Physiological, Cytological and Transcriptome Analysis of a Yellow–Green Leaf Mutant in Magnolia sinostellata
Abstract
1. Introduction
2. Results
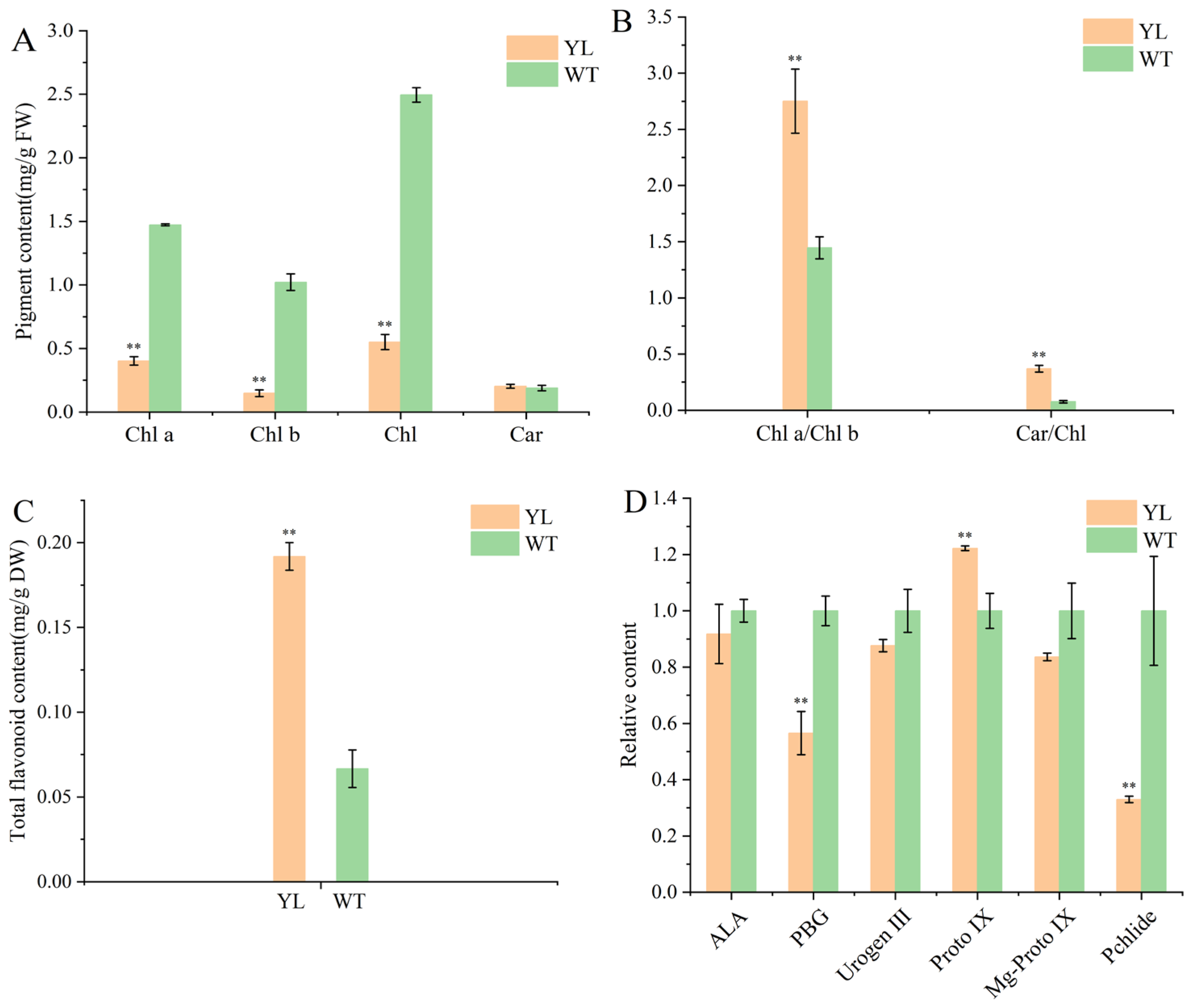
2.1. Pigment Contents of Leaves
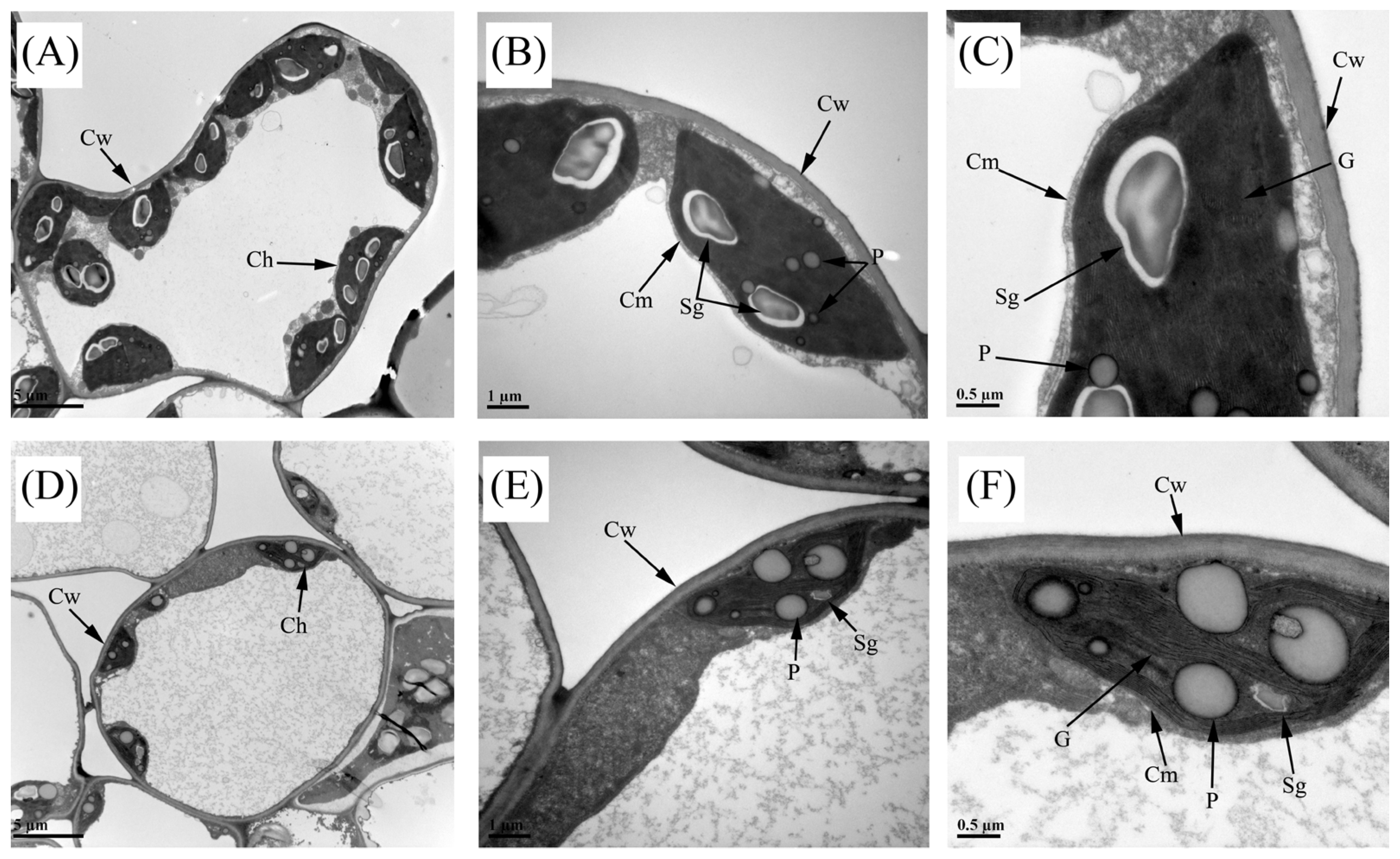
2.2. Cytological Observations
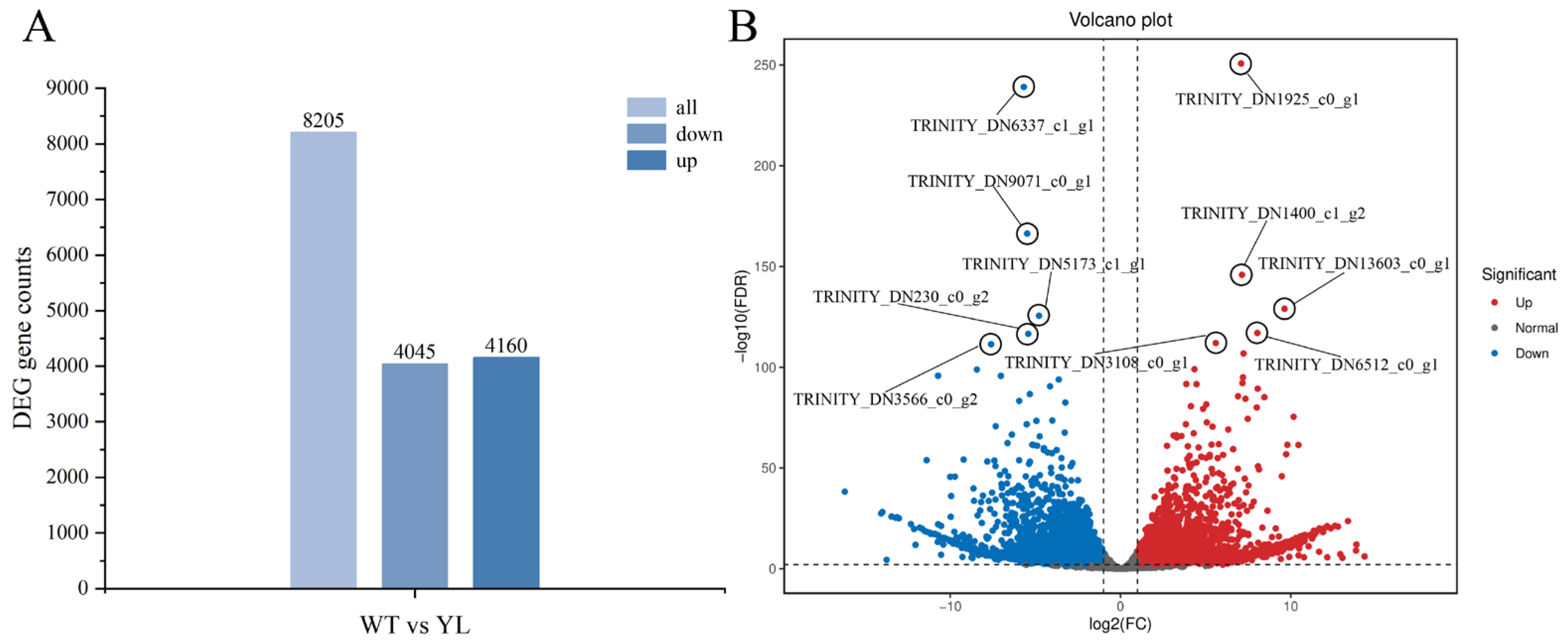
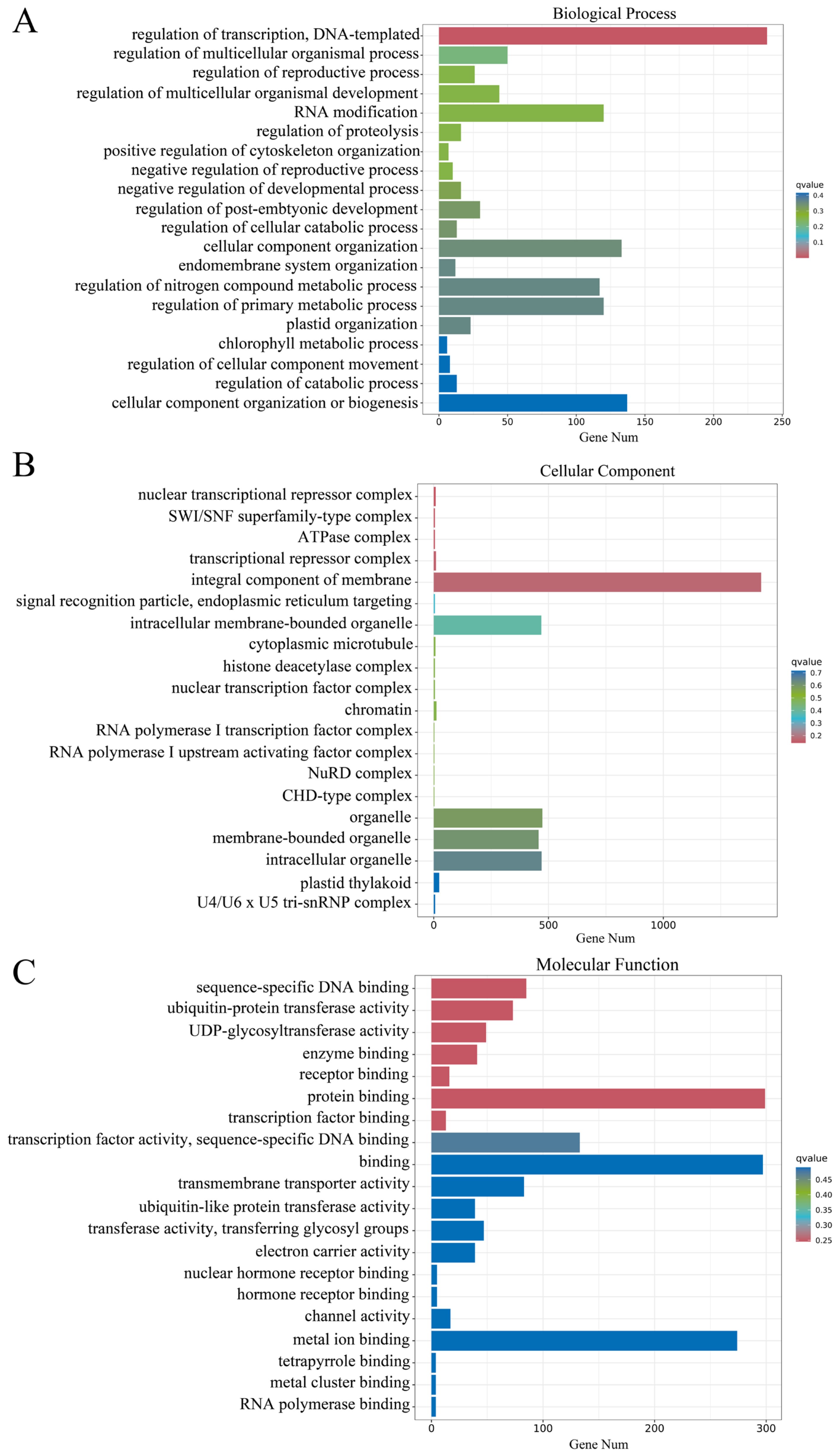
2.3. Transcriptomics Analysis
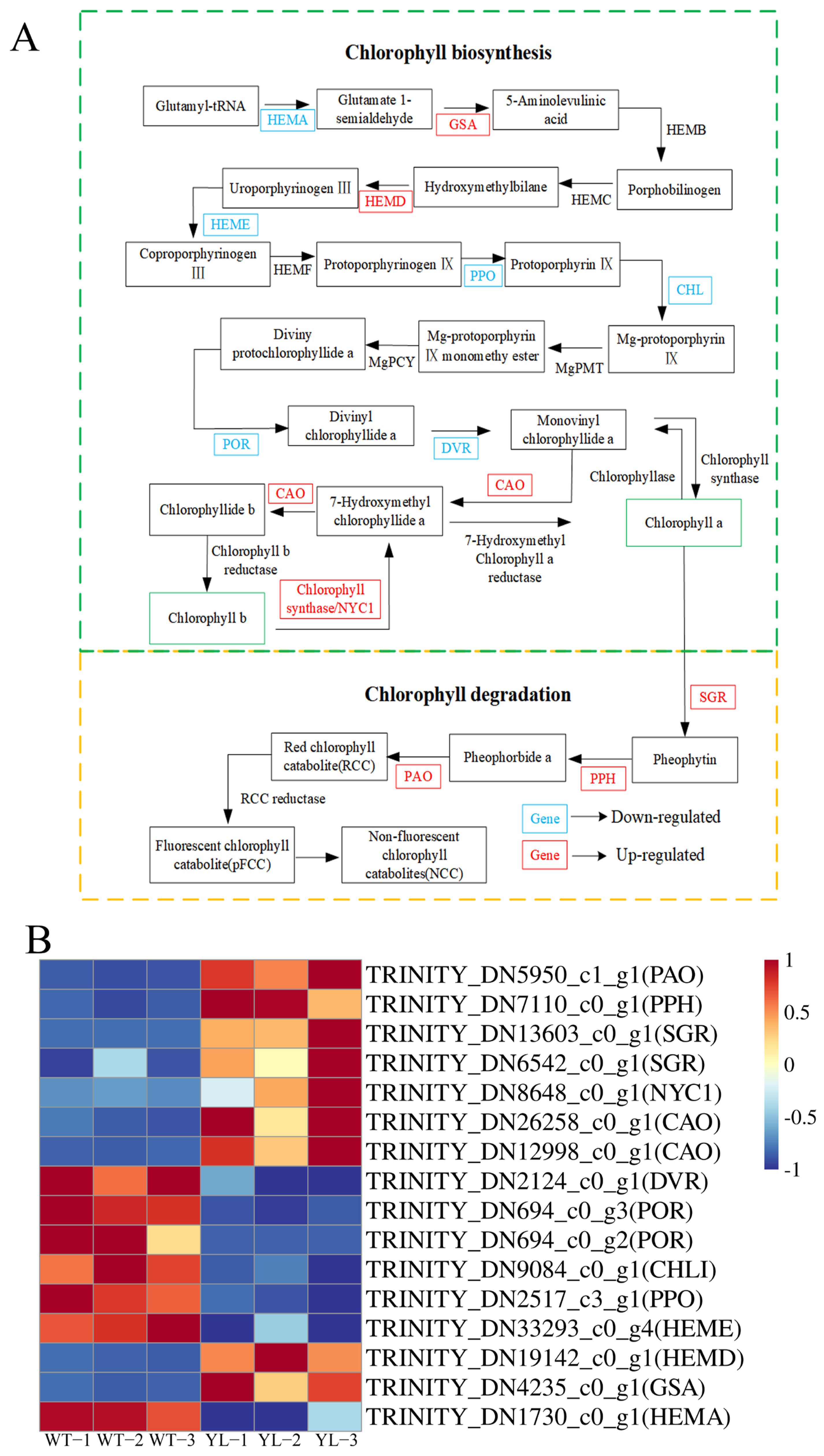
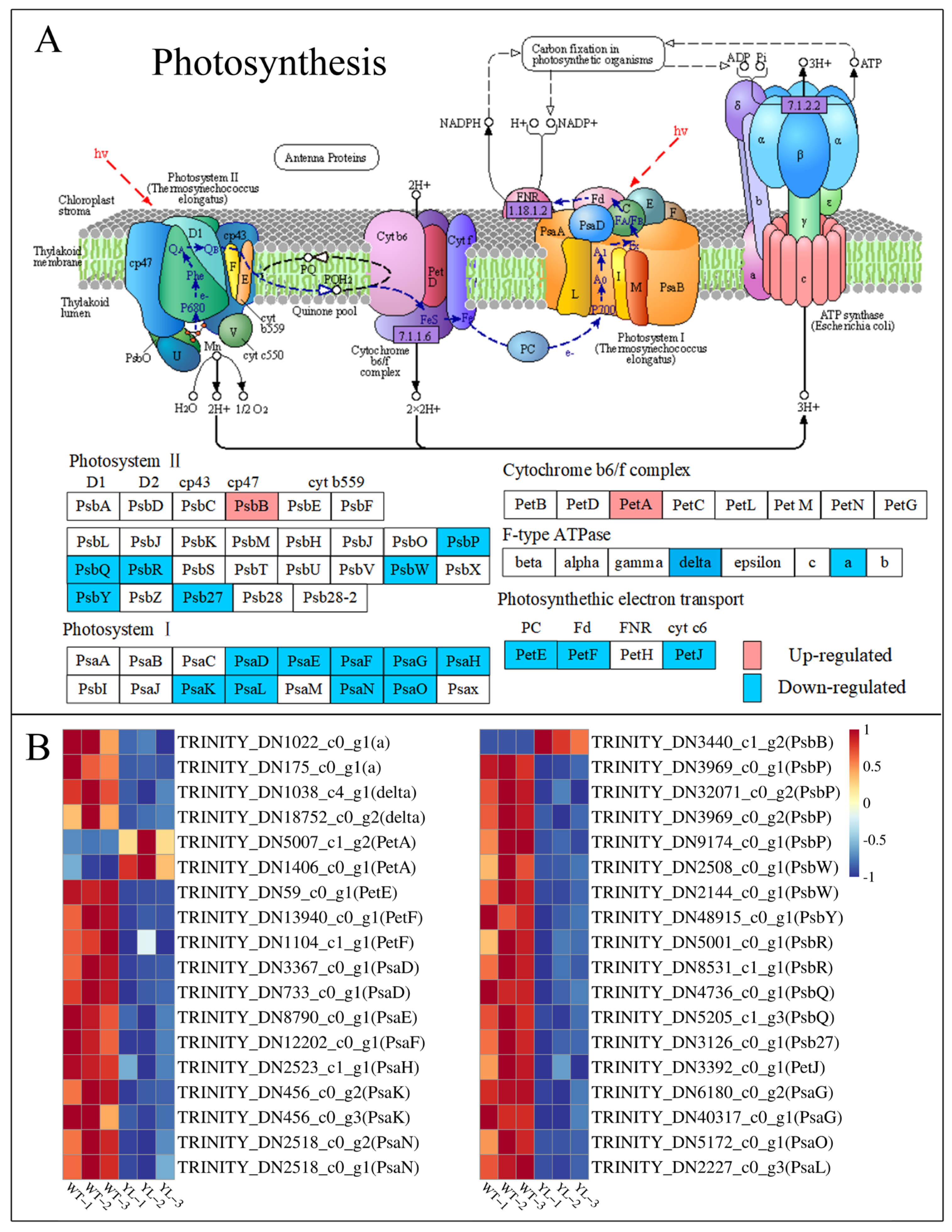
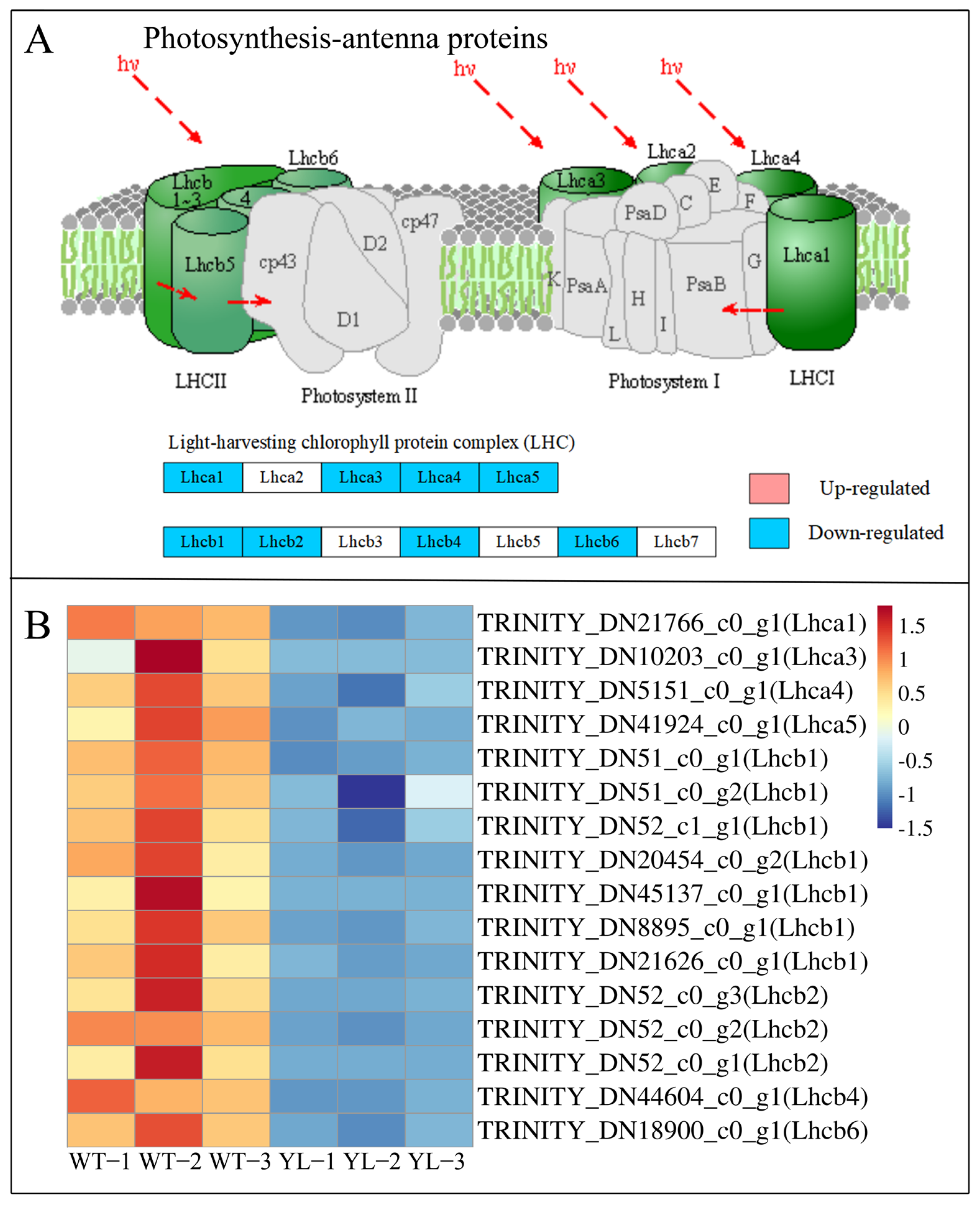
2.3.1. Differentially Expressed Genes Involved in Chlorophyll Anabolism and Photosynthesis
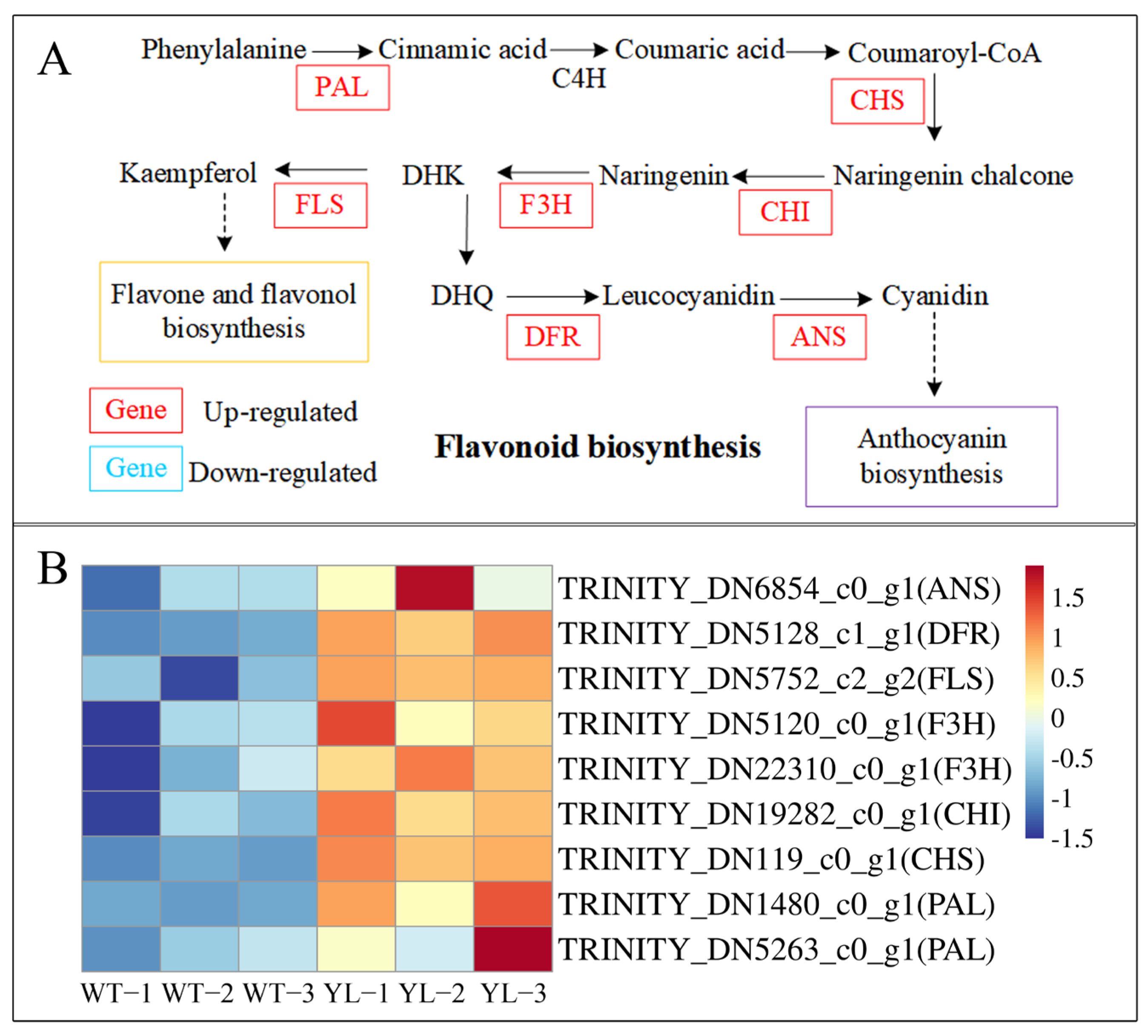
2.3.2. Differentially Expressed Genes Involved in Flavonoid Biosynthesis
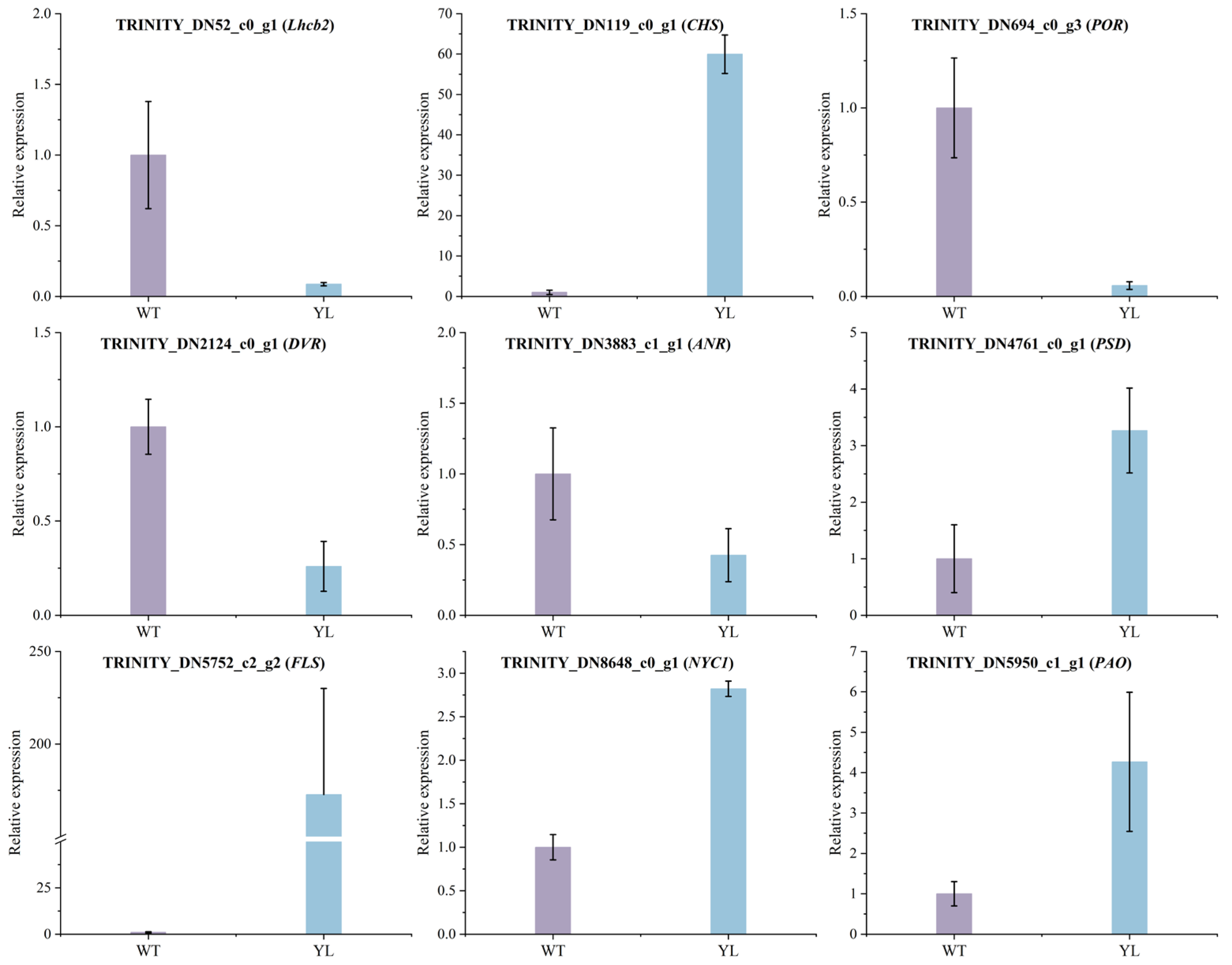
2.4. Validation of Gene Expression by qRT–PCR
3. Discussion
3.1. Leaf Yellowing Due to Changes in Leaf Pigmentation Ratios
3.2. Aberrant Structure of Chloroplasts Affects Chlorophyll Synthesis
3.3. Signaling Pathway Abnormalities
4. Materials and Methods
4.1. Plant Material
4.2. Pigment Content
4.2.1. Chlorophylls and Carotenoids
4.2.2. Chlorophyll Precursor
4.2.3. Total Flavonoids
4.3. Ultrastructure Microscopy
4.4. RNA Extraction, Library Construction, and RNA–Seq
4.5. Gene Annotation and Differentially Expressed Genes (DEGs) Analysis
4.6. Quantitative qRT–PCR (qPCR)
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, Y.; Bai, P.P.; Gu, K.J.; Yang, S.Z.; Lin, H.Y.; Shi, C.G.; Zhao, Y.P. Dynamic transcriptome and network-based analysis of yellow leaf mutant Ginkgo biloba. BMC Plant Biol. 2022, 22, 456. [Google Scholar] [CrossRef] [PubMed]
- Li, W.X.; Yang, S.B.; Lu, Z.G.; He, Z.C.; Ye, Y.L.; Zhao, B.B.; Wang, L.; Jin, B. Cytological, physiological, and transcriptomic analyses of golden leaf coloratiosn in Ginkgo biloba L. Hortic. Res. 2018, 5, 12. [Google Scholar] [PubMed]
- Borek, M.; Baczek-Kwinta, R.; Rapacz, M. Photosynthetic activity of variegated leaves of Coleus × hybridus hort. cultivars characterised by chlorophyll fluorescence techniques. Photosynthetica 2016, 54, 331–339. [Google Scholar]
- Li, Y.N.; Zhou, Y.; Chen, H.; Chen, C.; Liu, Z.M.; Han, C.; Wu, Q.K.; Yu, F.Y. Transcriptomic analyses reveal key genes involved in pigment biosynthesis related to leaf color change of Liquidambar formosana Hance. Molecules 2022, 27, 5433. [Google Scholar] [CrossRef]
- Xu, B.H.; Zhang, C.Y.; Gu, Y.; Cheng, R.; Huang, D.Y.; Liu, X.; Sun, Y.D. Physiological and transcriptomic analysis of a yellow leaf mutant in watermelon. Sci. Rep. 2023, 13, 9647. [Google Scholar]
- Han, H.W.; Zhou, Y.; Liu, H.F.; Chen, X.J.; Wang, Q.; Zhuang, H.M.; Sun, X.X.; Ling, Q.H.; Zhang, H.J.; Wang, B.K.; et al. Transcriptomics and metabolomics analysis provides insight into leaf color and photosynthesis variation of the yellow-green leaf mutant of Hami melon (Cucumis melo L.). Plants 2023, 12, 1623. [Google Scholar] [CrossRef]
- Zhang, S.Z.; Wu, X.L.; Cui, J.; Zhang, F.; Wan, X.Q.; Liu, Q.L.; Zhong, Y.; Lin, T.T. Physiological and transcriptomic analysis of yellow leaf coloration in Populus deltoides Marsh. PLoS ONE 2019, 14, e0216879. [Google Scholar]
- Huang, M.S.; Slewinski, T.L.; Baker, R.F.; Janick-Buckner, D.; Buckner, B.; Johal, G.S.; Braun, D.M. Camouflage patterning in maize leaves results from a defect in porphobilinogen deaminase. Mol. Plant 2009, 2, 773–789. [Google Scholar]
- Hung, C.Y.; Sun, Y.H.; Chen, J.J.; Darlington, D.E.; Williams, A.L.; Burkey, K.O.; Xie, J.H. Identification of a Mg-protoporphyrin IX monomethyl ester cyclase homologue, EaZIP, differentially expressed in variegated Epipremnum aureum ‘Golden Pothos’ is achieved through a unique method of comparative study using tissue regenerated plants. J. Exp. Bot. 2010, 61, 1483–1493. [Google Scholar]
- Kang, S.J.; Fang, Y.X.; Zou, G.X.; Ruan, B.P.; Zhao, J.; Dong, G.J.; Yan, M.X.; Gao, Z.Y.; Zhu, L. White-green leaf gene encoding protochlorophyllide oxidoreductase B is involved in chlorophyll synthesis of rice. Crop Sci. 2015, 55, 284–293. [Google Scholar] [CrossRef]
- Zhao, Y.H.; Huang, S.N.; Zhang, M.D.; Zhang, Y.; Feng, H. Mapping of a pale green mutant gene and its functional verification by allelic mutations in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Front. Plant Sci. 2021, 12, 699308. [Google Scholar] [CrossRef] [PubMed]
- Editorial Committee of Flora of China, Chinese Academy of Sciences. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1996; Volume 30, p. 198. [Google Scholar]
- Mei, X.; Zhang, K.; Lin, Y.; Su, H.; Lin, C.; Chen, B.; Yang, H.; Zhang, L. Metabolic and transcriptomic profiling reveals etiolated mechanism in Huangyu Tea (Camellia sinensis) leaves. Int. J. Mol. Sci. 2022, 23, 15044. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhang, J.; Zhang, Q.; Li, X.; Li, M.; Yang, Y.; Zhou, J.; Wei, Q.; Zhou, B. Methylome and transcriptome analyses of three different degrees of albinism in apple seedlings. BMC Genom. 2022, 23, 310. [Google Scholar] [CrossRef]
- Grimm, B.; Porra, R.J.; Rüdiger, W.; Scheer, H. Chlorophylls and Bacteriochlorophylls: Biochemistry, Biophysics, Functions and Applications; Springer: Dordrecht, The Netherlands, 2006; Volume 25, pp. 237–260. [Google Scholar]
- Motohashi, R.; Ito, T.; Kobayashi, M.; Taji, T.; Nagata, N.; Asami, T.; Yoshida, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Functional analysis of the 37 kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant J. 2003, 34, 719–731. [Google Scholar] [CrossRef]
- Masuda, T.; Fujita, Y. Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 2008, 7, 1131–1149. [Google Scholar]
- Beale, S.I. Green genes gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef]
- Pontoppidan, B.; Kannangara, C.G. Purification and partial characterisation of barley glutamyl-tRNAGlu reductase, the enzyme that directs glutamate to chlorophyll biosynthesis. Eur. J. Biochem. 1994, 225, 529–537. [Google Scholar]
- Zeng, Z.Q.; Lin, T.Z.; Zhao, J.Y.; Zheng, T.H.; Xu, L.F.; Wang, Y.H.; Liu, L.L.; Jiang, L.; Chen, S.H.; Wan, J.M. OsHemA gene, encoding glutamyl-tRNA reductase (GluTR) is essential for chlorophyll biosynthesis in rice (Oryza sativa). J. Integr. Agric. 2020, 19, 612–623. [Google Scholar]
- Jensen, P.E.; Reid, J.D.; Hunter, C.N. Modification of cysteine residues in the ChlI and ChlH subunits of magnesium chelatase results in enzyme inactivation. Biochem. J. 2000, 352, 435–441. [Google Scholar] [CrossRef]
- Pattanayak, G.K.; Tripathy, B.C. Overexpression of Protochlorophyllide Oxidoreductase C Regulates Oxidative Stress in Arabidopsis. PLoS ONE 2011, 6, e26532. [Google Scholar] [CrossRef]
- Long, W.H.; Long, S.F.; Jiang, X.; Xu, H.F.; Peng, Q.; Li, J.L.; Zhang, X.C.; Zhang, D.S.; Liu, X.W.; Zhu, S.S. A rice Yellow Green Leaf 219 mutant lacking the divinyl reductase affects chlorophyll biosynthesis and chloroplast development. J. Plant Growth Regul. 2022, 41, 3233–3242. [Google Scholar]
- Sato, R.; Ito, H.; Tanaka, A. Chlorophyll b degradation by chlorophyll b reductase under high-light conditions. Photosynthesis Res. 2015, 126, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Kimura, M.; Matsuura, H.; Tanaka, A.; Ito, H. Jasmonate production through chlorophyll a degradation by Stay-Green in Arabidopsis thaliana. J. Plant Physiol. 2019, 238, 53–62. [Google Scholar] [PubMed]
- Ren, M.J.; Ma, J.J.; Lu, D.Y.; Wu, C.; Zhu, S.Y.; Chen, X.J.; Wu, Y.F.; Shen, Y.M. STAY-GREEN accelerates chlorophyll degradation in Magnolia sinostellata under the condition of light deficiency. Int. J. Mol. Sci. 2023, 24, 8510. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Int. Plant Biol. 2020, 63, 180–209. [Google Scholar]
- Liu, G.F.; Han, Z.X.; Feng, L.; Gao, L.P.; Gao, M.J.; Gruber, M.Y.; Zhang, Z.L.; Xia, T.; Wan, X.C.; Wei, S. Metabolic flux redirection and transcriptomic reprogramming in the albino tea cultivar ‘Yu-Jin-Xiang’ with an emphasis on catechin production. Sci. Rep. 2017, 7, 45062. [Google Scholar]
- Feng, S.X.; Liu, Y.C.; Yan, S.F.; Dai, S.H.; Chen, L.Y.; Fan, Y.C.; Bai, Y.; Huang, Y.R. The potential mechanism of UpCrtR-b regulates leaf color change in Ulmus pumila by multi-omics and functional analysis. Sentia Hortic. 2024, 324, 112616. [Google Scholar]
- Wu, Y.Q.; Guo, J.; Wang, T.L.; Cao, F.L.; Wang, G.B. Metabolomic and transcriptomic analyses of mutant yellow leaves provide insights into pigment synthesis and metabolism in Ginkgo biloba. BMC Genom. 2020, 21, 858. [Google Scholar]
- Jensen, R.G.; Bassham, J.A. Photosynthesis by isolated chloroplasts. Proc. Natl. Acad. Sci. USA 1966, 56, 1095–1101. [Google Scholar]
- Kirchhoff, H. Chloroplast ultrastructure in plants. New Phytol. 2019, 223, 565–574. [Google Scholar] [CrossRef]
- Chang, Q.S.; Chen, S.M.; Chen, Y.; Deng, Y.M.; Chen, F.D.; Zhang, F.; Wang, S.W. Anatomical and physiological differences and differentially expressed genes between the green and yellow leaf tissue in a variegated chrysanthemum variety. Mol. Biotechnol. 2013, 54, 393–411. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, K.L.; Li, L.; Wang, M.N.; Fang, L.; Zeng, S.J. Cytological, biochemical, and transcriptomic analyses of a novel yellow leaf variation in a Paphiopedilum (Orchidaceae) SCBG COP15. Genes 2022, 13, 71. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shem, A.; Frolow, F.; Nelson, N. Crystal structure of plant photosystem I. Nature 2003, 426, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Nugent, J.H. Oxygenic photosynthesis. Electron transfer in photosystem I and photosystem II. Eur. J. Biochem. 1996, 237, 519–531. [Google Scholar] [CrossRef]
- Goral, T.K.; Johnson, M.P.; Duffy, C.D.P.; Brain, A.P.R.; Ruban, A.V.; Mullineaux, C.W. Light-harvesting antenna composition controls the macrostructure and dynamics of thylakoid membranes in Arabidopsis. Plant J. 2012, 69, 289–301. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, T.; Feng, B.; Zhang, C.; Peng, S.; Zhang, X.; Fu, G.; Tao, L. Non-photochemical quenching plays a key role in light acclimation of rice plants differing in leaf color. Front. Plant Sci. 2017, 7, 1968. [Google Scholar] [CrossRef]
- Gebremeskel, H.; Umer, M.J.; Hongju, Z.; Li, B.; Shengjie, Z.; Xuqiang, L.; Nan, H.; Yuan, P.; Wenge, L. Genetic mapping and molecular characterization of the delayed green gene dg in watermelon (Citrullus lanatus). Front. Plant Sci. 2023, 14, 1152644. [Google Scholar] [CrossRef]
- Li, S.; Wang, S.; Wang, P.; Gao, L.; Yang, R.; Li, Y. Label-free comparative proteomic and physiological analysis provides insight into leaf color variation of the golden-yellow leaf mutant of Lagerstroemia indica. J. Proteom. 2020, 228, 103942. [Google Scholar] [CrossRef]
- Nott, A.; Jung, H.S.; Koussevitzky, S.; Chory, J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006, 57, 739–759. [Google Scholar] [CrossRef]
- Zhang, M.M.; Zhang, S.Q. Mitogen-activated protein kinase cascades in plant signaling. J. Int. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef]
- Guo, H.L.; Feng, P.Q.; Chi, W.; Sun, X.W.; Xu, X.M.; Li, Y.; Ren, D.T.; Lu, C.M.; Rochaix, J.D.; Leister, D. Plastid-nucleus communication involves calcium-modulated MAPK signaling. Nat. Commun. 2016, 7, 12173. [Google Scholar] [PubMed]
- Liang, J.Y.; Zhang, Q.X.; Liu, Y.R.; Zhang, J.J.; Wang, W.Y.; Zhang, Z.M. Chlorosis seedling lethality 1 encoding a MAP3K protein is essential for chloroplast development in rice. BMC Plant Biol. 2022, 22, 20. [Google Scholar]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Köhler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar]
- Qiu, K.; Li, Z.P.; Yang, Z.; Chen, J.Y.; Wu, S.X.; Zhu, X.Y.; Gao, S.; Gao, J.; Ren, G.D.; Kuai, B.K. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 2004, 16, 3386–3399. [Google Scholar]
- Joo, S.; Liu, Y.; Lueth, A.; Zhang, S. MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 2008, 54, 129–140. [Google Scholar]
- Jagodzik, P.; Tajdel-Zielinska, M.; Ciesla, A.; Marczak, M.; Ludwikow, A. Mitogen-Activated Protein Kinase Cascades in Plant Hormone Signaling. Front. Plant Sci. 2018, 9, 1387. [Google Scholar]
- Wellburn, A.R.; Lichtenthaler, H. Formulae and program to determine total carotenoids and chlorophylls a and b of leaf extracts in different solvents. In Advances in Photosynthesis Research Advances in Agricultural Biotechnology; Springer: Dordrecht, The Netherlands, 1984; Volume 2, pp. 9–12. [Google Scholar]
- Roger, L.; Hideo, T. Effect of Benzyladenine Treatment Duration on B-Aminolevuliniic Acid Accumulation in the Dark, Chlorophyll Lag Phase Abolition, and Long-Term Chlorophyll Production in Excised Cotyledons of Dark-Grown Cucumber Seedlings. Plant Physiol. 1982, 69, 663–667. [Google Scholar]
- Bogorad, L. Porphyrin synthesis. Methods Enzymol. 1962, 5, 885–895. [Google Scholar]
- Yang, H.Y. Study on Mechanism of Leaf Color Variation of Pseudosasa japonica f. akebonosuji. Ph.D. Thesis, Beijing Forestry University, Beijing, China, 2015. [Google Scholar]
- Ying, C.; Wan, D.R. Quantitative determination of total and individual flavonoids in stems and leaves of Buddleja davidii and Buddleja albiflora. Pharmacogn. Mag. 2012, 8, 273–279. [Google Scholar]
- Wang, Q.Y.; Chang, P.J.; Shen, Y.M.; Zhang, C.; Dong, B.; Shi, B.Z. Reference genes for quantitative PCR in Magnolia sinostellata with heat stress. J. Zhejiang AF Univ. 2019, 36, 935–942. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Yang, S.; Zhou, F.; Xu, L.; Shi, C.; He, Q. Physiological, Cytological and Transcriptome Analysis of a Yellow–Green Leaf Mutant in Magnolia sinostellata. Plants 2025, 14, 1037. https://doi.org/10.3390/plants14071037
Zhou X, Yang S, Zhou F, Xu L, Shi C, He Q. Physiological, Cytological and Transcriptome Analysis of a Yellow–Green Leaf Mutant in Magnolia sinostellata. Plants. 2025; 14(7):1037. https://doi.org/10.3390/plants14071037
Chicago/Turabian StyleZhou, Xiawen, Shaozong Yang, Fangwei Zhou, Liang Xu, Congguang Shi, and Qiuling He. 2025. "Physiological, Cytological and Transcriptome Analysis of a Yellow–Green Leaf Mutant in Magnolia sinostellata" Plants 14, no. 7: 1037. https://doi.org/10.3390/plants14071037
APA StyleZhou, X., Yang, S., Zhou, F., Xu, L., Shi, C., & He, Q. (2025). Physiological, Cytological and Transcriptome Analysis of a Yellow–Green Leaf Mutant in Magnolia sinostellata. Plants, 14(7), 1037. https://doi.org/10.3390/plants14071037




