Characterization and Functional Analysis of RhHsfA7, a Heat Stress Transcription Factor in Roses (Rosa hybrid ‘Samantha’)
Abstract
1. Introduction
2. Results
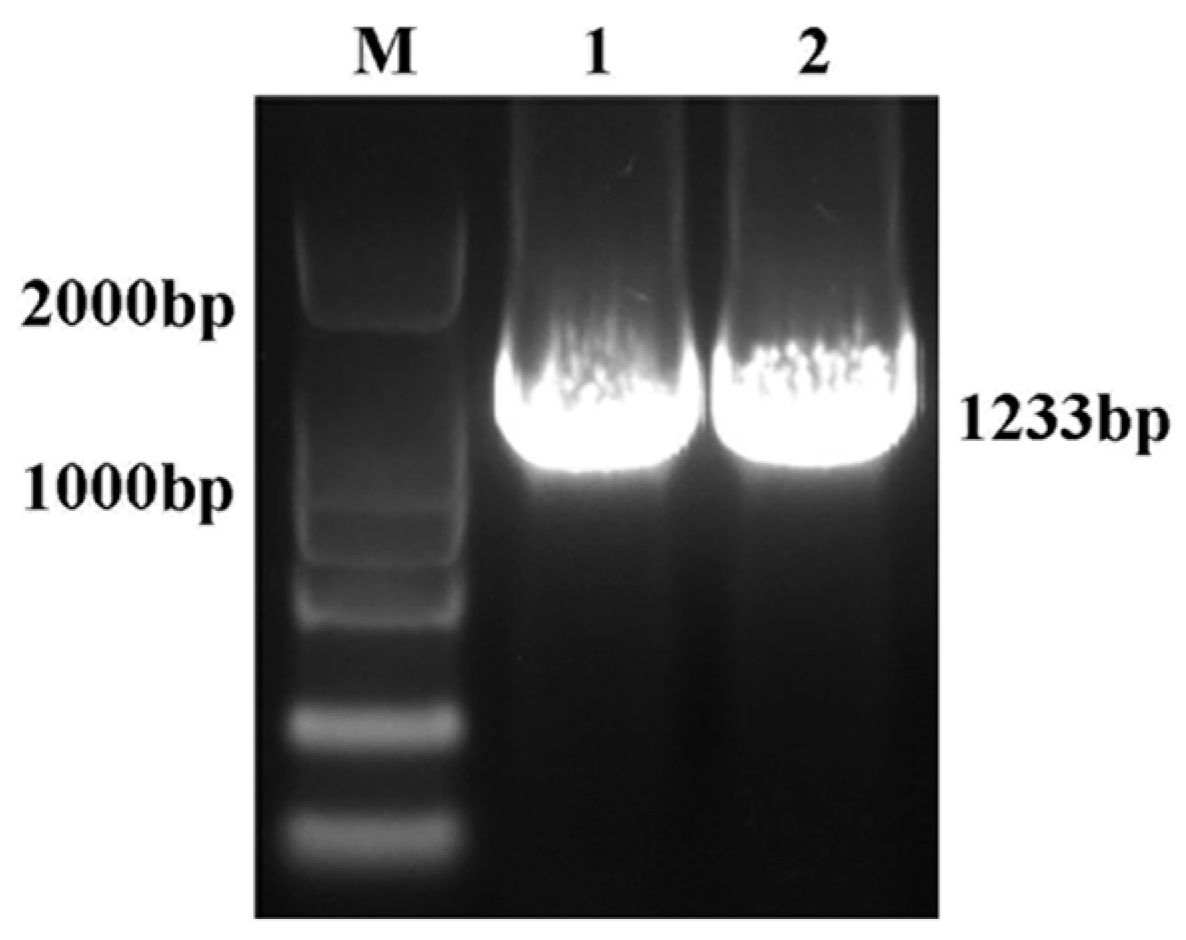
2.1. Cloning and Bioinformatics Analyses of RhHsfA7
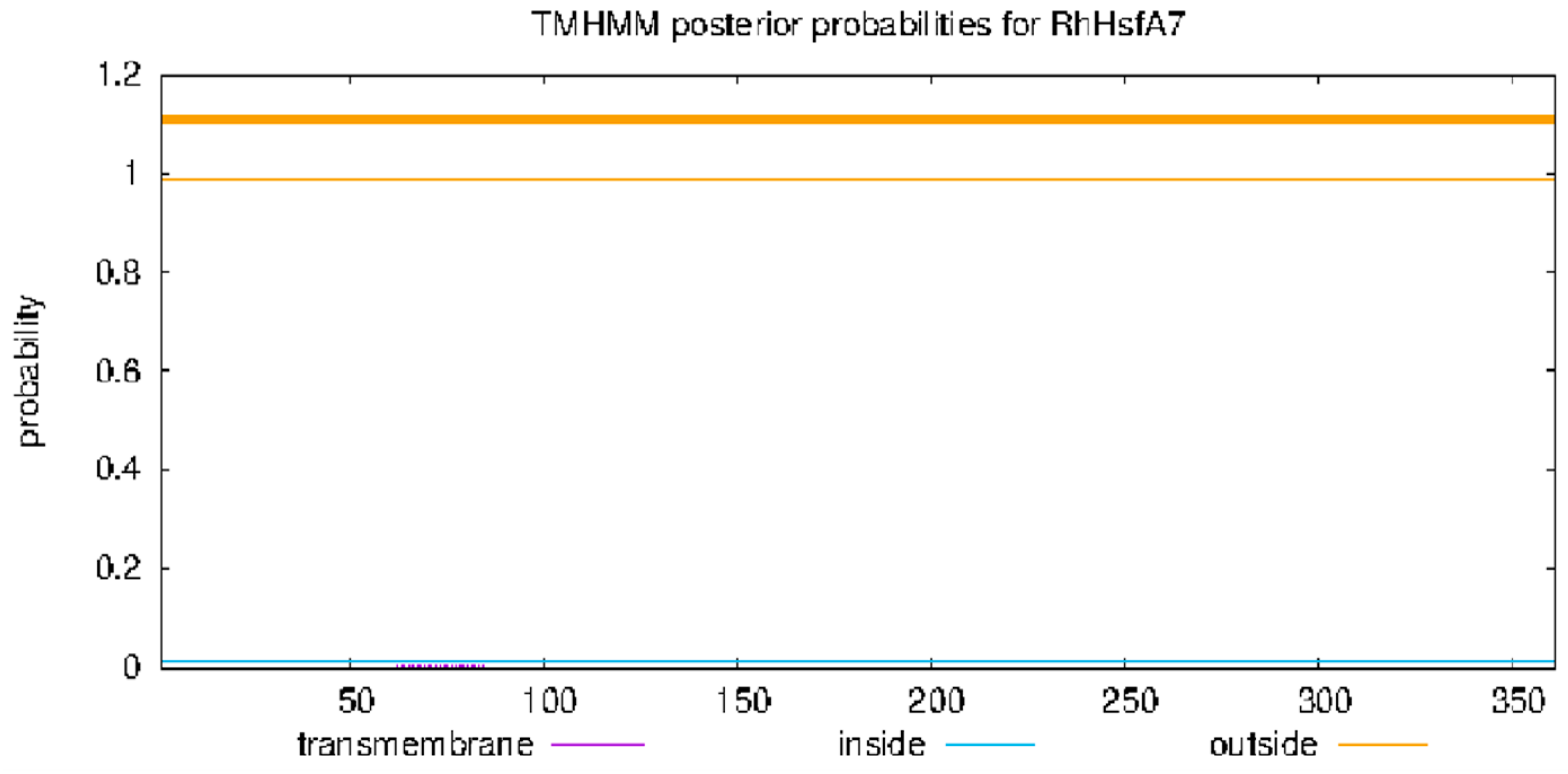
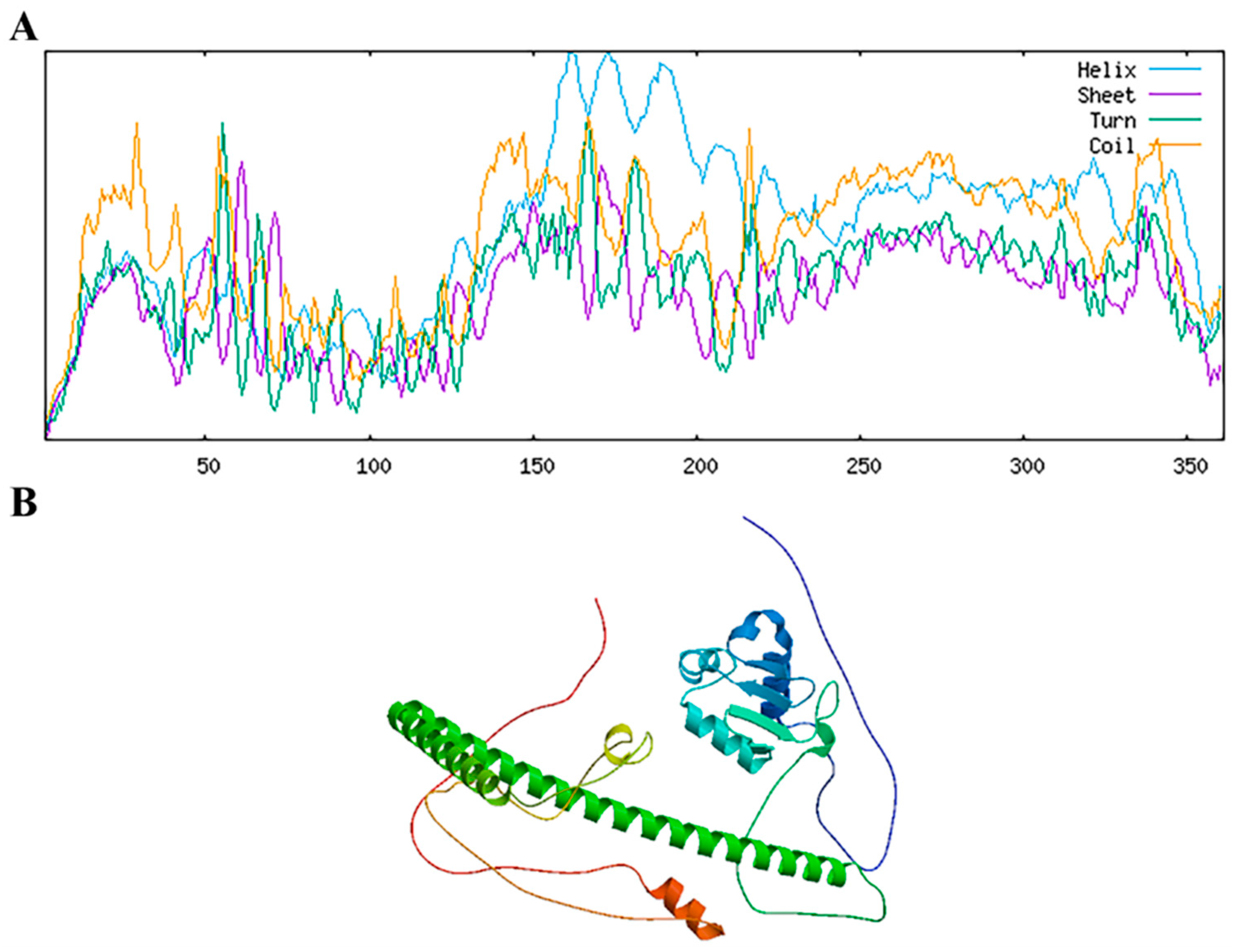
2.2. Structural Prediction of RhHsfA7 Protein
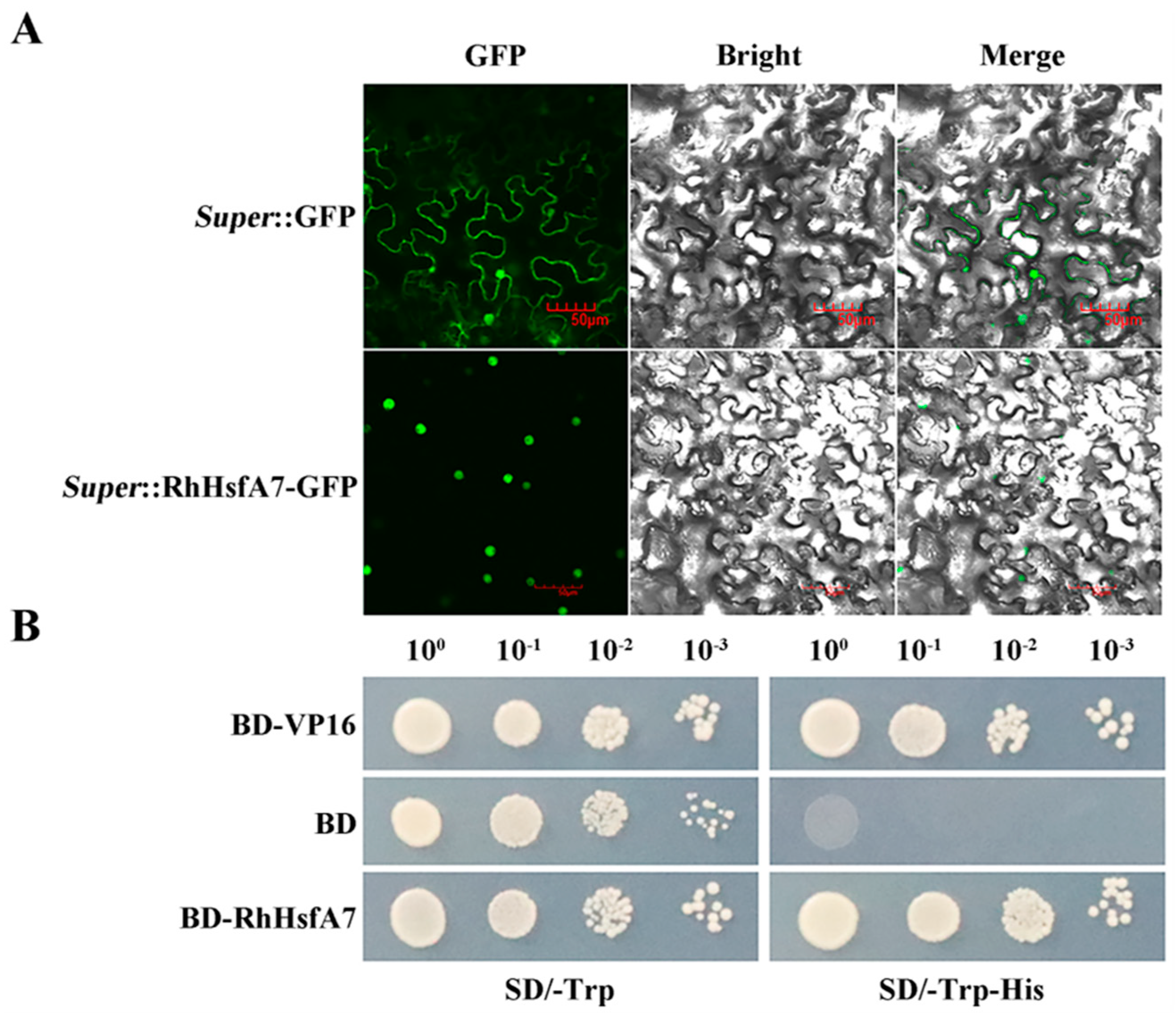
2.3. Subcellular Localization and Transactivation Activity of RhHsfA7 Protein
2.4. Expression Analyses of RhHsfA7 in R. hybrida
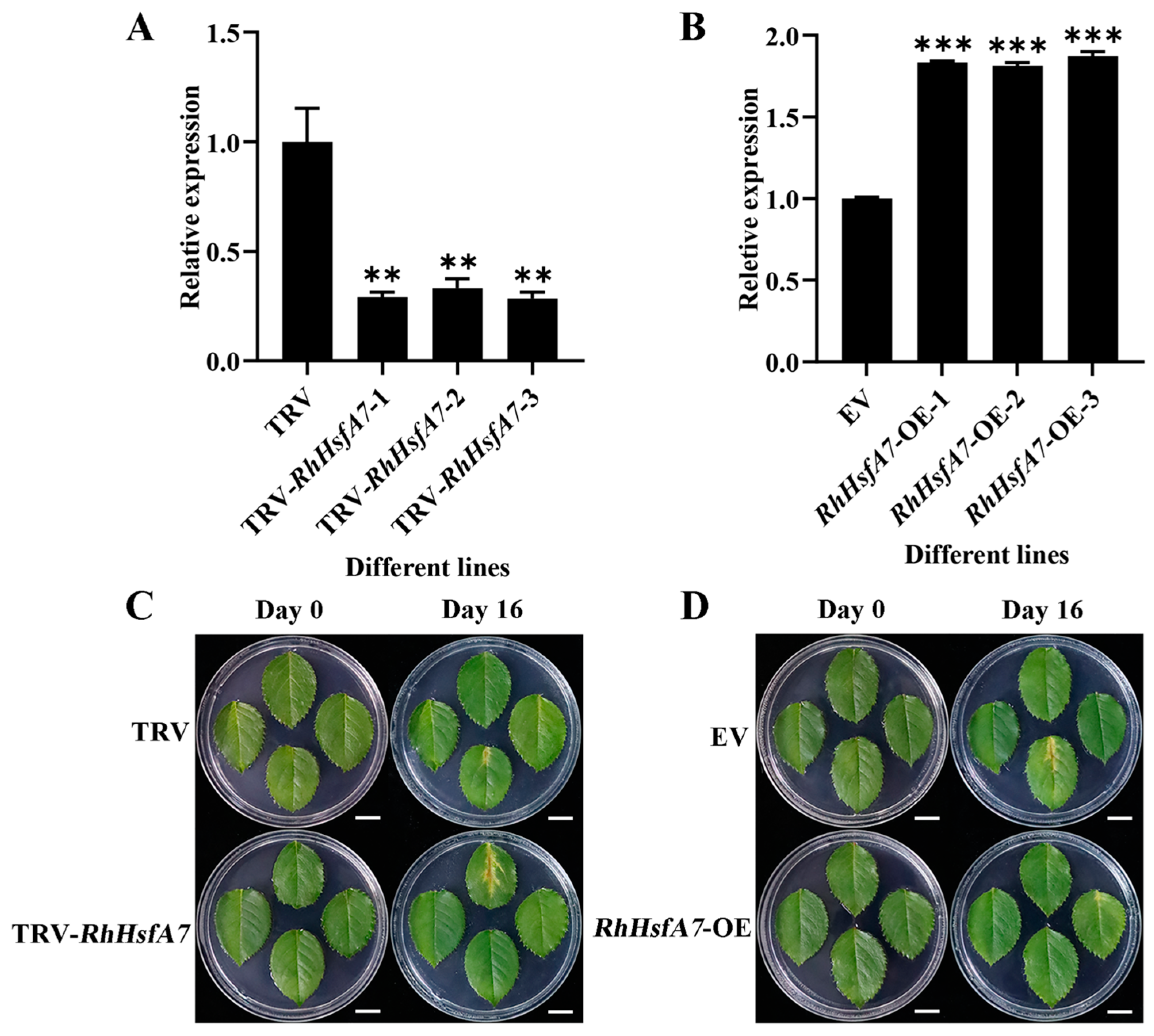
2.5. RhHsfA7 Silencing Decreased Heat Tolerance in Rose
2.6. Overexpressing RhHsfA7 Improved Heat Tolerance in Rose
3. Discussion
4. Materials and Methods
4.1. Plant Material and Treatments
4.2. Cloning and Sequence Analysis of RhHsfA7
4.3. Subcellular Localization of RhHsfA7
4.4. Transcription Activation Assay
4.5. Quantitative Real-Time PCR Analyses
4.6. VIGS and Transient Overexpression of RhHsfA7 in Rose Leaves
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Raymond, O.; Gouzy, J.; Just, J.; Badouin, H.; Verdenaud, M.; Lemainque, A.; Vergne, P.; Moja, S.; Choisne, N.; Pont, C.; et al. The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 2018, 50, 772–777. [Google Scholar] [CrossRef]
- Chen, C.; Hussain, N.; Wang, Y.; Li, M.; Liu, L.; Qin, M.; Ma, N.; Gao, J.; Sun, X. An Ethylene-inhibited NF-YC Transcription Factor RhNF-YC9 Regulates Petal Expansion in Rose. Hortic. Plant J. 2020, 6, 419–427. [Google Scholar] [CrossRef]
- Qi, W.; Zhang, C.; Wang, W.; Cao, Z.; Li, S.; Li, H.; Zhu, W.; Huang, Y.; Bao, M.; He, Y.; et al. Comparative transcriptome analysis of different heat stress responses between self-root grafting line and heterogeneous grafting line in rose. Hortic. Plant J. 2021, 7, 243–255. [Google Scholar] [CrossRef]
- Zaccai, M.; Ackerman, R.; Genis, O.; Riov, J.; Zik, M. The bent peduncle phenomenon in roses is a developmental process involving auxin. Plant Sci. 2009, 176, 736–743. [Google Scholar] [CrossRef]
- Li, Z.Q.; Xing, W.; Luo, P.; Zhang, F.J.; Jin, X.L.; Zhang, M.H. Comparative transcriptome analysis of Rosa chinensis ‘Slater’s crimson China’ provides insights into the crucial factors and signaling pathways in heat stress response. Plant Physiol. Biochem. 2019, 142, 312–331. [Google Scholar] [CrossRef] [PubMed]
- Qian, R.; Hu, Q.; Ma, X.; Zhang, X.; Ye, Y.; Liu, H.; Gao, H.; Zheng, J. Comparative transcriptome analysis of heat stress responses of Clematis lanuginosa and Clematis crassifolia. BMC Plant Biol. 2022, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Changwal, C.; Thapa, B.; Tanpure, R.S.; Hada, A.; Singh, P.K.; Ghuge, S.A. Transcription factors: A tool box for countering the effect of abiotic stresses. In Stress Tolerance in Horticultural Crops; Elsevier: Amsterdam, The Netherlands, 2021; pp. 169–192. [Google Scholar]
- Yan, Y.; Zhao, J.; Lin, S.; Li, M.; Liu, J.; Raymond, O.; Vergne, P.; Kong, W.; Wu, Q.; Zhang, X.; et al. Light-mediated anthocyanin biosynthesis in rose petals involves a balanced regulatory module comprising transcription factors RhHY5, RhMYB114a, and RhMYB3b. J. Exp. Bot. 2023, 74, 5783–5804. [Google Scholar] [PubMed]
- Fan, K.; Mao, Z.; Ye, F.; Pan, X.; Li, Z.; Lin, W.; Zhang, Y.; Huang, J.; Lin, W. Genome-wide identification and molecular evolution analysis of the heat shock transcription factor (HSF) gene family in four diploid and two allopolyploid Gossypium species. Genomics 2021, 113, 3112–3127. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.; Ma, X.; Luo, D.; Gong, Z.; Lu, M. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jiang, J.; Han, X.; Zhang, Y.; Zhuo, R. Identification, expression analysis of the hsf family, and characterization of class A4 in Sedum Alfredii hance under cadmium stress. Int. J. Mol. Sci. 2018, 19, 1216. [Google Scholar] [CrossRef]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-C.; Niu, C.-Y.; Yang, C.-R.; Jinn, T.-L. The heat-stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 2016, 172, 1182–1199. [Google Scholar] [CrossRef]
- Larkindale, J.; Hall, J.D.; Knight, M.R.; Vierling, E. Heat Stress Phenotypes of Arabidopsis Mutants Implicate Multiple Signaling Pathways in the Acquisition of Thermotolerance. Plant Physiol. 2005, 138, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Zang, D.; Wang, J.; Zhang, X.; Liu, Z.; Wang, Y. Arabidopsis heat shock transcription factor HSFA7b positively mediates salt stress tolerance by binding to an E-box-like motif to regulate gene expression. J. Exp. Bot. 2019, 70, 5355–5374. [Google Scholar] [CrossRef]
- Mesihovic, A.; Ullrich, S.; Rosenkranz, R.R.; Gebhardt, P.; Bublak, D.; Eich, H.; Weber, D.; Berberich, T.; Scharf, K.-D.; Schleiff, E.; et al. HsfA7 coordinates the transition from mild to strong heat stress response by controlling the activity of the master regulator HsfA1a in tomato. Cell Rep. 2022, 38, 110224. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, R.; Zhou, J.; Du, J.; Li, X.; Zhang, Y.; Chen, Q.; Wang, Y.; Lin, Y.; Zhang, Y.; et al. Comprehensive analysis of HSF genes from celery (Apium graveolens L.) and functional characterization of AgHSFa6-1 in response to heat stress. Front. Plant Sci. 2023, 14, 1132307. [Google Scholar] [CrossRef] [PubMed]
- Mou, S.; He, W.; Jiang, H.; Meng, Q.; Zhang, T.; Liu, Z.; Qiu, A.; He, S. Transcription factor CaHDZ15 promotes pepper basal thermotolerance by activating HEAT SHOCK FACTORA6a. Plant Physiol. 2024, 195, 812–831. [Google Scholar] [CrossRef]
- Xin, H.; Zhang, H.; Chen, L.; Li, X.; Lian, Q.; Yuan, X.; Hu, X.; Cao, L.; He, X.; Yi, M. Cloning and characterization of HsfA2 from Lily (Lilium longiflorum). Plant Cell Rep. 2010, 29, 875–885. [Google Scholar] [CrossRef]
- Wang, Y.; Song, C.; Tong, S.; Guo, Y.; Yang, X.; Li, C.; Shao, Y.; Yi, M.; He, J. The diversity in interaction between HsfA2 and ACTIN leads to differences in heat stress responses among different lily varieties. Ornam. Plant Res. 2024, 4, e011. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, W.; Ni, D.; Wang, M.; Guo, G. Genome-wide characterization of tea plant (Camellia sinensis) Hsf transcription factor family and role of CsHsfA2 in heat tolerance. BMC Plant Biol. 2020, 20, 244. [Google Scholar] [CrossRef]
- Ding, M.; Xing, W.; Li, Z.; Jin, X.; Yu, Q.; Sun, J. The class B heat shock factor RcHsf17 from Rosa chinensis enhances basal thermotolerance in Rosa rugosa. Environ. Exp. Bot. 2024, 225, 105832. [Google Scholar] [CrossRef]
- Kang, Y.; Sun, P.; Yang, Y.; Li, M.; Wang, H.; Sun, X.; Jin, W. Genome-Wide Analysis of the Hsf Gene Family in Rosa chinensis and RcHsf17 Function in Thermotolerance. Int. J. Mol. Sci. 2024, 26, 287. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sun, Y.; Hu, Z.; Dong, F.; Zhu, J.; Cao, M.; Wang, C. Cloning and expression analysis of RhHsf24 gene in Rose (Rosa hybrida). Sci. Rep. 2025, 15, 8182. [Google Scholar] [CrossRef]
- Tian, J.; Pei, H.; Zhang, S.; Chen, J.; Chen, W.; Yang, R.; Meng, Y.; You, J.; Gao, J.; Ma, N. TRV–GFP: A modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. J. Exp. Bot. 2014, 65, 311–322. [Google Scholar] [CrossRef]
- Huang, H.; Chang, K.; Wu, S. High irradiance sensitive phenotype of Arabidopsis hit2/xpo1a mutant is caused in part by nuclear confinement of AtHsfA4a. Biol. Plant. 2018, 62, 69–79. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Röth, S.; Schleiff, E.; Scharf, K. Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ. 2015, 38, 1881–1895. [Google Scholar] [CrossRef] [PubMed]
- Maheswari, U.; Jabbari, K.; Petit, J.-L.; Porcel, B.M.; E Allen, A.; Cadoret, J.-P.; De Martino, A.; Heijde, M.; Kaas, R.; La Roche, J.; et al. Digital expression profiling of novel diatom transcripts provides insight into their biological functions. Genome Biol. 2010, 11, R85. [Google Scholar] [CrossRef]
- Qian, G.; Meng, X.; Wang, S.; Mi, Y.; Qin, Z.; Liu, T.; Zhang, Y.; Wan, H.; Chen, W.; Sun, W.; et al. Genome-wide identification of HSF gene family and their expression analysis in vegetative tissue of young seedlings of hemp under different light treatments. Ind. Crops Prod. 2023, 204, 117375. [Google Scholar] [CrossRef]
- Xu, Y.; Jin, Y.; He, D.; Di, H.; Liang, Y.; Xu, Y. A Genome-Wide Analysis and Expression Profile of Heat Shock Transcription Factor (Hsf) Gene Family in Rhododendron simsii. Plants 2023, 12, 3917. [Google Scholar] [CrossRef]
- Hu, Y.; Han, Y.-T.; Wei, W.; Li, Y.-J.; Zhang, K.; Gao, Y.-R.; Zhao, F.-L.; Feng, J.-Y. Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 2015, 6, 736. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, X.; Wang, D.; Li, L.; Zhou, W.; Zhang, Q.; Wang, X.; Ye, S.; Wang, Z. Genome-wide identification, classification and expression profile analysis of the HSF gene family in Hypericum perforatum. PeerJ 2021, 9, e11345. [Google Scholar] [CrossRef]
- Rao, S.; Das, J.R.; Balyan, S.; Verma, R.; Mathur, S. Cultivar-biased regulation of HSFA7 and HSFB4a govern high-temperature tolerance in tomato. Planta 2022, 255, 31. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/ NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Zhao, Q.; Jing, W.; Fu, X.; Yang, R.; Zhu, C.; Zhao, J.; Choisy, P.; Xu, T.; Ma, N.; Zhao, L.; et al. TSPO-induced degradation of the ethylene receptor RhETR3 promotes salt tolerance in rose (Rosa hybrida). Hortic. Res. 2024, 11, uhae040. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Li, S.; Wu, X.; Zhu, J.; Dong, F.; Pei, Z.; Li, Z.; Zhao, S.; Wang, C. Characterization and Functional Analysis of RhHsfA7, a Heat Stress Transcription Factor in Roses (Rosa hybrid ‘Samantha’). Plants 2025, 14, 1155. https://doi.org/10.3390/plants14081155
Sun Y, Li S, Wu X, Zhu J, Dong F, Pei Z, Li Z, Zhao S, Wang C. Characterization and Functional Analysis of RhHsfA7, a Heat Stress Transcription Factor in Roses (Rosa hybrid ‘Samantha’). Plants. 2025; 14(8):1155. https://doi.org/10.3390/plants14081155
Chicago/Turabian StyleSun, Yaqi, Sudan Li, Xiang Wu, Jiao Zhu, Fei Dong, Zhaoshun Pei, Zhenguo Li, Shanxing Zhao, and Chengpeng Wang. 2025. "Characterization and Functional Analysis of RhHsfA7, a Heat Stress Transcription Factor in Roses (Rosa hybrid ‘Samantha’)" Plants 14, no. 8: 1155. https://doi.org/10.3390/plants14081155
APA StyleSun, Y., Li, S., Wu, X., Zhu, J., Dong, F., Pei, Z., Li, Z., Zhao, S., & Wang, C. (2025). Characterization and Functional Analysis of RhHsfA7, a Heat Stress Transcription Factor in Roses (Rosa hybrid ‘Samantha’). Plants, 14(8), 1155. https://doi.org/10.3390/plants14081155




