Expression of the Nicotiana benthamiana Retrozyme 1 (NbRZ1) Genomic Locus
Abstract
1. Introduction
2. Results
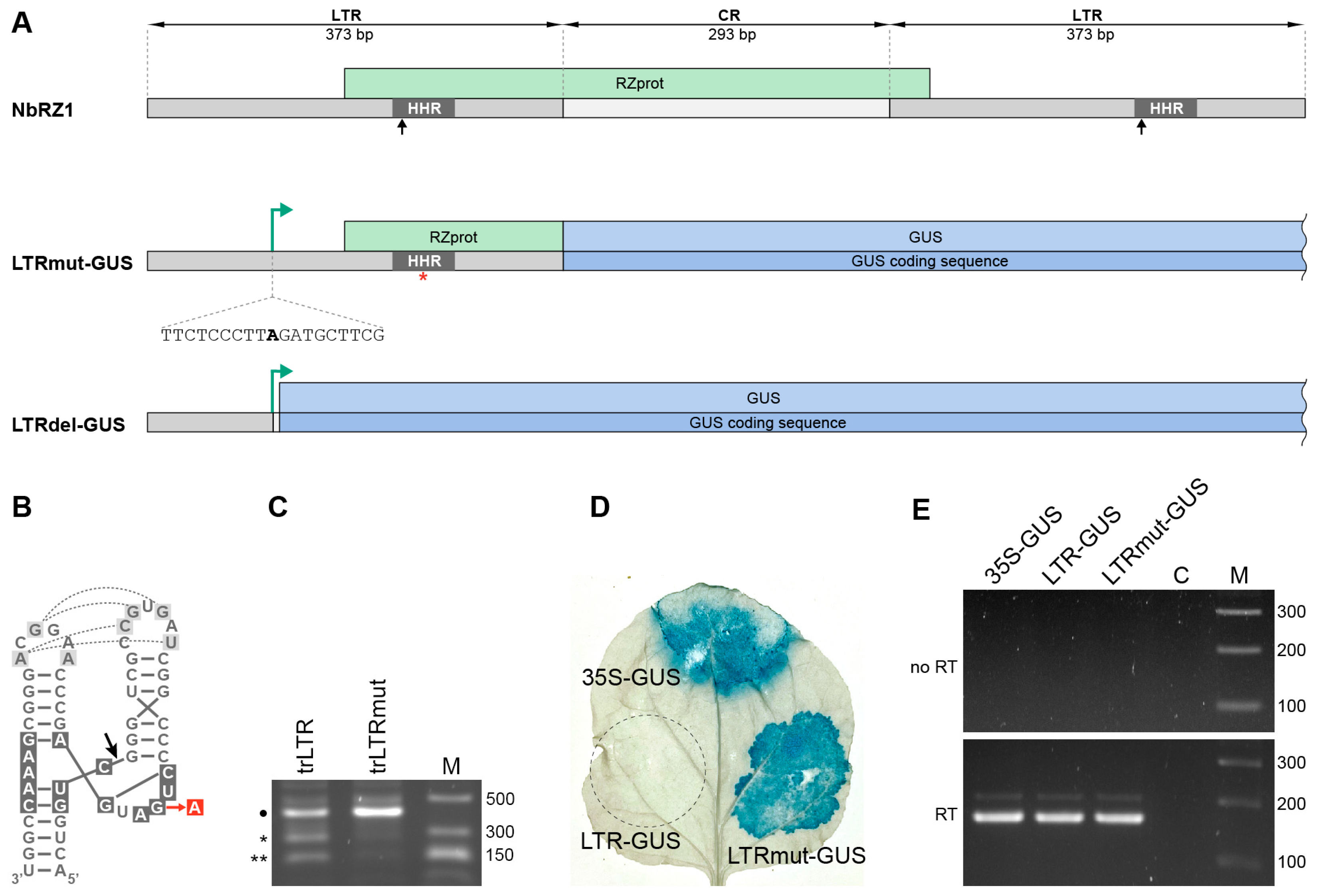
2.1. Mapping of the NbRZ1 Transcription Start Site
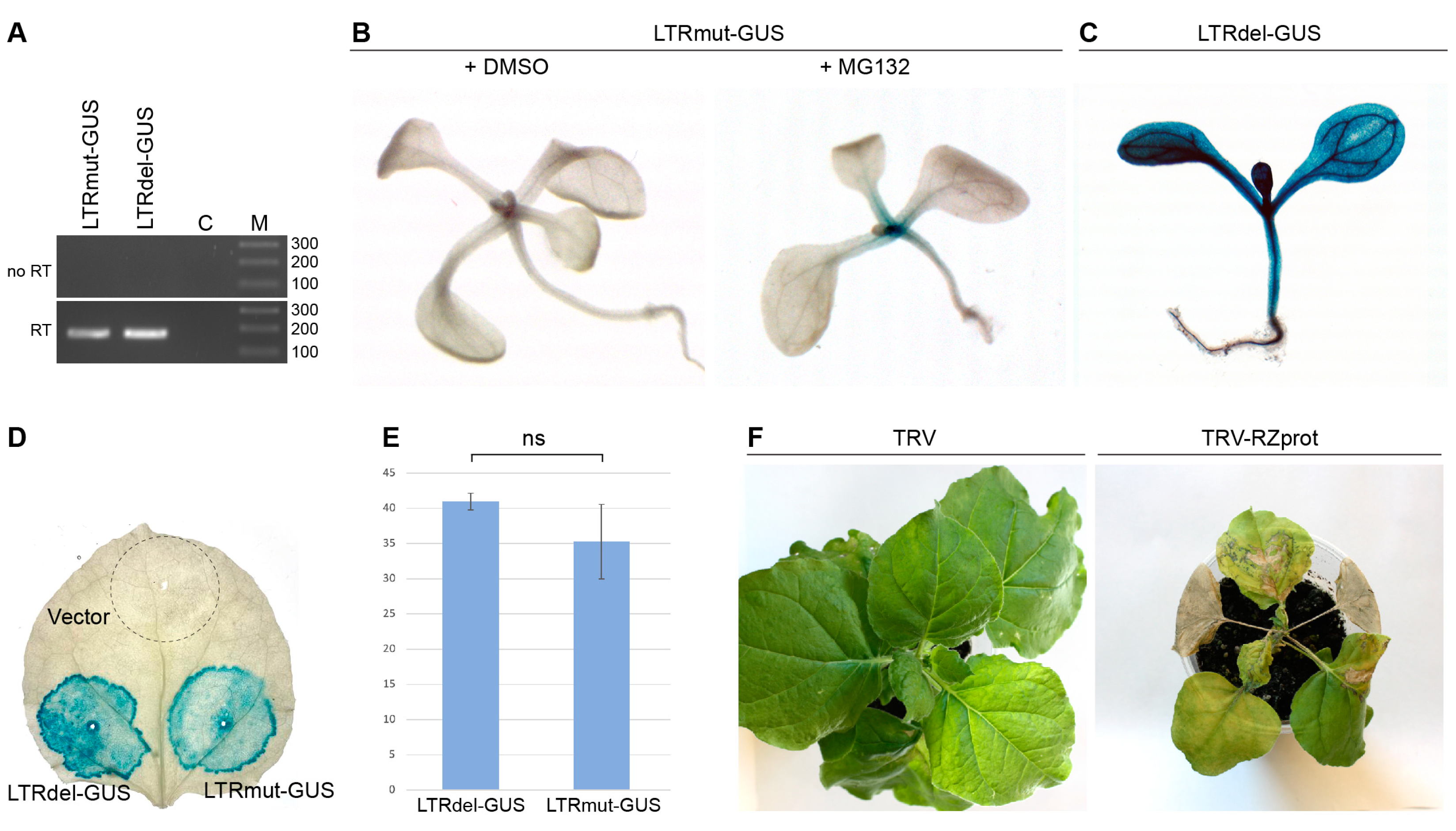
2.2. Analysis of Retrozyme Expression in Plants
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Plant Agroinfiltration
4.3. Molecular Cloning and Recombinant Constructs
4.4. 5′-RACE
4.5. GUS Staining
4.6. Transgenic Lines Generation
4.7. 26S Proteasome Inhibition
4.8. In Vitro Transcription and Ribozyme Processing
4.9. Total RNA Extraction, Reverse Transcription and Quantitative PCR
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, R.; Ma, Y.; Guo, T.; Li, G. Identification, Biogenesis, Function, and Mechanism of Action of Circular RNAs in Plants. Plant Commun. 2023, 4, 100430. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-Splicing Yields Circular RNA Molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A CircRNA from SEPALLATA3 Regulates Splicing of Its Cognate mRNA through R-Loop Formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Dinger, M.E. Endogenous MicroRNA Sponges: Evidence and Controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient MicroRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Pollock, A.; Bian, S.; Zhang, C.; Chen, Z.; Sun, T. Growth of the Developing Cerebral Cortex Is Controlled by MicroRNA-7 through the P53 Pathway. Cell Rep. 2014, 7, 1184–1196. [Google Scholar] [CrossRef]
- Liao, X.; Li, X.J.; Zheng, G.T.; Chang, F.R.; Fang, L.; Yu, H.; Huang, J.; Zhang, Y.F. Mitochondrion-Encoded Circular RNAs Are Widespread and Translatable in Plants. Plant Physiol. 2022, 189, 1482–1500. [Google Scholar] [CrossRef]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.K.; Wender, P.A.; Chang, H.Y. Engineering Circular RNA for Enhanced Protein Production. Nat. Biotechnol. 2022, 41, 262–272. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive Translation of Circular RNAs Driven by N6-Methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Diallo, L.H.; Tatin, F.; David, F.; Godet, A.C.; Zamora, A.; Prats, A.C.; Garmy-Susini, B.; Lacazette, E. How Are CircRNAs Translated by Non-Canonical Initiation Mechanisms? Biochimie 2019, 164, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, C.M.; Sehgal, R.; Tain, L.S.; Cheng, J.; Eßer, J.; Pahl, A.; Dieterich, C.; Grönke, S.; Partridge, L. An Insulin-Sensitive Circular RNA That Regulates Lifespan in Drosophila. Mol. Cell 2020, 79, 268-279.e5. [Google Scholar] [CrossRef] [PubMed]
- Cervera, A.; Urbina, D.; de la Peña, M. Retrozymes Are a Unique Family of Non-Autonomous Retrotransposons with Hammerhead Ribozymes That Propagate in Plants through Circular RNAs. Genome Biol. 2016, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Cervera, A.; De La Peña, M. Small CircRNAs with Self-Cleaving Ribozymes Are Highly Expressed in Diverse Metazoan Transcriptomes. Nucleic Acids Res. 2020, 48, 5054–5064. [Google Scholar] [CrossRef]
- Lezzhov, A.A.; Tolstyko, E.A.; Atabekova, A.K.; Chergintsev, D.A.; Morozov, S.Y.; Solovyev, A.G. In-Plant Persistence and Systemic Transport of Nicotiana Benthamiana Retrozyme RNA. Int. J. Mol. Sci. 2022, 23, 13890. [Google Scholar] [CrossRef]
- Lezzhov, A.A.; Atabekova, A.K.; Chergintsev, D.A.; Lazareva, E.A.; Solovyev, A.G.; Morozov, S.Y. Viroids and Retrozymes: Plant Circular RNAs Capable of Autonomous Replication. Plants 2024, 14, 61. [Google Scholar] [CrossRef]
- Cavrak, V.V.; Lettner, N.; Jamge, S.; Kosarewicz, A.; Bayer, L.M.; Mittelsten Scheid, O. How a Retrotransposon Exploits the Plant’s Heat Stress Response for Its Activation. PLoS Genet. 2014, 10, e1004115. [Google Scholar] [CrossRef]
- Zervudacki, J.; Yu, A.; Amesefe, D.; Wang, J.; Drouaud, J.; Navarro, L.; Deleris, A. Transcriptional Control and Exploitation of an Immune-Responsive Family of Plant Retrotransposons. EMBO J. 2018, 37, e98482. [Google Scholar] [CrossRef]
- Papolu, P.K.; Ramakrishnan, M.; Wei, Q.; Vinod, K.K.; Zou, L.H.; Yrjala, K.; Kalendar, R.; Zhou, M. Long Terminal Repeats (LTR) and Transcription Factors Regulate PHRE1 and PHRE2 Activity in Moso Bamboo under Heat Stress. BMC Plant Biol. 2021, 21, 585. [Google Scholar] [CrossRef]
- Ruffner, D.E.; Uhlenbeck, O.C.; Stormo, G.D. Sequence Requirements of the Hammerhead RNA Self-Cleavage Reaction. Biochemistry 1990, 29, 10695–10702. [Google Scholar] [CrossRef]
- Maruri-López, I.; Rodríguez-Kessler, M.; Rodríguez-Hernández, A.A.; Becerra-Flora, A.; Olivares-Grajales, J.E.; Jiménez-Bremont, J.F. A Maize Spermine Synthase 1 PEST Sequence Fused to the GUS Reporter Protein Facilitates Proteolytic Degradation. Plant Physiol. Biochem. 2014, 78, 80–87. [Google Scholar] [CrossRef]
- Gray, W.M.; Kepinski, S.; Rouse, D.; Leyser, O.; Estelle, M. Auxin Regulates SCF(TIR1)-Dependent Degradation of AUX/IAA Proteins. Nature 2001, 414, 271–276. [Google Scholar] [CrossRef]
- Deng, F.; Guo, T.; Lefebvre, M.; Scaglione, S.; Antico, C.J.; Jing, T.; Yang, X.; Shan, W.; Ramonell, K.M. Expression and Regulation of ATL9, an E3 Ubiquitin Ligase Involved in Plant Defense. PLoS ONE 2017, 12, e0188458. [Google Scholar] [CrossRef]
- Lee, D.H.; Goldberg, A.L. Proteasome Inhibitors: Valuable New Tools for Cell Biologists. Trends Cell Biol. 1998, 8, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Mantis, J.; Tague, B.W. Comparing the Utility of β-Glucuronidase and Green Fluorescent Protein for Detection of Weak Promoter Activity in Arabidopsis Thaliana. Plant Mol. Biol. Report. 2000, 18, 319–330. [Google Scholar] [CrossRef]
- Abrahamian, P.; Hammond, R.W.; Hammond, J. Plant Virus-Derived Vectors: Applications in Agricultural and Medical Biotechnology. Annu. Rev. Virol. 2020, 7, 513–535. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C.; Jiang, N.; Wessler, S.R. Plant Transposable Elements: Where Genetics Meets Genomics. Nat. Rev. Genet. 2002, 3, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Hurst, T.P.; Magiorkinis, G. Epigenetic Control of Human Endogenous Retrovirus Expression: Focus on Regulation of Long-Terminal Repeats (LTRs). Viruses 2017, 9, 130. [Google Scholar] [CrossRef]
- Sabot, F.; Schulman, A.H. Parasitism and the Retrotransposon Life Cycle in Plants: A Hitchhiker’s Guide to the Genome. Heredity 2006, 97, 381–388. [Google Scholar] [CrossRef]
- Ichino, L.; Boone, B.A.; Strauskulage, L.; Harris, C.J.; Kaur, G.; Gladstone, M.A.; Tan, M.; Feng, S.; Jami-Alahmadi, Y.; Duttke, S.H.; et al. MBD5 and MBD6 Couple DNA Methylation to Gene Silencing through the J-Domain Protein SILENZIO. Science 2021, 372, 1434–1439. [Google Scholar] [CrossRef]
- Wang, S.; Wang, M.; Ichino, L.; Boone, B.A.; Zhong, Z.; Papareddy, R.K.; Lin, E.K.; Yun, J.; Feng, S.; Jacobsen, S.E. MBD2 Couples DNA Methylation to Transposable Element Silencing during Male Gametogenesis. Nat. Plants 2024, 10, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Yang, Y.; Chen, C.; Wang, Z. Pervasive Translation of Circular RNAs Driven by Short IRES-like Elements. Nat. Commun. 2022, 13, 3751. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-Methyladenosine Alters RNA Structure to Regulate Binding of a Low-Complexity Protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef]
- Hernández, C.; Flores, R. Plus and Minus RNAs of Peach Latent Mosaic Viroid Self-Cleave in Vitro via Hammerhead Structures. Proc. Natl. Acad. Sci. USA 1992, 89, 3711–3715. [Google Scholar] [CrossRef]
- Flores, R.; Daròs, J.A.; Hernández, C. Avsunviroidae Family: Viroids Containing Hammerhead Ribozymes. Adv. Virus Res. 2000, 55, 271–323. [Google Scholar] [CrossRef]
- López-Galiano, M.J.; Chiba, S.; Forgia, M.; Navarro, B.; Cervera, A.; Babaian, A.; Di Serio, F.; Turina, M.; Peña, M. de la Self-Cleaving Ribozymes Conserved in RNA Viruses Unveil a New Role in Protein Translation. bioRxiv 2024. [Google Scholar] [CrossRef]
- Carbonell, A.; De la Peña, M.; Flores, R.; Gago, S. Effects of the Trinucleotide Preceding the Self-Cleavage Site on Eggplant Latent Viroid Hammerheads: Differences in Co- and Post-Transcriptional Self-Cleavage May Explain the Lack of Trinucleotide AUC in Most Natural Hammerheads. Nucleic Acids Res. 2006, 34, 5613. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, A.G.; Minina, E.A.; Makarova, S.S.; Erokhina, T.N.; Makarov, V.V.; Kaplan, I.B.; Kopertekh, L.; Schiemann, J.; Richert-Pöggeler, K.R.; Morozov, S.Y. Subcellular Localization and Self-Interaction of Plant-Specific Nt-4/1 Protein. Biochimie 2013, 95, 1360–1370. [Google Scholar] [CrossRef]
- MacFarlane, S.A.; Popovich, A.H. Efficient Expression of Foreign Proteins in Roots from Tobravirus Vectors. Virology 2000, 267, 29–35. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Marathe, R.; Dinesh-Kumar, S.P. Tobacco Rar1, EDS1 and NPR1/NIM1 like Genes Are Required for N-Mediated Resistance to Tobacco Mosaic Virus. Plant J. 2002, 30, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Matz, M.V.; Alieva, N.O.; Chenchik, A.; Lukyanov, S. Amplification of cDNA Ends Using PCR Suppression Effect and Step-out PCR. Methods Mol. Biol. 2003, 221, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-Mediated Transformation of Arabidopsis Thaliana Using the Floral Dip Method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lezzhov, A.A.; Atabekova, A.K.; Chergintsev, D.A.; Solovyev, A.G.; Morozov, S.Y. Expression of the Nicotiana benthamiana Retrozyme 1 (NbRZ1) Genomic Locus. Plants 2025, 14, 1205. https://doi.org/10.3390/plants14081205
Lezzhov AA, Atabekova AK, Chergintsev DA, Solovyev AG, Morozov SY. Expression of the Nicotiana benthamiana Retrozyme 1 (NbRZ1) Genomic Locus. Plants. 2025; 14(8):1205. https://doi.org/10.3390/plants14081205
Chicago/Turabian StyleLezzhov, Alexander A., Anastasia K. Atabekova, Denis A. Chergintsev, Andrey G. Solovyev, and Sergey Y. Morozov. 2025. "Expression of the Nicotiana benthamiana Retrozyme 1 (NbRZ1) Genomic Locus" Plants 14, no. 8: 1205. https://doi.org/10.3390/plants14081205
APA StyleLezzhov, A. A., Atabekova, A. K., Chergintsev, D. A., Solovyev, A. G., & Morozov, S. Y. (2025). Expression of the Nicotiana benthamiana Retrozyme 1 (NbRZ1) Genomic Locus. Plants, 14(8), 1205. https://doi.org/10.3390/plants14081205






