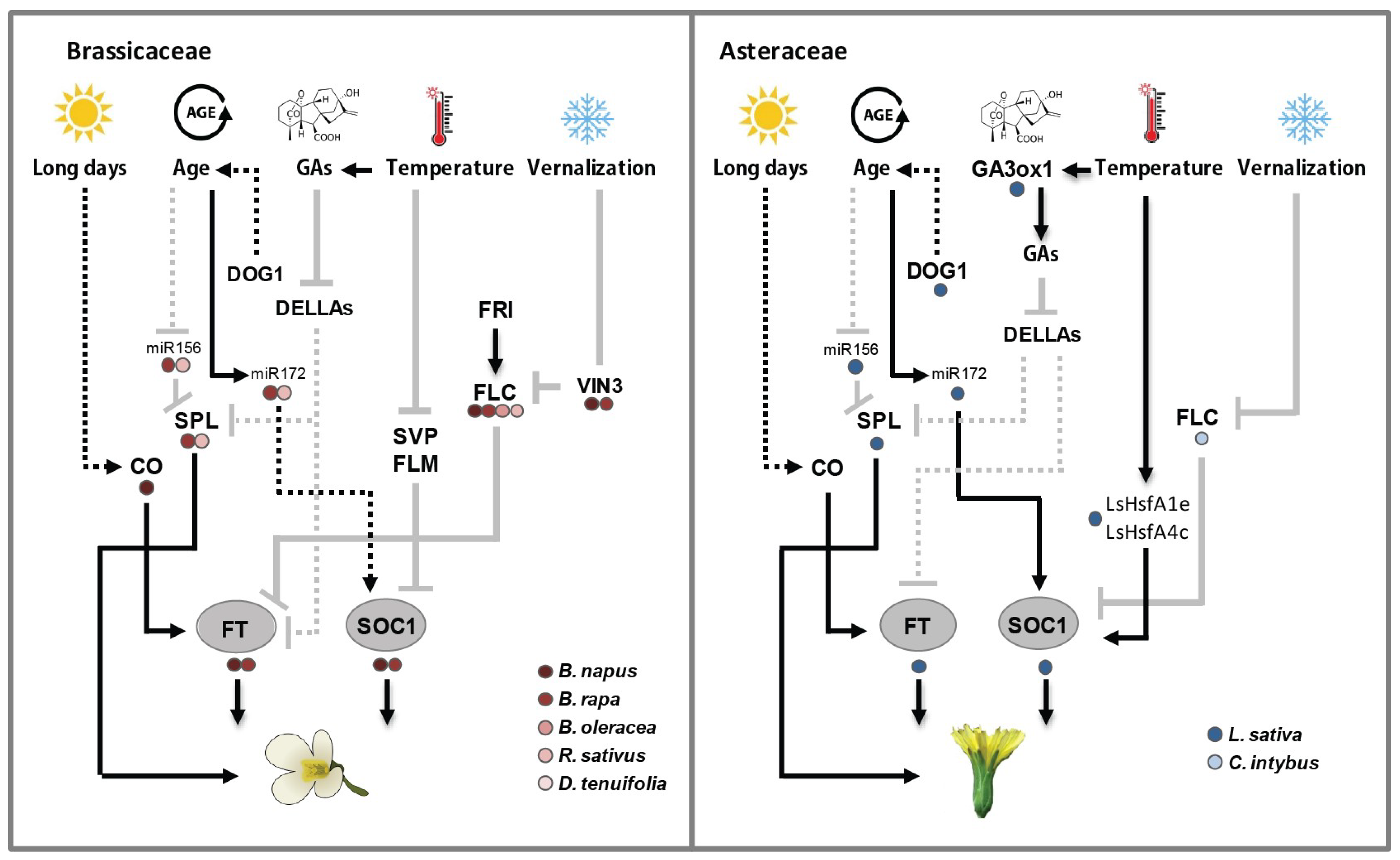
4. Conserved and Divergent Flowering Time Genes in Brassicaceae and Asteraceae
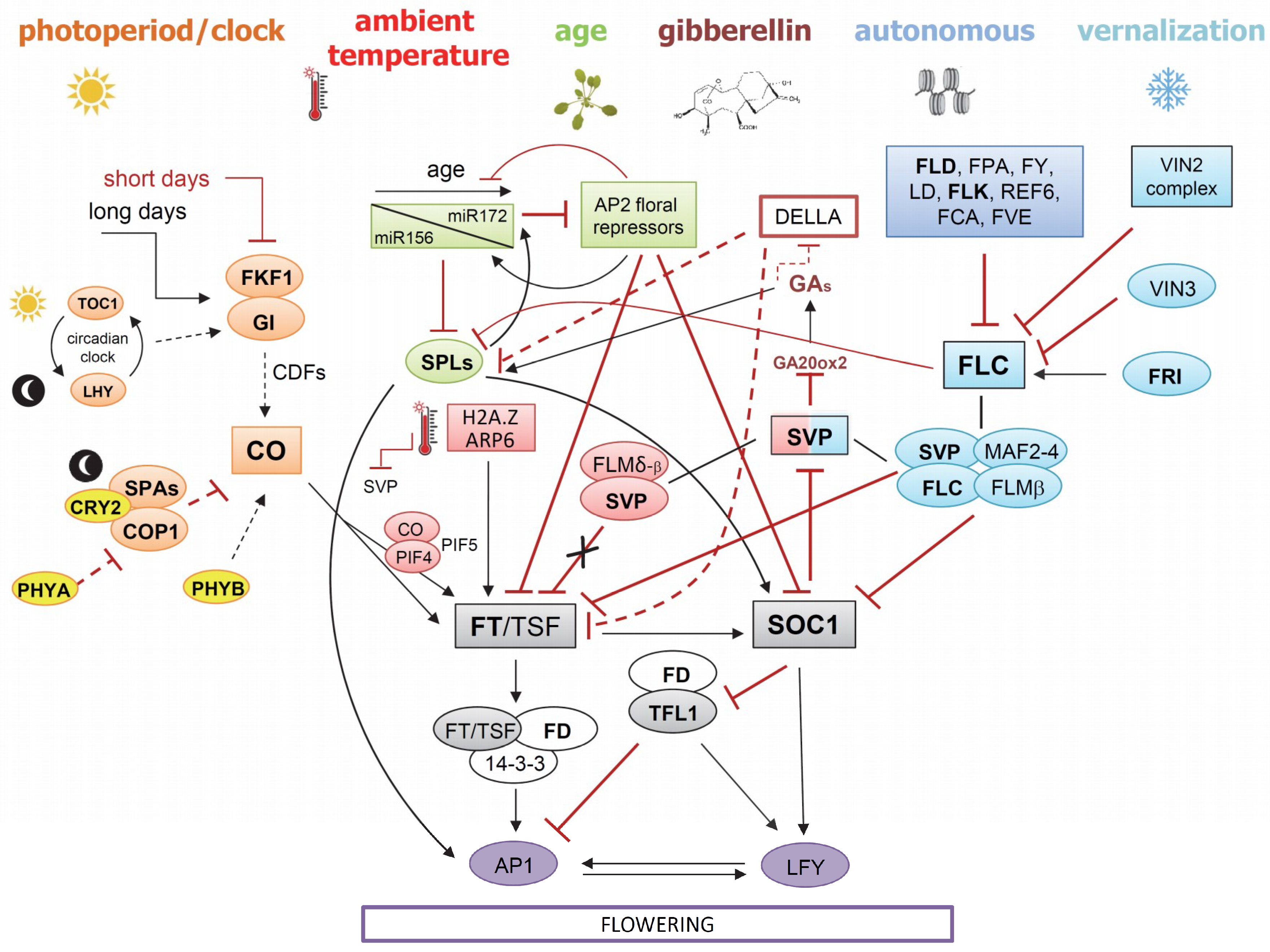
4.1. Floral Integrator Genes: An Overview
4.1.1. Floral Integrator Genes in Brassicaceae
4.1.2. Floral Integrator Genes in Asteraceae
4.2. Overview of the Vernalization and Autonomous Pathways
4.2.1. Vernalization and Autonomous Pathway in Brassicaceae
4.2.2. Vernalization and Autonomous Pathway in Asteraceae
4.3. Overview of the Ambient Temperature Pathways
4.3.1. Ambient Temperature Pathways in Brassicaceae
4.3.2. Ambient Temperature Pathways in Asteraceae
4.4. Overview of the Photoperiodic Pathway
4.4.1. The Photoperiodic Pathway in Brassicaceae
4.4.2. The Photoperiodic Pathway in Asteraceae
4.5. Overview of the Age Pathway
4.5.1. Age Pathway in Brassicaceae
4.5.2. Age Pathway in Asteraceae
4.6. Overview of the Hormonal Pathway
4.6.1. Hormonal Pathway in Brassicaceae
4.6.2. Hormonal Pathway in Asteraceae
5. Quantitative Trait Loci (QTL)
6. Perspectives for Breeding Strategies
6.1. Environmental Changes
6.2. Yield Increase
6.3. Genetic Resources
6.4. Speeding up Breeding
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blackman, B.K. Changing Responses to Changing Seasons: Natural Variation in the Plasticity of Flowering Time. Plant Physiol. 2017, 173, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Muller, A.E. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009, 14, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Liu, T.; Hou, X. Isolation and Functional Characterization of a Floral Repressor, BcMAF1, from Pak-choi (Brassica rapa ssp. Chinensis). Front. Plant Sci. 2018, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- Schiessl, S.; Huettel, B.; Kuehn, D.; Reinhardt, R.; Snowdon, R.J. Targeted deep sequencing of flowering regulators in Brassica napus reveals extensive copy number variation. Sci. Data 2017, 4, 170013. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; L, R.; Qiu, D.; Jiang, C.; Long, Y.; Morgan, C.; Bancroft, I.; Zhao, J.; Meng, J. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 2009, 182, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Bluemel, M.; Dally, N.; Jung, C. Flowering time regulation in crops—What did we learn from Arabidopsis? Curr. Opin. Biotechnol. 2015, 32, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, Y. Florigen and anti-florigen: Flowering regulation in horticultural crops. Breed. Sci. 2018, 68, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Amasino, R. Seasonal and developmental timing of flowering. Plant J. 2010, 61, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell 2010, 141, 550-550.e2. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, M.; Alonso-Blanco, C.; Vreugdenhil, D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant. Biol. 2004, 55, 141–172. [Google Scholar] [CrossRef] [PubMed]
- Gulden, R.H.; Warwick, S.I.; Thomas, A.G. The biology of Canadian weeds. 137. Brassica napus L. and B. rapa L. Can. J. Plant Sci. 2008, 88, 951–996. [Google Scholar] [CrossRef]
- Wang, N.; Qian, W.; Suppanz, I.; Wei, L.; Mao, B.; Long, Y.; Meng, J.; Müller, A.E.; Jung, C. Flowering time variation in oilseed rape (Brassica napus L.) is associated with allelic variation in the FRIGIDA homologue BnaA. FRI. a. J. Exp. Bot. 2011, 62, 5641–5658. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.-H.; Bancroft, I.; Cheng, F. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Parkin, I.A.; Koh, C.; Tang, H.; Robinson, S.J.; Kagale, S.; Clarke, W.E.; Town, C.D.; Nixon, J.; Krishnakumar, V.; Bidwell, S.L. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Boil. 2014, 15, R77. [Google Scholar] [CrossRef] [PubMed]
- Kitashiba, H.; Li, F.; Hirakawa, H.; Kawanabe, T.; Zou, Z.; Hasegawa, Y.; Tonosaki, K.; Shirasawa, S.; Fukushima, A.; Yokoi, S. Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res. 2014, 21, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, S.; Xu, W.; Liu, X. Genome-wide transcriptome profiling of radish (Raphanus sativus L.) in response to vernalization. PLoS ONE 2017, 12, e0177594. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.L.; Massiah, A.; Kennedy, S.; Hong, Y.; Jackson, S.D. FLC expression is down-regulated by cold treatment in Diplotaxis tenuifolia (wild rocket), but flowering time is unaffected. J. Plant Physiol. 2017, 214, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Chin-Wo, S.; Wang, Z.; Yang, X.; Kozik, A.; Arikit, S.; Song, C.; Xia, L.; Froenicke, L.; Lavelle, D.O.; Truco, M.J.; et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017, 8, 14953. [Google Scholar] [CrossRef] [PubMed]
- Gonthier, L.; Bellec, A.; Blassiau, C.; Prat, E.; Helmstetter, N.; Rambaud, C.; Huss, B.; Hendriks, T.; Bergès, H.; Quillet, M.-C. Construction and characterization of two BAC libraries representing a deep-coverage of the genome of chicory (Cichorium intybus L., Asteraceae). BMC Res. Notes 2010, 3, 225. [Google Scholar] [CrossRef] [PubMed]
- Périlleux, C.; Pieltain, A.; Jacquemin, G.; Bouché, F.; Detry, N.; D’aloia, M.; Thiry, L.; Aljochim, P.; Delansnay, M.; Mathieu, A.S. A root chicory MADS box sequence and the Arabidopsis flowering repressor FLC share common features that suggest conserved function in vernalization and de-vernalization responses. Plant J. 2013, 75, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, N.; Nishikawa, K.; Tanimura, Y.; Urushibara, S.; Matsuura, T.; Yokoi, S.; Takahata, Y.; Yui, S. Development of late-bolting F1 hybrids of Chinese cabbage (Brassica rapa L.) allowing early spring cultivation without heating. Euphytica 2017, 213, 292. [Google Scholar] [CrossRef]
- Rosen, A.; Hasan, Y.; Briggs, W.; Uptmoor, R. Genome-based prediction of time to curd induction in cauliflower. Front. Plant Sci. 2018, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, N.; Yui, S.; Nishikawa, K.; Takahata, Y.; Yokoi, S. A naturally occurring long insertion in the first intron in the Brassica rapaFLC2 gene causes delayed bolting. Euphytica 2014, 196, 213–223. [Google Scholar] [CrossRef]
- Simonne, A.; Simonne, E.; Eitenmiller, R.; Coker, C.H. Bitterness and composition of lettuce varieties grown in the southeastern United States. HortTechnology 2002, 12, 721–726. [Google Scholar] [CrossRef]
- Street, R.A.; Sidana, J.; Prinsloo, G. Cichorium intybus: Traditional uses, phytochemistry, pharmacology, and toxicology. Evid.-Based Complement. Altern. Med. 2013, 2013, 579319. [Google Scholar] [CrossRef] [PubMed]
- Renna, M.; Gonnella, M.; Giannino, D.; Santamaria, P. Quality evaluation of cook-chilled chicory stems (Cichorium intybus L., Catalogna group) by conventional and sous vide cooking methods. J. Sci. Food Agric. 2014, 94, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Barcaccia, G.; Ghedina, A.; Lucchin, M. Current advances in genomics and breeding of leaf Chicory (Cichorium intybus L.). Agriculture 2016, 6, 50. [Google Scholar] [CrossRef]
- Kwiatkowska, D. Flowering and apical meristem growth dynamics. J. Exp. Bot. 2008, 59, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Kobayashi, Y.; Goto, K.; Abe, M.; Araki, T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005, 46, 1175–1189. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Miller, D.; Winter, V.J.; Banfield, M.J.; Lee, J.H.; Yoo, S.Y.; Henz, S.R.; Brady, R.L.; Weigel, D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006, 25, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Flowering Time Pathway (Arabidopsis thaliana). Available online: https://www.wikipathways.org/index.php/Pathway:WP2312 (accessed on 19 September 2018).
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: Functional evolution and molecular mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Schiessl, S.V.; Huettel, B.; Kuehn, D.; Reinhardt, R.; Snowdon, R.J. Flowering time gene variation in Brassica species shows evolutionary principles. Front. Plant Sci. 2017, 8, 1742. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, W.; Ge, D.; Han, Y.; Ning, K.; Luo, C.; Wang, S.; Liu, R.; Zhang, X.; Wang, Q. LCM-seq reveals the crucial role of Ls SOC 1 in heat-promoted bolting of lettuce (Lactuca sativa L.). Plant J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, C.; Hou, X.; Xi, W.; Shen, L.; Tao, Z.; Wang, Y.; Yu, H. FTIP1 is an essential regulator required for florigen transport. PLoS Boil. 2012, 10, e1001313. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Suárez-López, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Ying, H.; Helliwell, C.A.; Taylor, J.M.; Peacock, W.J.; Dennis, E.S. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 6680–6685. [Google Scholar] [CrossRef] [PubMed]
- Mateos, J.L.; Madrigal, P.; Tsuda, K.; Rawat, V.; Richter, R.; Romera-Branchat, M.; Fornara, F.; Schneeberger, K.; Krajewski, P.; Coupland, G. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Boil. 2015, 16, 31. [Google Scholar] [CrossRef] [PubMed]
- Taoka, K.-i.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, H.; Er, H.L.; Soo, H.M.; Kumar, P.P.; Han, J.-H.; Liou, Y.C.; Yu, H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 2008, 135, 1481–1491. [Google Scholar] [CrossRef] [PubMed]
- Hanzawa, Y.; Money, T.; Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 2005, 102, 7748–7753. [Google Scholar] [CrossRef] [PubMed]
- Conti, L.; Bradley, D. TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 2007, 19, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Hanano, S.; Goto, K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 2011, 23, 3172–3184. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hopkins, C.J.; Hou, J.; Zou, X.; Wang, C.; Long, Y.; Kurup, S.; King, G.J.; Meng, J. Promoter variation and transcript divergence in Brassicaceae lineages of FLOWERING LOCUS T. PLoS ONE 2012, 7, e47127. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Long, Y.; Wu, B.; Liu, J.; Jiang, C.; Shi, L.; Zhao, J.; King, G.J.; Meng, J. The evolution of Brassica napus FLOWERING LOCUST paralogues in the context of inverted chromosomal duplication blocks. BMC Evol. Boil. 2009, 9, 271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Meng, L.; Liu, B.; Hu, Y.; Cheng, F.; Liang, J.; Aarts, M.G.; Wang, X.; Wu, J. A transposon insertion in FLOWERING LOCUS T is associated with delayed flowering in Brassica rapa. Plant Sci. 2015, 241, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Raman, H.; Raman, R.; Coombes, N.; Song, J.; Prangnell, R.; Bandaranayake, C.; Tahira, R.; Sundaramoorthi, V.; Killian, A.; Meng, J. Genome-wide association analyses reveal complex genetic architecture underlying natural variation for flowering time in canola. Plant Cell Environ. 2016, 39, 1228–1239. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Hans, H.; Christian, J.; Molina, C. Mutations in single FT-and TFL1-paralogs of rapeseed (Brassica napus L.) and their impact on flowering time and yield components. Front. Plant Sci. 2014, 5, 282. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Weinholdt, C.; Jedrusik, N.; Molina, C.; Zou, J.; Große, I.; Schiessl, S.; Jung, C.; Emrani, N. Whole transcriptome analysis reveals genetic factors underlying flowering time regulation in rapeseed (Brassica napus L.). Plant Cell Environ. 2018. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhao, W.; Li, D.; Chao, H.; Zhao, X.; Ta, N.; Li, Y.; Guan, Z.; Guo, L.; Zhang, L. Genetic dissection of the mechanism of flowering time based on an environmentally stable and specific QTL in Brassica napus. Plant Sci. 2018, 277, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Duan, W.; Huang, Z.; Liu, G.; Wu, P.; Liu, T.; Li, Y.; Hou, X. Comprehensive analysis of the flowering genes in Chinese cabbage and examination of evolutionary pattern of CO-like genes in plant kingdom. Sci. Rep. 2015, 5, 14631. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.K.; Kim, S.-Y.; Kim, K.-S.; Kwon, S.-J.; Kim, J.S.; Kim, J.A.; Lee, S.I.; Lee, Y.-H. Overexpression of a Brassica rapa MADS-box gene, BrAGL20, induces early flowering time phenotypes in Brassica napus. Plant. Biotechnol. Rep. 2013, 7, 231–237. [Google Scholar] [CrossRef]
- Franks, S.J.; Perez-Sweeney, B.; Strahl, M.; Nowogrodzki, A.; Weber, J.J.; Lalchan, R.; Jordan, K.P.; Litt, A. Variation in the flowering time orthologs BrFLC and BrSOC1 in a natural population of Brassica rapa. PeerJ 2015, 3, e1339. [Google Scholar] [CrossRef] [PubMed]
- Cavaiuolo, M.; Cocetta, G.; Spadafora, N.D.; Müller, C.T.; Rogers, H.J.; Ferrante, A. Gene expression analysis of rocket salad under pre-harvest and postharvest stresses: A transcriptomic resource for Diplotaxis tenuifolia. PLoS ONE 2017, 12, e0178119. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Matsuo, S.; Kikuchi, K.; Kawazu, Y.; Fujiyama, R.; Honda, I. Isolation and functional characterization of the FLOWERING LOCUS T homolog, the LsFT gene, in lettuce. J. Plant Physiol. 2011, 168, 1602–1607. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Han, Y.; Ning, K.; Ding, Y.; Zhao, W.; Yan, S.; Luo, C.; Jiang, X.; Wang, Q.; Zhang, X. Inflorescence development and the role of LsFT in regulating bolting in lettuce (Lactuca sativa L.). Front. Plant Sci. 2017, 8, 2248. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Yanai, Y.; Nakano, Y.; Sasaki, H.; Uragami, A.; Okada, K. Isolation and Gene Expression Analysis of Flowering-related Genes in Lettuce (Lactuca sativa L.). Hortic. J. 2017, 86, 340–348. [Google Scholar] [CrossRef]
- Han, Y.; Chen, Z.; Lv, S.; Ning, K.; Ji, X.; Liu, X.; Wang, Q.; Liu, R.; Fan, S.; Zhang, X. MADS-box genes and gibberellins regulate bolting in Lettuce (Lactuca sativa L.). Front. Plant Sci. 2016, 7, 1889. [Google Scholar] [CrossRef] [PubMed]
- Chouard, P. Vernalization and its relations to dormancy. Annu. Rev. Plant Physiol. 1960, 11, 191–238. [Google Scholar] [CrossRef]
- Albani, M.C.; Coupland, G. Comparative analysis of flowering in annual and perennial plants. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 91, pp. 323–348. [Google Scholar]
- Hepworth, J.; Antoniou-Kourounioti, R.L.; Bloomer, R.H.; Selga, C.; Berggren, K.; Cox, D.; Collier Harris, B.R.; Irwin, J.A.; Holm, S.; Sall, T.; et al. Absence of warmth permits epigenetic memory of winter in Arabidopsis. Nat. Commun 2018, 9, 639. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Blanco, C.; Andrade, J.; Becker, C.; Bemm, F.; Bergelson, J.; Borgwardt, K.M.; Cao, J.; Chae, E.; Dezwaan, T.M.; Ding, W. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 2016, 166, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Suppanz, I.; Raman, H.; Hou, J.; Wang, J.; Long, Y.; Jung, C.; Meng, J. Comparative analysis of FLC homologues in Brassicaceae provides insight into their role in the evolution of oilseed rape. PLoS ONE 2012, 7, e45751. [Google Scholar] [CrossRef] [PubMed]
- Schranz, M.E.; Quijada, P.; Sung, S.-B.; Lukens, L.; Amasino, R.; Osborn, T.C. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 2002, 162, 1457–1468. [Google Scholar] [PubMed]
- Razi, H.; Howell, E.C.; Newbury, H.J.; Kearsey, M.J. Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor. Appl. Genet. 2008, 116, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, C.; Severing, E.; Karl, R.; Bergonzi, S.; Koch, M.; Tresch, A.; Coupland, G. Divergence of annual and perennial species in the Brassicaceae and the contribution of cis-acting variation at FLC orthologues. Mol. Ecol. 2017, 26, 3437–3457. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, C.C.; Burn, J.E.; Perez, P.P.; Metzger, J.; Edwards, J.A.; Peacock, W.J.; Dennis, E.S. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 1999, 11, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Johanson, U.; West, J.; Lister, C.; Michaels, S.; Amasino, R.; Dean, C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 2000, 290, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Werner, J.D.; Borevitz, J.O.; Uhlenhaut, N.H.; Ecker, J.R.; Chory, J.; Weigel, D. FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 2005, 170, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Marquardt, S.; Lister, C.; Swiezewski, S.; Dean, C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 2010, 327, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Howard, M.; Dean, C. Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr. Boil. 2014, 24, 1793–1797. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Sung, S. Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell 2017, 40, 302.e4–312.e4. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Xi, Y.; Sung, S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 2017, 13, e1006939. [Google Scholar] [CrossRef] [PubMed]
- Bastow, R.; Mylne, J.S.; Lister, C.; Lippman, Z.; Martienssen, R.A.; Dean, C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 2004, 427, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.; Amasino, R.M. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 2004, 427, 159. [Google Scholar] [CrossRef] [PubMed]
- Ausín, I.; Alonso-Blanco, C.; Jarillo, J.A.; Ruiz-García, L.; Martínez-Zapater, J.M. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 2004, 36, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Gendall, A.R.; Levy, Y.Y.; Wilson, A.; Dean, C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 2001, 107, 525–535. [Google Scholar] [CrossRef]
- Levy, Y.Y.; Mesnage, S.; Mylne, J.S.; Gendall, A.R.; Dean, C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 2002, 297, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Le, C.; Wang, Y.; Li, Z.; Jiang, D.; Wang, Y.; He, Y. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 2013, 4, 1947. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.H.; Hill, C.B. Rapid-cycling populations of Brassica. Science 1986, 232, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, K.; Sakamoto, K.; Kikuchi, R.; Saito, A.; Togashi, E.; Kuginuki, Y.; Matsumoto, S.; Hirai, M. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor. Appl. Genet. 2007, 114, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Al-Shahrour, F.; Minguez, P.; Marqués-Bonet, T.; Gazave, E.; Navarro, A.; Dopazo, J. Selection upon genome architecture: Conservation of functional neighborhoods with changing genes. PLoS Comput. Boil. 2010, 6, e1000953. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Sonnhammer, E.L. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 2003, 13, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.A.; Lister, C.; Soumpourou, E.; Zhang, Y.; Howell, E.C.; Teakle, G.; Dean, C. Functional alleles of the flowering time regulator FRIGIDA in the Brassica oleracea genome. BMC Plant Boil. 2012, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wei, K.; Cheng, F.; Li, S.; Wang, Q.; Zhao, J.; Bonnema, G.; Wang, X. A naturally occurring InDel variation in BraA. FLC. b (BrFLC2) associated with flowering time variation in Brassica rapa. BMC Plant Boil. 2012, 12, 151. [Google Scholar]
- Xiao, D.; Zhao, J.J.; Hou, X.L.; Basnet, R.K.; Carpio, D.P.; Zhang, N.W.; Bucher, J.; Lin, K.; Cheng, F.; Wang, X.W. The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co-expression networks. J. Exp. Bot. 2013, 64, 4503–4516. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.-X.; Wu, J.; Sun, R.-F.; Zhang, X.-W.; Xu, D.-H.; Bonnema, G.; Wang, X.-W. A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 2009, 60, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Kulkarni, V.; Liu, N.; Pino Del Carpio, D.; Bucher, J.; Bonnema, G. BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J. Exp. Bot. 2010, 61, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Kawanabe, T.; Osabe, K.; Itabashi, E.; Okazaki, K.; Dennis, E.S.; Fujimoto, R. Development of primer sets that can verify the enrichment of histone modifications, and their application to examining vernalization-mediated chromatin changes in Brassica rapa L. Genes Genet. Syst. 2016, 91, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Wei, K.; Gao, B.; Liu, J.; Liang, J.; Cheng, F.; Wang, X.; Wu, J. BrFLC5: A weak regulator of flowering time in Brassica rapa. Theor. Appl. Genet. 2018, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Liu, T.; Wang, J.; Hou, X. Isolation and functional characterization of a floral repressor, BcFLC2, from Pak-choi (Brassica rapa ssp. chinensis). Planta 2018, 248, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, S.; Bai, J.; He, Y. Tuning growth cycles of Brassica crops via natural antisense transcripts of Br FLC. Plant. Biotechnol. J. 2016, 14, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Zhang, H.; Zhang, B.; Wu, X.; Shao, S.; Li, Y.; Hou, X.; Liu, T. Role of vernalization-mediated demethylation in the floral transition of Brassica rapa. Planta 2017, 245, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Matschegewski, C.; Zetzsche, H.; Hasan, Y.; Leibeguth, L.; Briggs, W.; Ordon, F.; Uptmoor, R. Genetic variation of temperature-regulated curd induction in cauliflower: Elucidation of floral transition by genome-wide association mapping and gene expression analysis. Front. Plant Sci. 2015, 6, 720. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.A.; Soumpourou, E.; Lister, C.; Ligthart, J.D.; Kennedy, S.; Dean, C. Nucleotide polymorphism affecting FLC expression underpins heading date variation in horticultural brassicas. Plant J. 2016, 87, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Fadina, O.; Pankin, A.; Khavkin, E. Molecular characterization of the flowering time gene FRIGIDA in Brassica genomes A and C. Russ. J. Plant Physiol. 2013, 60, 279–289. [Google Scholar] [CrossRef]
- Yi, G.; Park, H.; Kim, J.-S.; Chae, W.B.; Park, S.; Huh, J.H. Identification of three FLOWERING LOCUS C genes responsible for vernalization response in radish (Raphanus sativus L.). Hortic. Environ. Biotechnol. 2014, 55, 548–556. [Google Scholar] [CrossRef]
- Jung, W.Y.; Park, H.J.; Lee, A.; Lee, S.S.; Kim, Y.-S.; Cho, H.S. Identification of flowering-related genes responsible for differences in bolting time between two radish inbred lines. Front. Plant Sci. 2016, 7, 1844. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, Y.; Xu, L.; Nie, S.; Chen, Y.; Liang, D.; Sun, X.; Karanja, B.K.; Luo, X.; Liu, L. Genome-wide characterization of the MADS-Box gene family in Radish (Raphanus sativus L.) and assessment of its roles in flowering and floral organogenesis. Front. Plant Sci. 2016, 7, 1390. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.L. Delayed Bolting in Rocket for Improved Quality and GREATER Sustainability. Ph.D. Thesis, University of Warwick, Coventry, UK, 2015. [Google Scholar]
- Lee, J.H.; Park, S.H.; Lee, J.S.; Ahn, J.H. A conserved role of SHORT VEGETATIVE PHASE (SVP) in controlling flowering time of Brassica plants. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 2007, 1769, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Lutz, U.; Nussbaumer, T.; Spannagl, M.; Diener, J.; Mayer, K.F.; Schwechheimer, C. Natural haplotypes of FLM non-coding sequences fine-tune flowering time in ambient spring temperatures in Arabidopsis. eLife 2017, 6, e22114. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Sureshkumar, S.; Lempe, J.; Weigel, D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006, 2, e106. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Weigel, D. Temperature induced flowering in Arabidopsis thaliana. Plant Signal. Behav. 2006, 1, 227–228. [Google Scholar] [CrossRef] [PubMed]
- Capovilla, G.; Schmid, M.; Pose, D. Control of flowering by ambient temperature. J. Exp. Bot 2015, 66, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Wigge, P.A. H2A. Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Lucyshyn, D.; Jaeger, K.E.; Alós, E.; Alvey, E.; Harberd, N.P.; Wigge, P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.C.; Youn, Y.; Duarte, M.I.; Harmon, F.G. The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. J. Exp. Bot. 2014, 65, 1141–1151. [Google Scholar] [CrossRef] [PubMed]
- Fernández, V.; Takahashi, Y.; Le Gourrierec, J.; Coupland, G. Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J. 2016, 86, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Capovilla, G.; Pajoro, A.; Immink, R.G.; Schmid, M. Role of alternative pre-mRNA splicing in temperature signaling. Curr. Opin. Plant Boil. 2015, 27, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.; Shen, L.; Wu, Y.; Chen, H.; Robertson, M.; Helliwell, C.A.; Ito, T.; Meyerowitz, E.; Yu, H. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 2008, 15, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Ryu, H.-S.; Chung, K.S.; Posé, D.; Kim, S.; Schmid, M.; Ahn, J.H. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 2013, 342, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Meyerowitz, E.M. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Boil. 2010, 6, 419. [Google Scholar] [CrossRef] [PubMed]
- Posé, D.; Verhage, L.; Ott, F.; Yant, L.; Mathieu, J.; Angenent, G.C.; Immink, R.G.; Schmid, M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 2013, 503, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Verhage, L.; Severing, E.I.; Bucher, J.; Lammers, M.; Busscher-Lange, J.; Bonnema, G.; Rodenburg, N.; Proveniers, M.C.; Angenent, G.C.; Immink, R.G. Splicing-related genes are alternatively spliced upon changes in ambient temperatures in plants. PLoS ONE 2017, 12, e0172950. [Google Scholar] [CrossRef] [PubMed]
- Capovilla, G.; Symeonidi, E.; Wu, R.; Schmid, M. Contribution of major FLM isoforms to temperature-dependent flowering in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 5117–5127. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Vigo, B.; Martínez-Zapater, J.M.; Alonso-Blanco, C. The flowering repressor SVP underlies a novel Arabidopsis thaliana QTL interacting with the genetic background. PLoS Genet. 2013, 9, e1003289. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Chen, C.; Yin, S.; Li, H.; Li, Z.; Wang, B.; King, G.J.; Wang, J.; Liu, K. Sequence variation and functional analysis of a FRIGIDA orthologue (BnaA3. FRI) in Brassica napus. BMC Plant Boil. 2018, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, A.-S.; Lutts, S.; Vandoorne, B.; Descamps, C.; Périlleux, C.; Dielen, V.; Van Herck, J.-C.; Quinet, M. High temperatures limit plant growth but hasten flowering in root chicory (Cichorium intybus) independently of vernalisation. J. Plant Physiol. 2014, 171, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Garner, W.W.; Allard, H.A. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. Mon. Weather. Rev. 1920, 48, 415. [Google Scholar] [CrossRef]
- Hernando, C.E.; Romanowski, A.; Yanovsky, M.J. Transcriptional and post-transcriptional control of the plant circadian gene regulatory network. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2017, 1860, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Valverde, F.; Mouradov, A.; Soppe, W.; Ravenscroft, D.; Samach, A.; Coupland, G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Ito, S.; Imaizumi, T. Similarities in the circadian clock and photoperiodism in plants. Curr. Opin. Plant Biol. 2010, 13, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Higashi, T.; Aoki, K.; Nagano, A.J.; Honjo, M.N.; Fukuda, H. Circadian Oscillation of the Lettuce Transcriptome under Constant Light and Light–Dark Conditions. Front. Plant Sci. 2016, 7, 1114. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi, T.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Smith, R.W.; To, B.J.; Millar, A.J.; Imaizumi, T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 2012, 336, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Estrada, D.A.; Johnson, R.S.; Kim, S.K.; Lee, S.Y.; MacCoss, M.J.; Imaizumi, T. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2014, 111, 17672–17677. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Marchal, V.; Panigrahi, K.C.; Wenkel, S.; Soppe, W.; Deng, X.W.; Valverde, F.; Coupland, G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008, 27, 1277–1288. [Google Scholar] [CrossRef] [PubMed]
- Laubinger, S.; Marchal, V.; Gentilhomme, J.; Wenkel, S.; Adrian, J.; Jang, S.; Kulajta, C.; Braun, H.; Coupland, G.; Hoecker, U. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 2006, 133, 3213–3222. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-J.; Zhang, Y.-C.; Li, Q.-H.; Sang, Y.; Mao, J.; Lian, H.-L.; Wang, L.; Yang, H.-Q. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 2008, 20, 292–306. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Boil. 2008, 9, R130. [Google Scholar] [CrossRef] [PubMed]
- Nohales, M.A.; Kay, S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Boil. 2016, 23, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; Knowles, S.M.; Andronis, C.; Ong, M.S.; Tobin, E.M. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009, 150, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Pérez-García, P.; Pokhilko, A.; Millar, A.; Antoshechkin, I.; Riechmann, J.L.; Mas, P. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 2012, 336, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Kamioka, M.; Takao, S.; Suzuki, T.; Taki, K.; Higashiyama, T.; Kinoshita, T.; Nakamichi, N. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED 1 in Arabidopsis circadian clock. Plant Cell 2016, 28, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; Webb, C.J.; Knowles, S.M.; Kim, S.H.; Wang, Z.-Y.; Tobin, E.M. CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012, 158, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Farré, E.M.; Harmer, S.L.; Harmon, F.G.; Yanovsky, M.J.; Kay, S.A. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Boil. 2005, 15, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Devisetty, U.K.; Harmer, S.L. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2013, 2, e00473. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.-H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Herrero, E.; Kolmos, E.; Bujdoso, N.; Yuan, Y.; Wang, M.; Berns, M.C.; Uhlworm, H.; Coupland, G.; Saini, R.; Jaskolski, M. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 2012, 24, 428–443. [Google Scholar] [CrossRef] [PubMed]
- Chow, B.Y.; Helfer, A.; Nusinow, D.A.; Kay, S.A. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal. Behav. 2012, 7, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef] [PubMed]
- Robert, L.S.; Robson, F.; Sharpe, A.; Lydiate, D.; Coupland, G. Conserved structure and function of the Arabidopsis flowering time gene CONSTANS in Brassica napus. Plant Mol. Boil. 1998, 37, 763–772. [Google Scholar] [CrossRef]
- Kim, J.A.; Jung, H.-E.; Hong, J.K.; Hermand, V.; McClung, C.R.; Lee, Y.-H.; Kim, J.Y.; Lee, S.I.; Jeong, M.-J.; Kim, J. Reduction of GIGANTEA expression in transgenic Brassica rapa enhances salt tolerance. Plant Cell Rep. 2016, 35, 1943–1954. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Lou, P.; Hermand, V.; Aman, R.; Park, H.J.; Yun, D.-J.; Kim, W.Y.; Salmela, M.J.; Ewers, B.E.; Weinig, C. Allelic polymorphism of GIGANTEA is responsible for naturally occurring variation in circadian period in Brassica rapa. Proc. Natl. Acad. Sci. USA 2015, 112, 3829–3834. [Google Scholar] [PubMed]
- Curtis, I.S.; Nam, H.G.; Yun, J.Y.; Seo, K.-H. Expression of an antisense GIGANTEA (GI) gene fragment in transgenic radish causes delayed bolting and flowering. Transgenic Res. 2002, 11, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Thiruvengadam, M.; Shih, C.-F.; Yang, C.-H. Expression of an antisense Brassica oleracea GIGANTEA (BoGI) gene in transgenic broccoli causes delayed flowering, leaf senescence, and post-harvest yellowing retardation. Plant Mol. Boil. Rep. 2015, 33, 1499–1509. [Google Scholar] [CrossRef]
- Lou, P.; Wu, J.; Cheng, F.; Cressman, L.G.; Wang, X.; McClung, C.R. Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell 2012, 24, 2415–2426. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Wu, M.-F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.-W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Teotia, S.; Tang, G. To bloom or not to bloom: Role of microRNAs in plant flowering. Mol. Plant 2015, 8, 359–377. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.; Richter, R.; Coupland, G. Competence to flower: Age-controlled sensitivity to environmental cues. Plant Physiol. 2017, 173, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Xu, L.; Wang, Y.; Huang, D.; Muleke, E.M.; Sun, X.; Wang, R.; Xie, Y.; Gong, Y.; Liu, L. Identification of bolting-related microRNAs and their targets reveals complex miRNA-mediated flowering-time regulatory networks in radish (Raphanus sativus L.). Sci. Rep. 2015, 5, 14034. [Google Scholar] [CrossRef] [PubMed]
- Huo, H.; Wei, S.; Bradford, K.J. DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. USA 2016, 113, E2199–E2206. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Boil. 2008, 67, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.-Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Seo, Y.-H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.-H.; Park, C.-M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Yant, L.J.; Mürdter, F.; Küttner, F.; Schmid, M. Repression of flowering by the miR172 target SMZ. PLoS Boil. 2009, 7, e1000148. [Google Scholar] [CrossRef] [PubMed]
- Yant, L.; Mathieu, J.; Dinh, T.T.; Ott, F.; Lanz, C.; Wollmann, H.; Chen, X.; Schmid, M. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 2010, 22, 2156–2170. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.-Y.; Zhou, Y.; He, F.; Dong, X.; Liu, L.-Y.; Coupland, G.; Turck, F.; de Meaux, J. miR824-regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell 2014, 26, 2024–2037. [Google Scholar] [CrossRef] [PubMed]
- Bergonzi, S.; Albani, M.C.; van Themaat, E.V.L.; Nordström, K.J.V.; Wang, R.; Schneeberger, K.; Moerland, P.D.; Coupland, G. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 2013, 340, 1094–1097. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Luo, H. MicroRNA-mediated gene regulation: Potential applications for plant genetic engineering. Plant Mol. Boil. 2013, 83, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.; Zou, J.; Hubertus Behrens, F.; Chen, L.; Ye, C.; Dai, S.; Li, R.; Ni, M.; Jiang, X.; Qiu, J. Identification, evolution, and expression partitioning of miRNAs in allopolyploid Brassica napus. J. Exp. Bot. 2015, 66, 7241–7253. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Hao, M.; Wang, W.; Mei, D.; Tong, C.; Wang, H.; Liu, J.; Fu, L.; Hu, Q. Genomic identification, characterization and differential expression analysis of SBP-box gene family in Brassica napus. BMC Plant Boil. 2016, 16, 196. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hou, X.; Yang, X. Identification of conserved microRNAs and their targets in Chinese cabbage (Brassica rapa subsp. pekinensis). Genome 2011, 54, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, X.; Xu, H.; Chi, X.; Zhang, M.; Hou, X. Identification and characterization of microRNAs and their target genes in Brassica oleracea. Gene 2012, 505, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Geng, M.; Li, H.; Jin, C.; Liu, Q.; Chen, C.; Song, W.; Wang, C. Genome-wide identification and characterization of miRNAs in the hypocotyl and cotyledon of cauliflower (Brassica oleracea L. var. botrytis) seedlings. Planta 2014, 239, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhu, B.; Luan, F.; Zhu, H.; Shao, Y.; Chen, A.; Lu, C.; Luo, Y. Conserved miRNAs and their targets identified in lettuce (Lactuca) by EST analysis. Gene 2010, 463, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Footitt, S.; Huang, Z.; Clay, H.A.; Mead, A.; Finch-Savage, W.E. Temperature, light and nitrate sensing coordinate A rabidopsis seed dormancy cycling, resulting in winter and summer annual phenotypes. Plant J. 2013, 74, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Singh, N.; Srivastava, G.; Sharma, A. MiRNA mediated gene regulatory network analysis of Cichorium intybus (chicory). Agric. Gene 2017, 3, 37–45. [Google Scholar] [CrossRef]
- Bernier, G.; Havelange, A.E.; Houssa, C.; Petitjean, A.; Lejeune, P. Physiological signals that induce flowering. Plant Cell 1993, 5, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.N.; Heckman, J.W.; Somerville, C.R. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 1992, 100, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Duclos, D.V.; Björkman, T. Gibberellin Control of Reproductive Transitions in Brassica oleracea Curd Development. J. Am. Soc. Hortic. Sci. 2015, 140, 57–67. [Google Scholar]
- Bernier, G. The control of floral evocation and morphogenesis. Annu. Rev. Plant Physiol. Plant Mol. Boil. 1988, 39, 175–219. [Google Scholar] [CrossRef]
- Eriksson, S.; Böhlenius, H.; Moritz, T.; Nilsson, O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 2006, 18, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Bernier, G. My favourite flowering image: The role of cytokinin as a flowering signal. J. Exp. Bot. 2011, 64, 5795–5799. [Google Scholar] [CrossRef] [PubMed]
- Besnard-Wibaut, C. Effectiveness of gibberellins and 6-benzyladenine on flowering of Arabidopsis thaliana. Physiol. Plant. 1981, 53, 205–212. [Google Scholar] [CrossRef]
- D’aloia, M.; Bonhomme, D.; Bouché, F.; Tamseddak, K.; Ormenese, S.; Torti, S.; Coupland, G.; Périlleux, C. Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J. 2011, 65, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Michniewicz, M.; Kamieńska, A. Studies on the role of kinetin and vitamin E in the flowering of the cold requiring plant Cichorium intybus) and the long-day plant (Arabidopsis thaliana) grown in non-inductive. Acta Soc. Bot. Pol. 1967, 36, 67–72. [Google Scholar] [CrossRef]
- King, R.W.; Evans, L.T. Gibberellins and flowering of grasses and cereals: Prizing open the lid of the “florigen” black box. Annu. Rev. Plant. Boil. 2003, 54, 307–328. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Phillips, A.L. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Rieu, I.; Ruiz-Rivero, O.; Fernandez-Garcia, N.; Griffiths, J.; Powers, S.J.; Gong, F.; Linhartova, T.; Eriksson, S.; Nilsson, O.; Thomas, S.G. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008, 53, 488–504. [Google Scholar] [CrossRef] [PubMed]
- Plackett, A.R.; Powers, S.J.; Fernandez-Garcia, N.; Urbanova, T.; Takebayashi, Y.; Seo, M.; Jikumaru, Y.; Benlloch, R.; Nilsson, O.; Ruiz-Rivero, O. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1,-2, and-3 are the dominant paralogs. Plant Cell 2012, 24, 941–960. [Google Scholar] [CrossRef] [PubMed]
- Mitchum, M.G.; Yamaguchi, S.; Hanada, A.; Kuwahara, A.; Yoshioka, Y.; Kato, T.; Tabata, S.; Kamiya, Y.; Sun, T.p. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006, 45, 804–818. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-L.; Su, X.; Xiong, W.; Liu, J.-F.; Wu, Y.; Feng, Y.-Q.; Yuan, B.-F. Assessing gibberellins oxidase activity by anion exchange/hydrophobic polymer monolithic capillary liquid chromatography-mass spectrometry. PLoS ONE 2013, 8, e69629. [Google Scholar] [CrossRef] [PubMed]
- Porri, A.; Torti, S.; Romera-Branchat, M.; Coupland, G. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 2012, 139, 2198–2209. [Google Scholar] [CrossRef] [PubMed]
- Rieu, I.; Eriksson, S.; Powers, S.J.; Gong, F.; Griffiths, J.; Woolley, L.; Benlloch, R.; Nilsson, O.; Thomas, S.G.; Hedden, P. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 2008, 20, 2420–2436. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Murase, K.; Sun, T.-p.; Steber, C.M. Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 2008, 20, 2447–2459. [Google Scholar] [CrossRef] [PubMed]
- Willige, B.C.; Ghosh, S.; Nill, C.; Zourelidou, M.; Dohmann, E.M.; Maier, A.; Schwechheimer, C. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 2007, 19, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, M.; Shimada, A.; Takashi, Y.; Kim, Y.C.; Park, S.H.; Ueguchi-Tanaka, M.; Suzuki, H.; Katoh, E.; Iuchi, S.; Kobayashi, M. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 2006, 46, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Galvão, V.C.; Horrer, D.; Küttner, F.; Schmid, M. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 2012, 139, 4072–4082. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Galvão, V.C.; Zhang, Y.-C.; Horrer, D.; Zhang, T.-Q.; Hao, Y.-H.; Feng, Y.-Q.; Wang, S.; Markus, S.; Wang, J.-W. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING–LIKE transcription factors. Plant Cell 2012, 24, 3320–3332. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Porri, A.; Torti, S.; Mateos, J.; Romera-Branchat, M.; García-Martínez, J.L.; Fornara, F.; Gregis, V.; Kater, M.M.; Coupland, G. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. USA 2014, 201409567. [Google Scholar]
- Galvão, V.C.; Collani, S.; Horrer, D.; Schmid, M. Gibberellic acid signaling is required for ambient temperature-mediated induction of flowering in Arabidopsis thaliana. Plant J. 2015, 84, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Bartrina, I.; Jensen, H.; Novak, O.; Strnad, M.; Werner, T.; Schmülling, T. Gain-of-function mutants of the cytokinin receptors AHK2 and AHK3 regulate plant organ size, flowering time and plant longevity. Plant Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Greenboim-Wainberg, Y.; Maymon, I.; Borochov, R.; Alvarez, J.; Olszewski, N.; Ori, N.; Eshed, Y.; Weiss, D. Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 2005, 17, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, H.; Li, J.; Wang, B.; Dai, C.; Wang, J.; Liu, K. Brassica napus DS-3, encoding a DELLA protein, negatively regulates stem elongation through gibberellin signaling pathway. Theor. Appl. Genet. 2017, 130, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Tarkowská, D.; Filek, M.; Biesaga-Kościelniak, J.; Marcińska, I.; Macháčková, I.; Krekule, J.; Strnad, M. Cytokinins in shoot apices of Brassica napus plants during vernalization. Plant Sci. 2012, 187, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Wang, X.; Zhang, J.; Qi, X.; Ping, A.; Hou, L.; Xing, G.; Li, G.; Li, M. Genetic Regulation of GA Metabolism during Vernalization, Floral Bud Initiation and Development in Pak Choi (Brassica rapa ssp. chinensis Makino). Front. Plant Sci. 2017, 8, 1533. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Matsuo, S.; Kikuchi, K.; Mitsuhashi, W.; Toyomasu, T.; Honda, I. The endogenous level of GA1 is upregulated by high temperature during stem elongation in lettuce through LsGA3ox1 expression. J. Plant Physiol. 2009, 166, 2077–2084. [Google Scholar] [CrossRef] [PubMed]
- Frugis, G.; Giannino, D.; Mele, G.; Nicolodi, C.; Chiappetta, A.; Bitonti, M.B.; Innocenti, A.M.; Dewitte, W.; Van Onckelen, H.; Mariotti, D. Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol. 2001, 126, 1370–1380. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, S.; Piazza, P.; Craft, J.; Hay, A.; Woolley, L.; Rieu, I.; Phillips, A.; Hedden, P.; Tsiantis, M. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Boil. 2005, 15, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Sablowski, R.; Sandberg, G.; Samach, A.; Ori, N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Boil. 2005, 15, 1566–1571. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, E.; Iannelli, M.A.; Frugis, G. TALE and shape: How to make a leaf different. Plants 2013, 2, 317–342. [Google Scholar] [CrossRef] [PubMed]
- Salomé, P.A.; Bomblies, K.; Laitinen, R.A.; Yant, L.; Mott, R.; Weigel, D. Genetic architecture of flowering time variation in Arabidopsis thaliana. Genetics 2011, 188, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Brachi, B.; Faure, N.; Horton, M.; Flahauw, E.; Vazquez, A.; Nordborg, M.; Bergelson, J.; Cuguen, J.; Roux, F. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 2010, 6, e1000940. [Google Scholar] [CrossRef] [PubMed]
- Shindo, C.; Aranzana, M.J.; Lister, C.; Baxter, C.; Nicholls, C.; Nordborg, M.; Dean, C. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 2005, 138, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Caicedo, A.L.; Stinchcombe, J.R.; Olsen, K.M.; Schmitt, J.; Purugganan, M.D. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. USA 2004, 101, 15670–15675. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Filiault, D.; Box, M.S.; Kerdaffrec, E.; van Oosterhout, C.; Wilczek, A.M.; Schmitt, J.; McMullan, M.; Bergelson, J.; Nordborg, M. Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev. 2014, 28, 1635–1640. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, C.; Balasubramanian, S.; Warthmann, N.; Michael, T.P.; Lempe, J.; Sureshkumar, S.; Kobayashi, Y.; Maloof, J.N.; Borevitz, J.O.; Chory, J. Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. Genetics 2009, 183, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Shi, J.; Qiu, D.; Li, R.; Zhang, C.; Wang, J.; Hou, J.; Zhao, J.; Shi, L.; Park, B.-S. Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics 2007, 177, 2433–2444. [Google Scholar] [PubMed]
- Li, L.; Long, Y.; Zhang, L.; Dalton-Morgan, J.; Batley, J.; Yu, L.; Meng, J.; Li, M. Genome wide analysis of flowering time trait in multiple environments via high-throughput genotyping technique in Brassica napus L. PLoS ONE 2015, 10, e0119425. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Long, Y.; Raman, H.; Zou, X.; Wang, J.; Dai, S.; Xiao, Q.; Li, C.; Fan, L.; Liu, B. A Tourist-like MITE insertion in the upstream region of the BnFLC. A10 gene is associated with vernalization requirement in rapeseed (Brassica napus L.). BMC Plant. Boil. 2012, 12, 238. [Google Scholar]
- Luo, Z.; Wang, M.; Long, Y.; Huang, Y.; Shi, L.; Zhang, C.; Liu, X.; Fitt, B.D.; Xiang, J.; Mason, A.S. Incorporating pleiotropic quantitative trait loci in dissection of complex traits: Seed yield in rapeseed as an example. Theor. Appl. Genet. 2017, 130, 1569–1585. [Google Scholar] [CrossRef] [PubMed]
- Hartman, Y.; Hooftman, D.A.; Schranz, M.E.; van Tienderen, P.H. QTL analysis reveals the genetic architecture of domestication traits in Crisphead lettuce. Genet. Resour. Crop. Evol. 2013, 60, 1487–1500. [Google Scholar] [CrossRef]
- Hartman, Y.; Hooftman, D.A.; Uwimana, B.; van de Wiel, C.; Smulders, M.J.; Visser, R.G.; van Tienderen, P.H. Genomic regions in crop–wild hybrids of lettuce are affected differently in different environments: Implications for crop breeding. Evol. Appl. 2012, 5, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Hartman, Y.; Uwimana, B.; Hooftman, D.A.; Schranz, M.E.; Wiel, C.; Smulders, M.J.; Visser, R.G.; Tienderen, P.H. Genomic and environmental selection patterns in two distinct lettuce crop–wild hybrid crosses. Evol. Appl. 2013, 6, 569–584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Su, W.; Tao, R.; Zhang, W.; Chen, J.; Wu, P.; Yan, C.; Jia, Y.; Larkin, R.M.; Lavelle, D. RNA sequencing provides insights into the evolution of lettuce and the regulation of flavonoid biosynthesis. Nat. Commun. 2017, 8, 2264. [Google Scholar] [CrossRef] [PubMed]
- Javed, N.; Geng, J.; Tahir, M.; McVetty, P.; Li, G.; Duncan, R.W. Identification of QTL influencing seed oil content, fatty acid profile and days to flowering in Brassica napus L. Euphytica 2016, 207, 191–211. [Google Scholar] [CrossRef]
- Rahman, H.; Bennett, R.A.; Kebede, B. Molecular mapping of QTL alleles of Brassica oleracea affecting days to flowering and photosensitivity in spring Brassica napus. PLoS ONE 2018, 13, e0189723. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Xiang, Y.; Xu, E.; Ge, X.; Li, Z. Major Co-localized QTL for Plant Height, Branch Initiation Height, Stem Diameter, and Flowering Time in an Alien Introgression Derived Brassica napus DH Population. Front. Plant Sci. 2018, 9, 390. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Hu, K.; Zhang, Z.; Guan, C.; Chen, S.; Hua, W.; Li, J.; Wen, J.; Yi, B.; Shen, J. Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Res. 2015, 23, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Chen, B.; Xu, K.; Gao, G.; Li, F.; Qiao, J.; Yan, G.; Li, J.; Li, H.; Wu, X. Association mapping of flowering time QTLs and insight into their contributions to rapeseed growth habits. Front. Plant Sci. 2016, 7, 338. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, B.; Liu, G.; Hong, B.; Xu, J.; Chen, X.; Wang, B.; Wu, Z.; Hou, F.; Yue, X. A comprehensive and precise set of intervarietal substitution lines to identify candidate genes and quantitative trait loci in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 2018, 131, 2117–2129. [Google Scholar] [CrossRef] [PubMed]
- Schiessl, S.; Iniguez-Luy, F.; Qian, W.; Snowdon, R.J. Diverse regulatory factors associate with flowering time and yield responses in winter-type Brassica napus. BMC Genom. 2015, 16, 737. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, W.; Wang, Z.; Li, K.; Lim, Y.P.; Piao, Z. Construction of chromosome segment substitution lines enables QTL mapping for flowering and morphological traits in Brassica rapa. Front. Plant Sci. 2015, 6, 432. [Google Scholar] [CrossRef] [PubMed]
- Lou, P.; Zhao, J.; Kim, J.S.; Shen, S.; Del Carpio, D.P.; Song, X.; Jin, M.; Vreugdenhil, D.; Wang, X.; Koornneef, M. Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J. Exp. Bot. 2007, 58, 4005–4016. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, L.; Ji, X.; Yan, J.; Liu, Y.; Lv, X.; Feng, H. Mapping of quantitative trait loci for the bolting trait in Brassica rapa under vernalizing conditions. Genet. Mol. Res. 2014, 13, 3927–3939. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, C.; Shi, X.; Feng, H.; Wang, Y. Identification of QTLs with additive, epistatic, and QTL× environment interaction effects for the bolting trait in Brassica rapa L. Euphytica 2016, 210, 427–439. [Google Scholar] [CrossRef]
- Dechaine, J.M.; Brock, M.T.; Weinig, C. QTL architecture of reproductive fitness characters in Brassica rapa. BMC Plant Boil. 2014, 14, 66. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Liu, Y.; Zhang, L.; Li, Z.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Lv, H. QTL-seq for rapid identification of candidate genes for flowering time in broccoli× cabbage. Theor. Appl. Genet. 2018, 131, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Hasan, Y.; Briggs, W.; Matschegewski, C.; Ordon, F.; Stützel, H.; Zetzsche, H.; Groen, S.; Uptmoor, R. Quantitative trait loci controlling leaf appearance and curd initiation of cauliflower in relation to temperature. Theor. Appl. Genet. 2016, 129, 1273–1288. [Google Scholar] [CrossRef] [PubMed]
- FitzJohn, R.G.; Armstrong, T.T.; Newstrom-Lloyd, L.E.; Wilton, A.D.; Cochrane, M. Hybridisation within Brassica and allied genera: Evaluation of potential for transgene escape. Euphytica 2007, 158, 209–230. [Google Scholar] [CrossRef]
- Shea, D.J.; Tomaru, Y.; Itabashi, E.; Nakamura, Y.; Miyazaki, T.; Kakizaki, T.; Naher, T.N.; Shimizu, M.; Fujimoto, R.; Fukai, E. The production and characterization of a BoFLC2 introgressed Brassica rapa by repeated backcrossing to an F1. Breed. Sci. 2018, 68, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Lebeda, A.; Doležalová, I.; Křístková, E.; Kitner, M.; Petrželová, I.; Mieslerová, B.; Novotná, A. Wild Lactuca germplasm for lettuce breeding: Current status, gaps and challenges. Euphytica 2009, 170, 15. [Google Scholar] [CrossRef]
- Van Cutsem, P.; Du Jardin, P.; Boutte, C.; Beauwens, T.; Jacqmin, S.; Vekemans, X. Distinction between cultivated and wild chicory gene pools using AFLP markers. Theor. Appl. Genet. 2003, 107, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Hodgins, K.A.; Lai, Z.; Oliveira, L.O.; Still, D.W.; Scascitelli, M.; Barker, M.S.; Kane, N.C.; Dempewolf, H.; Kozik, A.; Kesseli, R.V. Genomics of Compositae crops: Reference transcriptome assemblies and evidence of hybridization with wild relatives. Mol. Ecol. Resour. 2014, 14, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Testone, G.; Mele, G.; Di Giacomo, E.; Gonnella, M.; Renna, M.; Tenore, G.C.; Nicolodi, C.; Frugis, G.; Iannelli, M.A.; Arnesi, G. Insights into the sesquiterpenoid pathway by metabolic profiling and de novo transcriptome assembly of stem-chicory (Cichorium intybus cultigroup “Catalogna”). Front. Plant Sci. 2016, 7, 1676. [Google Scholar] [CrossRef] [PubMed]
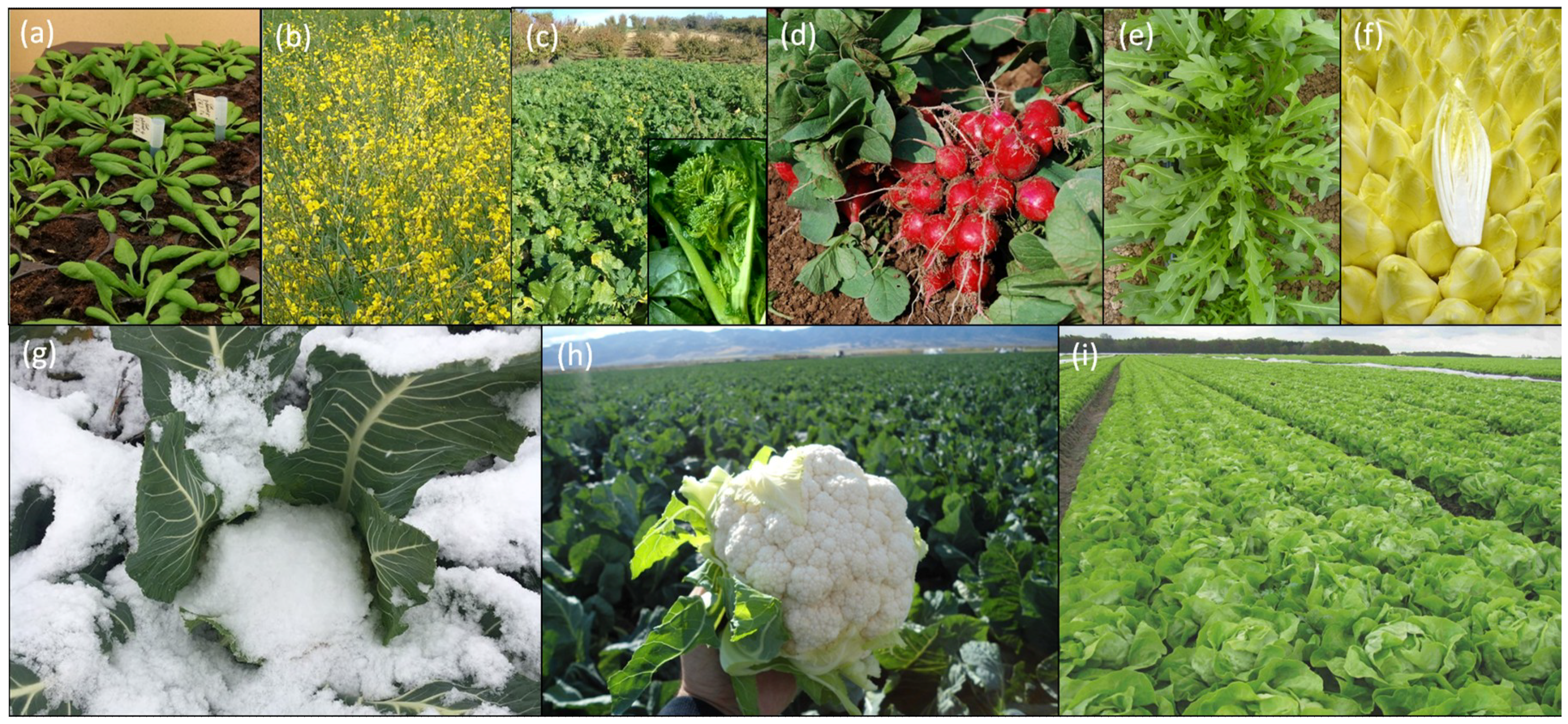
| Term | Definition |
|---|---|
| flowering time | the switch from plant vegetative growth to reproductive development |
| bolting | rapid elongation of the inflorescence/flowering stem |
| annuals | plants that complete their entire life cycle from seed to flower within one year and are characterized by short vegetative phase |
| biennials | plants which require two years to complete their life cycle, |
| perennials | plants that survive for several years and restrict the duration of reproduction by cycling between vegetative growth and flowering; perennials are characterized by prolonged vegetative phase that can last from a few weeks to several years |
| shoot apical meristem (SAM) | population of cells located at the tip of the shoot axis that produce lateral organs, stem tissue and regenerates itself |
| inflorescence meristem (IM) | a meristem that underwent transition from vegetative to reproductive fate and can produce floral meristems |
| floral meristem (FM) | group of cells responsible for the formation of floral organs |
| facultative photoperiod | plants that flower faster under a particular photoperiod but will eventually flower under all photoperiods (also called “quantitative”) |
| obligate photoperiod | plants that flower only under a particular photoperiod (also called “qualitative”) |
| long days | day length more than about 12 h, usually 16 h light and 8 h dark periods |
| short days | day length less than about 12 h, usually 8 h light and 16 h dark periods |
| Double Haploid (DH) | chromosome doubling of haploid cells to produce genetically homozygous plants |
| genome-wide association study (GWAS) | observational study of a genome-wide set of genetic variants in different individuals that occur more frequently in correlation with a specific trait, identifying inherited genetic variants associated with a trait |
| homolog | a gene related to a second gene by descent from a common ancestral DNA sequence |
| ortholog | genes in different species that evolved from a common ancestral gene by speciation; normally, orthologs retain the same function in the course of evolution |
| paralog | genes related by duplication within a genome that may evolve new functions |
| maturity | the state of being fully developed or full grown |
| uniformity | a state or condition of the plant in which everything is regular, homogeneous, or unvarying |
| predictable | always behaving or occurring in the way expected |
| robust | is a characteristic of being strong that, when transposed into a system, it refers to the ability of tolerating perturbations and remain effective |
| QTL | (or Quantitative Trait Locus), is a locus (section of DNA) which correlates with variation of a quantitative trait in the phenotype of a population of organisms |
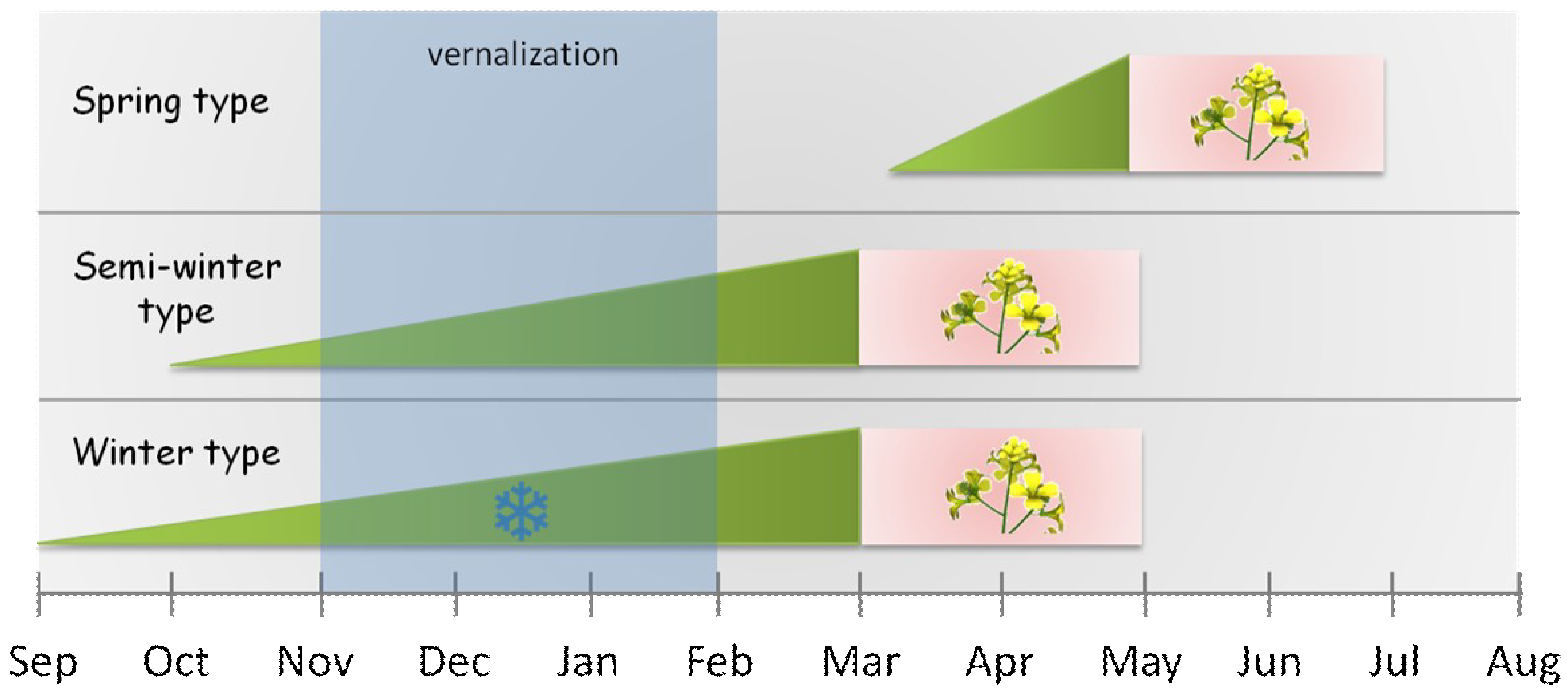
| vernalization | cold treatment needed to get many perennials to flower; usually the minimum period is six to twelve weeks at 4 °C |
| spring types | plants which flower early without vernalization |
| winter types | plants which have an obligate requirement for prolonged periods of cold temperatures |
| semi-winter types | plants which require mild vernalization and lack frost hardiness |
| Species | Chr. | Life Span | Vernalization | Types | Breeding Goal | Day Length 1 | Ref. |
|---|---|---|---|---|---|---|---|
| A. thaliana | 2n = 10 | Annual/biennial | Yes/no | Spring/semi-winter/winter | none | facultative LD | [12] |
| B. napus | 2n = 38 (AACC) | Annual/biennial | Yes/no | Spring/semi-winter/winter | flowering time adaptation | LD | [13,14] |
| B. rapa | 2n = 20 (AA) | Annual/biennial | Yes/no | Spring/semi-winter/winter | Late bolting | LD | [15] |
| B. oleracea | 2n = 18 (CC) | Annual/biennial | Yes/no | Spring/semi-winter/winter | Predictable harvest time | LD | [16] |
| R. sativus | 2n = 18 | Annual | No | Late bolting | facultative LD | [17,18] | |
| D. tenuifolia | 2n = 22 | Annual | No | Late bolting | LD | [19] | |
| L. sativa | 2n = 18 | Annual | No | Heat resistance | facultative LD | [20] | |
| C. intybus | 2n = 18 | Biennial/perennial | Yes | Resistance to bolting | LD | [21,22] |
| Pathway | Gene | Arabidopsis | B. napus gene ID | B. napus Chr position | B. rapa gene ID | B. rapa Chr position | B. oleracea gene ID | B. oleracea Chr position |
|---|---|---|---|---|---|---|---|---|
| Floral integrators | FT | AT1G65480 | GSBRNA2T00090951001 Bna.FT.A02 | A02:6375936.6379058 | Bra022475 BrFT1 | A02:8551268.8553758 | ||
| GSBRNA2T00030311001 Bna.FT.C02 | C02:996695.998788 | Bol045330 | Scaffold000001_P2: 1990327.1992083 | |||||
| TSF | AT4G20370 | GSBRNA2T00124448001 Bna.FT.A07 | A07:18855196.18857952 | Bra004117 BrFT2 | A07:20213069.20215397 | |||
| GSBRNA2T00146560001 | A07:22787807.22790354 | Bra015710 | A07:24515213.24516895 | |||||
| GSBRNA2T00077948001 | C02:20907503.20909228 | Bol039209 | C02:19450855.19452577 | |||||
| GSBRNA2T00113342001 | C04:12435074.12437644 | Bol017639 | C04:17148775.17151658 | |||||
| GSBRNA2T00067517001 Bna.FT.C06 | C06:28552966.28555216 | Bol012573 | C07:9349005.9351279 | |||||
| GSBRNA2T00050890001 | Cnn:48285424.48286397 | Bol027595 | C07:1423408.1425153 | |||||
| TFL1 | AT5G03840 | GSBRNA2T00136426001 | A10:16767409.16768474 | Bra009508 | A10:15774055.15775120 | |||
| GSBRNA2T00119620001 | Ann:609805.611005 | Bra028815 | A02:545667.546787 | |||||
| GSBRNA2T00078727001 | C02:1320757.1321835 | Bol005471 | C02:1447642.1448756 | |||||
| GSBRNA2T00134290001 | C03:673349.674628 | Bol015337 | C03:438359.439413 | |||||
| GSBRNA2T00073025001 | Cnn:9572005.9573076 | Bol010027 | C09:39511589.39512660 | |||||
| Bra005783 | A03:603455.604516 | |||||||
| SOC1 | AT2G45660 | GSBRNA2T00011646001 | A03:901877.905188 BnSOC1-A3 | Bra000393 | A03:10918286.10920672 | |||
| GSBRNA2T00063263001 | A04:18732428.18735897 | Bra039324 | A04:18723546.18725960 | |||||
| GSBRNA2T00116723001 | A05:2627051.2630394 | Bra004928 | A05:2530305.2532747 | |||||
| GSBRNA2T00037309001 | C04:48074887.48078345 | Bol021742 | C04:40413670.40414880 | |||||
| GSBRNA2T00083011001 | C04:867297.870707 | Bol030200 | C04:2998426.2999594 | |||||
| GSBRNA2T00029970001 | Cnn:35198162.35204681 | Bol029556 | C03:13421127.13422327 | |||||
| Vernalization | FLC | AT5G10140 | GSBRNA2T00143535001 Bna.FLC.A02 | A02:134362.138212 | Bra028599 BrFLC2 | A02:1524995.1528254 | ||
| GSBRNA2T00129741001 | A03:1360971.1364359 | Bra006051 BrFLC3 | A03:1764912.1767856 | |||||
| GSBRNA2T00142187001 | A03:6240056.6245305 | Bra022771 BrFLC5 | A03:6971946.6976797 | |||||
| GSBRNA2T00135921001 | A10:14998617.15003197 | Bra009055 BrFLC1 | A10:13856133.13860473 | |||||
| GSBRNA2T00068991001 Bna.FLC.C02 | C02:208562.212139 | Bol024642 BoFLC4 | C02:2720826.2721596 | |||||
| BoFLC2 | C02:2722189.2724345 | |||||||
| GSBRNA2T00134620001 | C03:2001058.2004665 | Bol008758 BoFLC3 | C03:1890867.1893743 | |||||
| GSBRNA2T00024568001 | C03:8403312.8410062 | BoFLC5 | C03:49708405.49709316 | |||||
| GSBRNA2T00016124001 | C09:46345350.46350092 | Bol043693 BoFLC1 | C09:37175182.37179020 | |||||
| GSBRNA2T00016119001 | C09:46366645.46371180 | |||||||
| FRI | AT4G00650 | GSBRNA2T00066686001 Bna.FRI.Xa | A03:6053113.6055294 | Bra029192 BrFRIa | A03:6784863.6787013 | |||
| GSBRNA2T00120967001 Bna.FRI.Xb | A10:4019556.4021675 | Bra035723 BrFRIb | A10:4133444.4134764 | |||||
| GSBRNA2T00052682001 Bna.FRI.Xd | C03:8149599.8151810 | Bol028107 BoFRIa | C03:7962008.7964180 | |||||
| GSBRNA2T00152364001 Bna.FRI.Xc | C09:29041826.29043953 | Bol004294BoFRIb | Scaffold000327:204688.206816 | |||||
| Ambient temperature | SVP | AT2G22540 | GSBRNA2T00032884001 | A04:10961147.10963402 | Bra030228 | A04:10192172.10194736 | ||
| GSBRNA2T00078179001 | A09:29590705.29594744 | Bra038511 | A09:33434743.33437921 | |||||
| GSBRNA2T00149752001 | C04:36478652.36481951 | Bol031759 | Scaffold000053:1406474.1408404 | |||||
| GSBRNA2T00127429001 | C08:32995398.32998881 | Bol044741 | C08:35213085.35214818 | |||||
| Photoperiod | CO | AT5G15840 | GSBRNA2T00135488001 | A10:13358777.13360064 | Bra008669 | A10:12117648.12118929 | ||
| GSBRNA2T00035272001 | C09:43745679.43747139 | Bol030488 | C09:33143053.33144339 | |||||
| GI | AT1G22770 | GSBRNA2T00015763001 | A09:22588149.22593013 | Bra024536 | A09:25756404.25760934 | |||
| GSBRNA2T00119480001 | C05:11778931.11784461 | Bol023541 | Scaffold000099_P1: 794479.799157 | |||||
| Age | SPL3 | AT2G33810 | GSBRNA2T00064576001 | A04:15462653.15463366 | Bra021880 | A04:15123762.15124274 | ||
| GSBRNA2T00095270001 | A05:5425249.5426076 | Bra005470 | A05:5668800.5669314 | |||||
| GSBRNA2T00132295001 | C03:9629272.9630113 | Bol036997 | C06:40526300.40526809 | |||||
| GSBRNA2T00020688001 | C04:44354526.44355241 | Bol037895 | C04:35540992.35541501 | |||||
| GSBRNA2T00038835001 | Cnn:4854484.4855112 | Bol027299 | C04:20510435.20510961 | |||||
| SPL9 | AT2G42200 | GSBRNA2T00123166001 | A04:17845227.17847617 | Bra016891 | A04:17839490.17841541 | |||
| GSBRNA2T00132740001 | A05:1443071.1445187 | Bra004674 | A05:1325605.1327587 | |||||
| GSBRNA2T00010840001 | C04:1886612.1888780 | Bol004847 | C04:966922.968875 | |||||
| GSBRNA2T00084688001 | C04:46904649.46905101 | |||||||
| GSBRNA2T00084692001 | C04:46915351.46917939 | Bol002678 | Scaffold000379:152205.154297 | |||||
| Bra015085 | A07:5833985.5836119 | |||||||
| SPL15 | AT3G57920 | GSBRNA2T00034335001 | A04:154881.156614 | Bra014599 | A04:1684031.1685273 | |||
| GSBRNA2T00098900001 | A07:14658857.14660105 | Bra003305 | A07:15783674.15784920 | |||||
| GSBRNA2T00087887001 | C04:25001142.25003655 | Bol011022 | C04:9176952.9178238 | |||||
| GSBRNA2T00105779001 | C06:19172101.19173360 | Bol007052 | C07:28887260.28888520 | |||||
| Gibberellin | GA20OX1 | AT4G25420 | Bra013890 | A01:8279446.8280885 | ||||
| Bra019165 | A03:25974634.25976038 | |||||||
| Bol039527 | C01:11622628.11624071 | |||||||
| Bol042237 | C06:43862307.43863764 | |||||||
| Bol041615 | Scaffold000009_P1: 317473.317667 | |||||||
| GA20OX2 | AT5G51810 | GSBRNA2T00036929001 | A02:5851980.5853392 | Bra022565 | A02:7878457.7879856 | |||
| GSBRNA2T00110217001 | A10:6243369.6244766 | Bra028277 | A10:4556457.4557854 | |||||
| GSBRNA2T00153037001 | C02:11109632.11111035 | Bol045266 | Scaffold000001_P2: 739593.740993 | |||||
| GSBRNA2T00108688001 | C09:30358041.30359437 | Bol029404 | C09:18392061.18393124 | |||||
| GSBRNA2T00108686001 | C09:30368578.30369971 | |||||||
| GSBRNA2T00025818001 | Cnn:77727013.77728046 | |||||||
| GA20OX3 | AT5G07200 | Bra028706 | A02:1065763.1067251 | |||||
| Bra005927 | A03:1251910.1253082 | |||||||
| Bra010064 | A06:14066314.14068214 | |||||||
| Bra009285 | A10:14501966.14503398 | |||||||
| Bol024532 | C02:2131323.2133012 | |||||||
| Bol008872 | C03:1262233.1263937 | |||||||
| Bol043862 | C09:38109369.38110750 | |||||||
| Bol041616 | Scaffold000009_P1: 331428.332030 | |||||||
| Bol024814 | Scaffold000091:1192479.1195919 | |||||||
| GA20OX4 | AT1G60980 | GSBRNA2T00070537001 | A09:7950969.7952898 | Bra027106 | A09:8952409.8954196 | |||
| GSBRNA2T00043758001 | Ann:21491303.21492435 | Bra039251 | Scaffold000162: 175188.176593 | |||||
| GSBRNA2T00043759001 | Ann:21493033.21494525 | Bra031467 | A01:17039613.17041240 | |||||
| GSBRNA2T00080857001 | Cnn:11274341.11276251 | Bol014320 | C03:52855996.52857792 | |||||
| GSBRNA2T00028142001 | Cnn:34189212.34190700 | Bol044153 | Scaffold000003_P1: 2530105.2531509 | |||||
| Bol007374 | Scaffold000262:280424.282063 | |||||||
| GA20OX5 | AT1G44090 | GSBRNA2T00097054001 | A08:969550.970698 | Bra014019 | A08:4525615.4527790 | |||
| GSBRNA2T00060328001 | C08:6199021.6199389 | |||||||
| GSBRNA2T00013490001 | Cnn:72340763.72341689 | Bol021441 | C07:30202821.30203904 |
| QTL Region | Species | Region 1 | Candidate Genes | References |
|---|---|---|---|---|
| At1 | A. thaliana | Chr1:24500000-29000000 | FT, FKF1, AP1, FLM | [216,217] |
| At2 | A. thaliana | Chr4:300000-1900000 | FRI | [216,217] |
| At3 | A. thaliana | Chr4:8000000-12000000 | VRN2, TSF, GA2ox | [216,217] |
| At4 | A. thaliana | Chr5:2700000-8100000 | FLC, CO, TFL2 | [216,217] |
| At5 | A. thaliana | Chr5:21500000-26000000 | VIN3, PRR3, TOE2, LFY, CDF1, MAF2-5 | [216,217] |
| Bn1 | B. napus | chrA02:114931.1575498 | Bna.FLC.A2, CO-like, RVE1 | [232,233] |
| Bn2 | B. napus | chrA02:1575449.4330821 | AP2-like, TOE2, PRR3 | [225,232,233] |
| Bn3 | B. napus | chrA02:5233136.8233310 | GA20ox, Bna.FT.A02 | [223,225,232,233,234,235] |
| Bn4 | B. napus | chrA02:8776742.9248051 | [5,222,234,236] | |
| Bn5 | B. napus | chrA03:5046910.6515058 | Bna.FRI.Xa, SPL13, CBF1, Bna.FLC.A03b | [66,222,234,236] |
| Bn6 | B. napus | chrA03:18872718.20131639 | AP2-like, FUL, TOC1 | [223,232,233,236] |
| Bn7 | B. napus | chrA04:257040.4734286 | AP2-like | [233,234,236] |
| Bn8 | B. napus | chrA04:7743947.10942653 | [233,234] | |
| Bn9 | B. napus | chrA04:11898475.13460703 | CO-like, ELF3 | [234,236] |
| Bn10 | B. napus | chrA06:23330530.23617143 | [232,236] | |
| Bn11 | B. napus | chrA07:14463578.18916565 | SPL15, AP2-like, GID1, AP1, Bna.FT.A07b | [232,233,234,236] |
| Bn12 | B. napus | chrA10:9835903.10695100 | PRR3, TOE2, AP2-like | [222,234] |
| Bn13 | B. napus | chrA10:13375104.15191366 | Bna.FLC.A10 | [66,222,224,233,234] |
| Bn14 | B. napus | chrC01:27417076.34893173 | FRI-like, VRN1 | [232,233] |
| Bn15 | B. napus | chrC02:6956919.13653054 | GA20ox, SPL | [222,232,234] |
| Bn16 | B. napus | chrC02:22287455.22560553 | [222,234] | |
| Bn17 | B. napus | chrC02:44366336.45788246 | FUL, MAF2, MAF3 | [225,232] |
| Bn18 | B. napus | chrC03:58161161.58296560 | [233,234] | |
| Bn19 | B. napus | chrC04:40003810.41181656 | [222,234] | |
| Bn20 | B. napus | chrC06:21784608.29654361 | ELF4, AP1 | [225,232,233,236] |
| Bn21 | B. napus | chrC07:26989258.31787256 | SEP4 | [225,232,234] |
| Bn22 | B. napus | chrC09:39312343.43429210 | SPL7, AP2-like, TFL2, RVE | [234,236] |
| Bn23 | B. napus | chrC09:45206288.47504024 | Bna.FLC.C09b, Ga20ox | [225,232] |
| Br1 | B. rapa | A01:81263.3282650 | AP2-like | [237,238] |
| Br2 | B. rapa | A02:1244721.4284193 | BrFLC2, AP2-like, CO-like, SPL7 | [49,92,237,238,239] |
| Br3 | B. rapa | A03:14357780.27239372 | CO-like, AP2-like, GA2ox, AGL24 | [237,238,239] |
| Br4 | B. rapa | A06:13769411.18840509 | LFY, GA20ox, CDF1, FLM, MAF4, VIN3-like, CO-like, | [238,240] |
| Br5 | B. rapa | A07:12545242.20240840 | AP2-like, SPL15, ELF4-like, AP1, BrFT2 | [49,238,240] |
| Br6 | B. rapa | A10:12936259.13856133 | BrFLC1 | [237,238,241] |
| Bo1 | B. oleracea | C02:900000.2900000 | GRF6, BoFLC4 | [101,242] |
| Bo2 | B. oleracea | C03:1800000.20000000 | BoFLC3, SOC1, BoFRIa, ELF4, GA20ox | [100,242,243] |
| Bo3 | B. oleracea | C04:10726862.16070000 | TOE2 | [100,243] |
| Bo4 | B. oleracea | C04:32446947.35540000 | [100,244] | |
| Bo5 | B. oleracea | C06:2396965.6360269 | TOE1, VIN3 | [242,243] |
| Bo6 | B. oleracea | C06:22550000.32446947 | [100,243] | |
| Ls1 | L. sativa | LG2:163353056.165477161 | CDF1, CO, FLC, PRR5, VRN1 | [226,227] |
| Ls2 | L. sativa | LG6:140450832.140481276 | [228] | |
| Ls3 | L. sativa | LG7:158780460.159063877 | [226,227] | |
| Ls4 | L. sativa | LG7:172306237.193636147 | [228] | |
| Ls5 | L. sativa | LG8:25874939.47456612 | PRR3, PRR5, PRR7, PRR9 | [228] |
| Ls6 | L. sativa | LG8:63537238.76202393 | FKF1 | [228] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leijten, W.; Koes, R.; Roobeek, I.; Frugis, G. Translating Flowering Time from Arabidopsis thaliana to Brassicaceae and Asteraceae Crop Species. Plants 2018, 7, 111. https://doi.org/10.3390/plants7040111
Leijten W, Koes R, Roobeek I, Frugis G. Translating Flowering Time from Arabidopsis thaliana to Brassicaceae and Asteraceae Crop Species. Plants. 2018; 7(4):111. https://doi.org/10.3390/plants7040111
Chicago/Turabian StyleLeijten, Willeke, Ronald Koes, Ilja Roobeek, and Giovanna Frugis. 2018. "Translating Flowering Time from Arabidopsis thaliana to Brassicaceae and Asteraceae Crop Species" Plants 7, no. 4: 111. https://doi.org/10.3390/plants7040111
APA StyleLeijten, W., Koes, R., Roobeek, I., & Frugis, G. (2018). Translating Flowering Time from Arabidopsis thaliana to Brassicaceae and Asteraceae Crop Species. Plants, 7(4), 111. https://doi.org/10.3390/plants7040111






