Genome-Wide Identification and Characterization of Basic Pentacysteine Transcription Factors in Brassica napus
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of BnaBPC Family Members
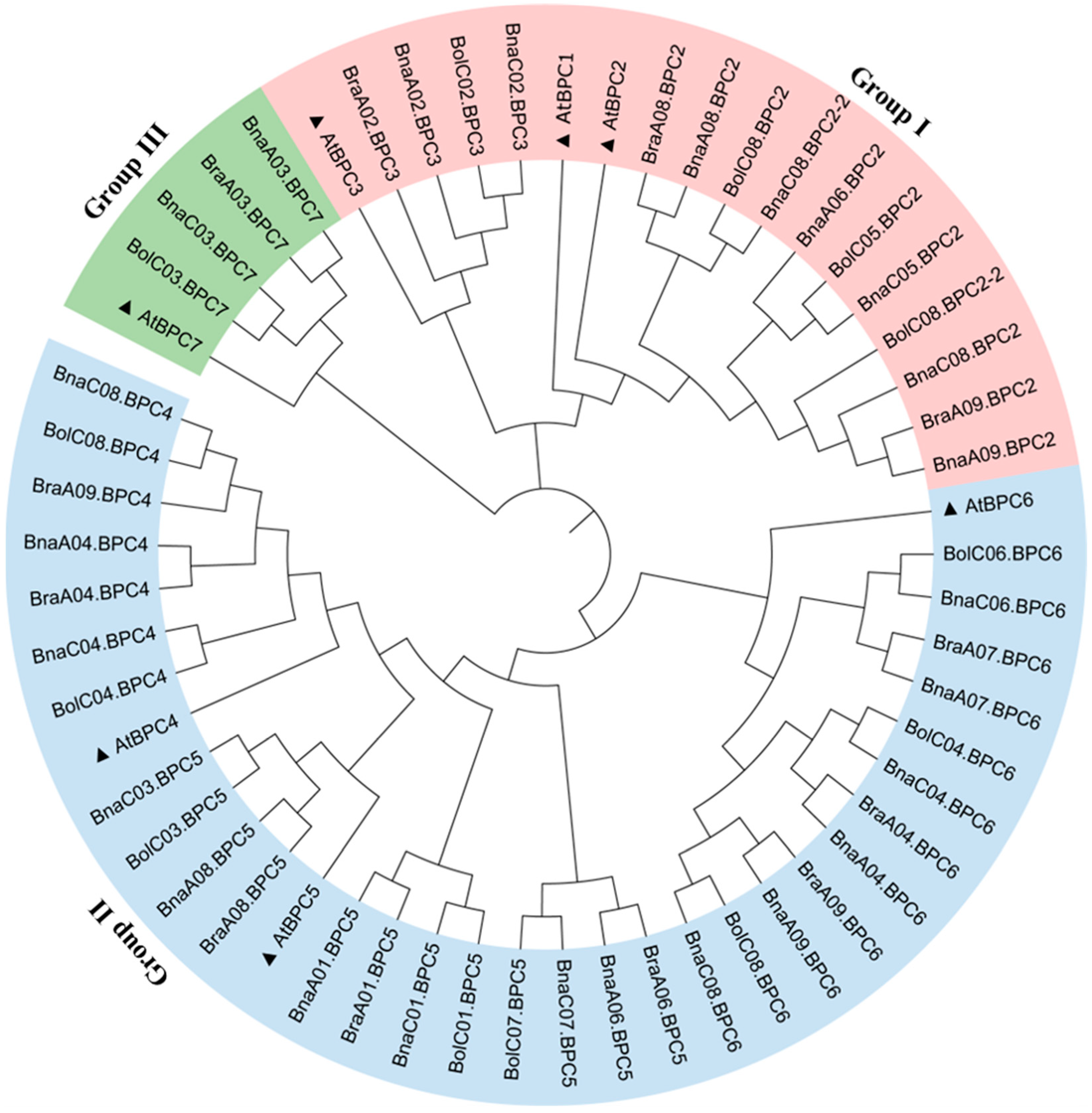
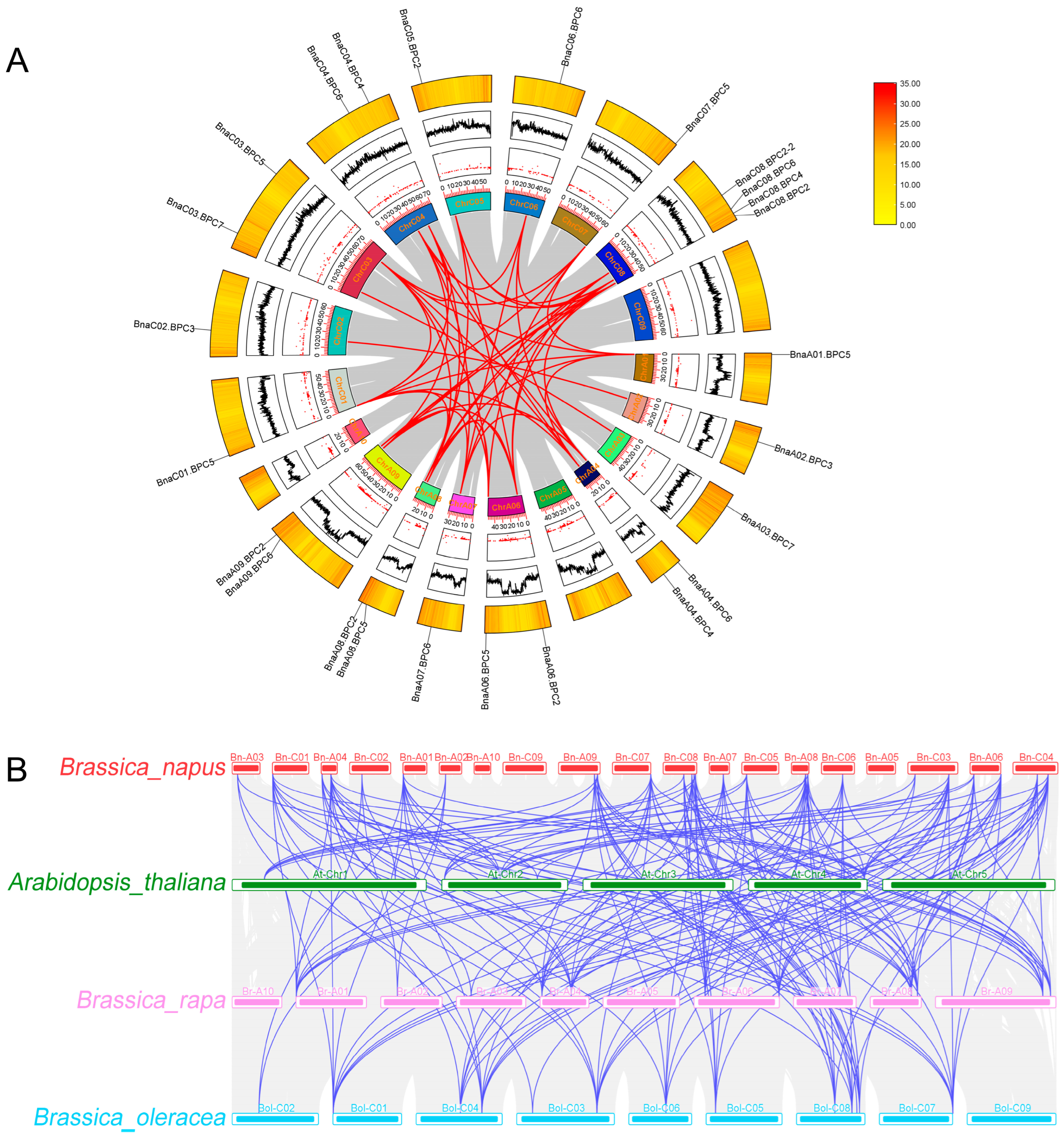
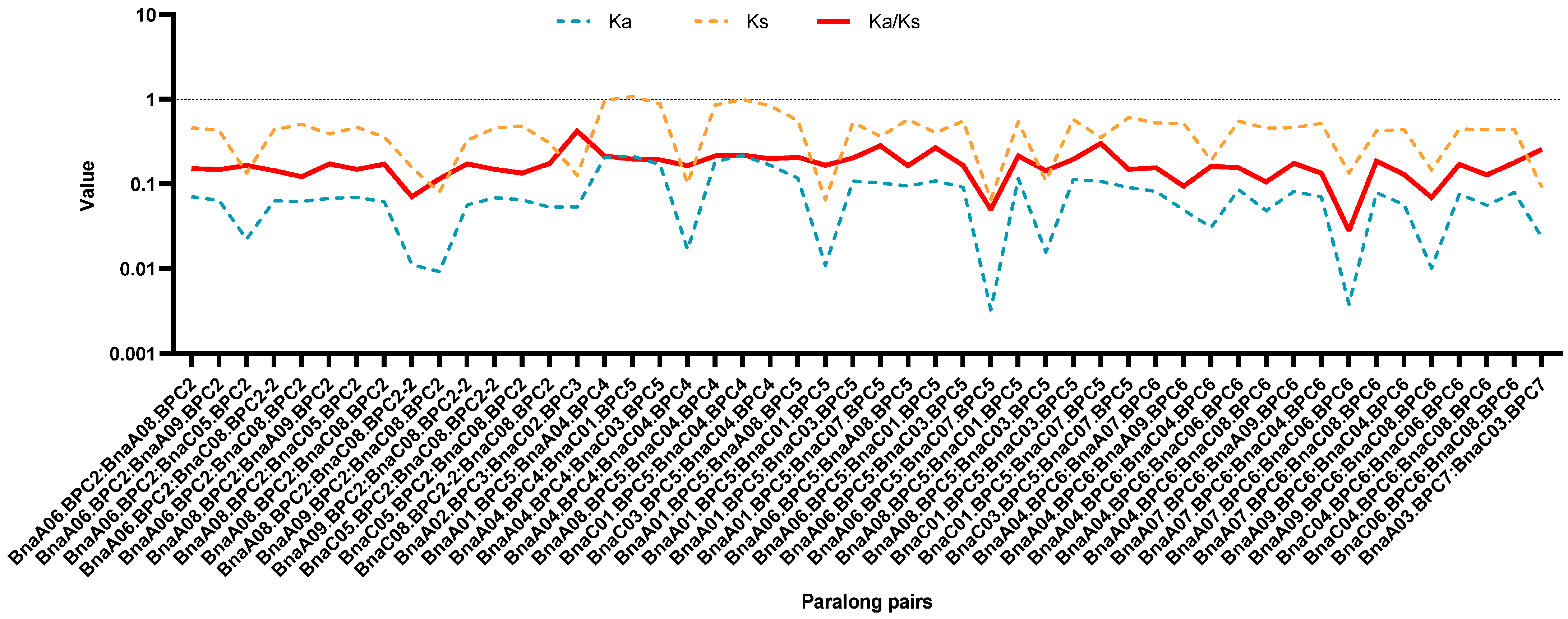
2.2. Phylogenetic and Genome Synteny Analysis of BnaBPC Proteins
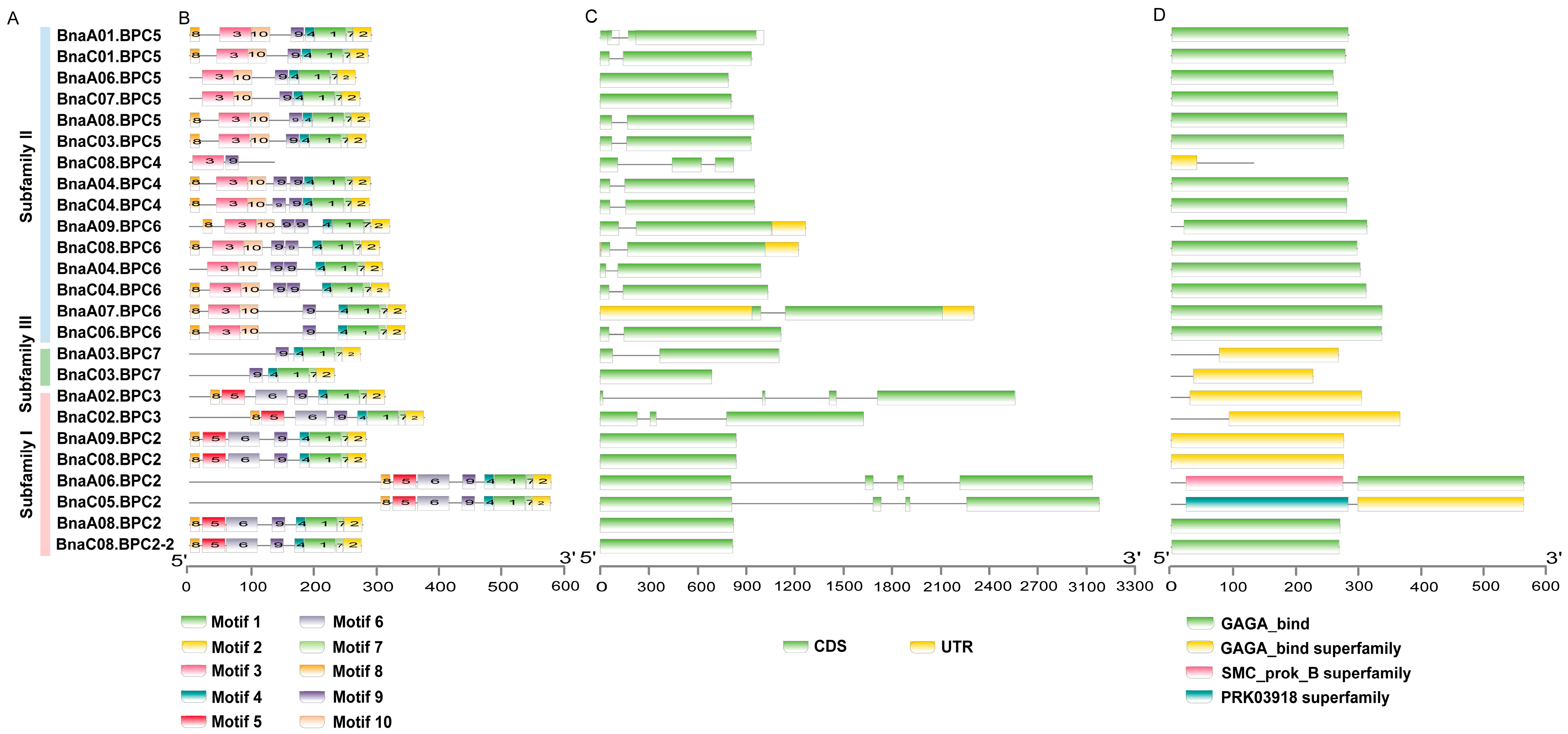
2.3. Gene Structures and Amino Acid-Conserved Structures of BnaBPC Genes
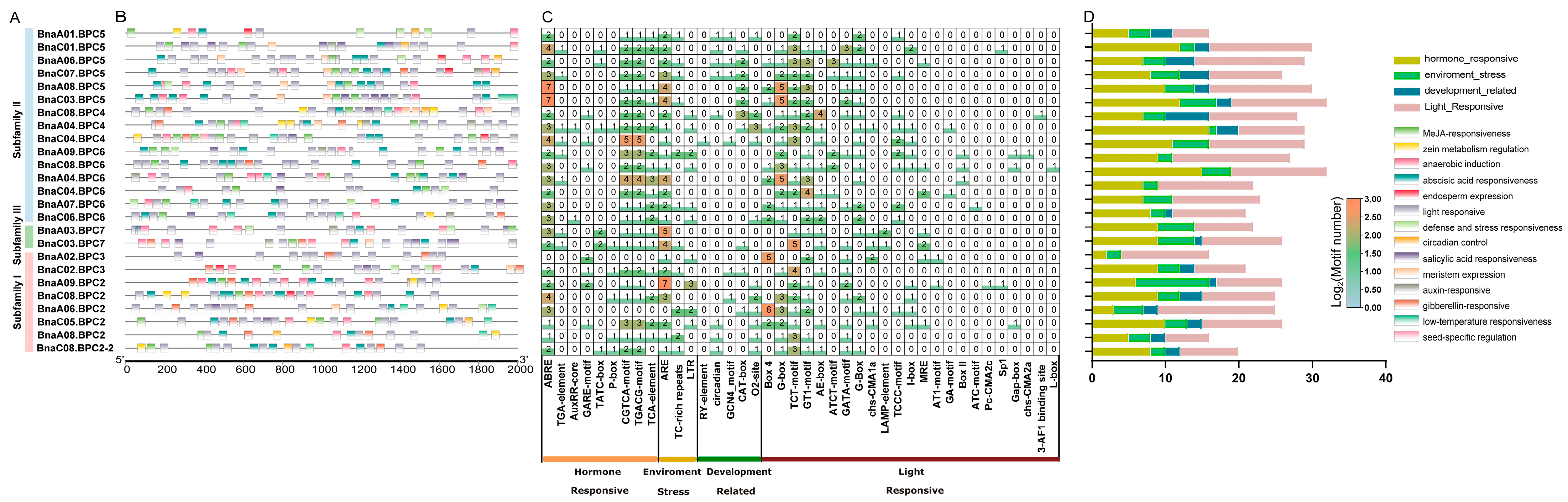
2.4. Cis-Element Analysis of BnaBPC Gene Promoters
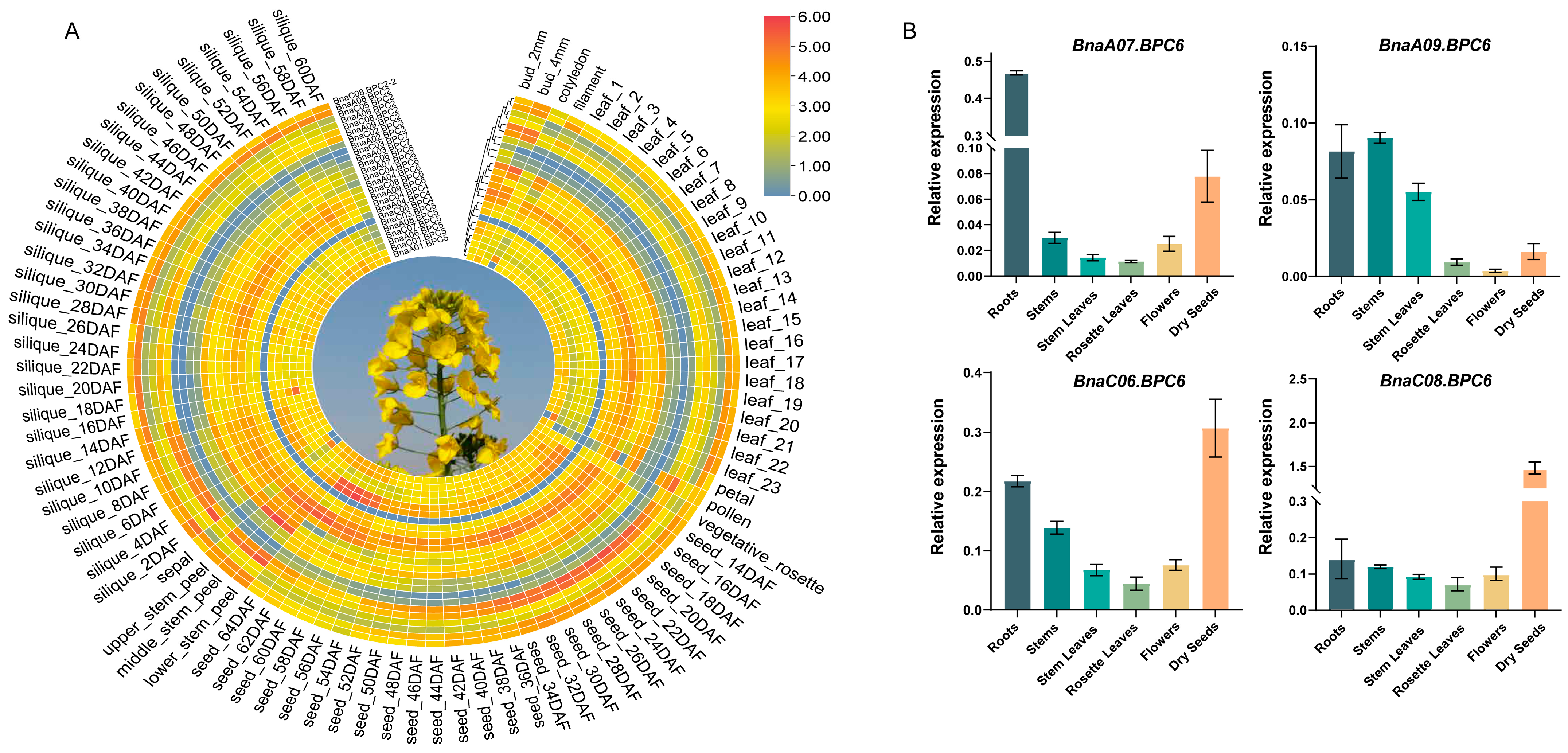
2.5. Expression Profiles of BnaBPC Genes in Various Tissues
3. Discussion
4. Materials and Methods
4.1. Identification of BnaBPCs in the B. napus Genome
4.2. Phylogenetic Analysis and Classification of the BnaBPC Family
4.3. Chromosomal Localization of BnaBPCs
4.4. Analysis of Gene Structures, Conserved Motifs, and Conserved Protein Domains of BnaBPCs
4.5. Cis-Element Analysis in the Promoters of BnaBPCs
4.6. Expression Patterns of BnaBPCs
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zandberg, J.D.; Fernandez, C.T.; Danilevicz, M.F.; Thomas, W.J.W.; Edwards, D.; Batley, J. The Global Assessment of Oilseed Brassica Crop Species Yield, Yield Stability and the Underlying Genetics. Plants 2022, 11, 2740. [Google Scholar] [CrossRef]
- Fu, D.H.; Jiang, L.Y.; Mason, A.S.; Xiao, M.L.; Zhu, L.R.; Li, L.Z.; Zhou, Q.H.; Shen, C.J.; Huang, C.H. Research progress and strategies for multifunctional rapeseed: A case study of China. J. Integr. Agric. 2016, 15, 1673–1684. [Google Scholar]
- Alotaibi, M. Climate change, its impact on crop production, challenges, and possible solutions. Not. Bot. Horti Agrobot. Cluj-Napoca 2023, 51, 13020. [Google Scholar]
- Chaudhry, S.; Sidhu, G.P.S. Climate change regulated abiotic stress mechanisms in plants: A comprehensive review. Plant Cell Rep. 2022, 41, 1–31. [Google Scholar] [PubMed]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of Climate Change on Crops Adaptation and Strategies to Tackle Its Outcome: A Review. Plants 2019, 8, 34. [Google Scholar] [CrossRef]
- Zhu, M.; Monroe, J.G.; Suhail, Y.; Villiers, F.; Mullen, J.; Pater, D.; Hauser, F.; Jeon, B.W.; Bader, J.S.; Kwak, J.M.; et al. Molecular and systems approaches towards drought-tolerant canola crops. New Phytol. 2016, 210, 1169–1189. [Google Scholar]
- Raza, A. Eco-physiological and Biochemical Responses of Rapeseed (Brassica napus L.) to Abiotic Stresses: Consequences and Mitigation Strategies. J. Plant Growth Regul. 2021, 40, 1368–1388. [Google Scholar]
- Young, L.W.; Wilen, R.W.; Bonham, P.C. High temperature stress of Brassica napus during flowering reduces micro- and megagametophyte fertility, induces fruit abortion, and disrupts seed production. J. Exp. Bot. 2004, 55, 485–495. [Google Scholar]
- Abelenda, J.A.; Trabanco, N.; Del Olmo, I.; Pozas, J.; Martín, M.D.M.; Gómez, J.; Esteve, A.; Pernas, M.; Jarillo, J.A.; Piñeiro, M. High ambient temperature impacts on flowering time in Brassica napus through both H2A.Z-dependent and independent mechanisms. Plant Cell Environ. 2023, 46, 1427–1441. [Google Scholar]
- Monfared, M.M.; Simon, M.K.; Meister, R.J.; Roig-Villanova, I.; Kooiker, M.; Colombo, L.; Fletcher, J.C.; Gasser, C.S. Overlapping and antagonistic activities of BASIC PENTACYSTEINE genes affect a range of developmental processes in Arabidopsis. Plant J. 2011, 66, 1020–1031. [Google Scholar]
- Meister, R.J.; Williams, L.A.; Monfared, M.M.; Gallagher, T.L.; Kraft, E.A.; Nelson, C.G.; Gasser, C.S. Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J. 2004, 37, 426–438. [Google Scholar] [CrossRef]
- Kooiker, M.; Airoldi, C.A.; Losa, A.; Manzotti, P.S.; Finzi, L.; Kater, M.M.; Colombo, L. BASIC PENTACYSTEINE1, a GA Binding Protein That Induces Conformational Changes in the Regulatory Region of the Homeotic Arabidopsis Gene SEEDSTICK. Plant Cell 2005, 17, 722–729. [Google Scholar] [CrossRef]
- Wu, J.; Mohamed, D.; Dowhanik, S.; Petrella, R.; Gregis, V.; Li, J.; Wu, L.; Gazzarrini, S. Spatiotemporal Restriction of FUSCA3 Expression by Class I BPCs Promotes Ovule Development and Coordinates Embryo and Endosperm Growth. Plant Cell 2020, 32, 1886–1904. [Google Scholar] [PubMed]
- Li, Q.; Wang, M.; Fang, L. BASIC PENTACYSTEINE2 negatively regulates osmotic stress tolerance by modulating LEA4-5 expression in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 168, 373–380. [Google Scholar] [PubMed]
- Lee, Y.C.; Tsai, P.T.; Huang, X.X.; Tsai, H.L. Family Members Additively Repress the Ectopic Expression of BASIC PENTACYSTEINE3 to Prevent Disorders in Arabidopsis Circadian Vegetative Development. Front. Plant Sci. 2022, 13, 919946. [Google Scholar]
- Mu, Y.; Zou, M.; Sun, X.; He, B.; Xu, X.; Liu, Y.; Zhang, L.; Chi, W. BASIC PENTACYSTEINE Proteins Repress ABSCISIC ACID INSENSITIVE4 Expression via Direct Recruitment of the Polycomb-Repressive Complex 2 in Arabidopsis Root Development. Plant Cell Physiol. 2017, 58, 607–621. [Google Scholar] [PubMed]
- Simonini, S.; Kater, M.M. Class I BASIC PENTACYSTEINE factors regulate HOMEOBOX genes involved in meristem size maintenance. J. Exp. Bot. 2014, 65, 1455–1465. [Google Scholar]
- Yan, J.; Liu, Y.; Yang, L.; He, H.; Huang, Y.; Fang, L.; Scheller, H.V.; Jiang, M.; Zhang, A. Cell wall β-1,4-galactan regulated by the BPC1/BPC2-GALS1 module aggravates salt sensitivity in Arabidopsis thaliana. Mol. Plant 2021, 14, 411–425. [Google Scholar] [CrossRef]
- Sangwan, I.; O’Brian, M.R. Identification of a soybean protein that interacts with GAGA element dinucleotide repeat DNA. Plant Physiol. 2002, 129, 1788–1794. [Google Scholar] [CrossRef]
- Santi, L.; Wang, Y.; Stile, M.R.; Berendzen, K.; Wanke, D.; Roig, C.; Pozzi, C.; Müller, K.; Müller, J.; Rohde, W.; et al. The GA octodinucleotide repeat binding factor BBR participates in the transcriptional regulation of the homeobox gene Bkn3. Plant J. 2003, 34, 813–826. [Google Scholar]
- Gong, R.; Cao, H.; Zhang, J.; Xie, K.; Wang, D.; Yu, S. Divergent functions of the GAGA-binding transcription factor family in rice. Plant J. 2018, 94, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Miao, L.; Huang, B.; Gao, L.; He, C.; Yan, Y.; Wang, J.; Yu, X.; Li, Y. Genome-Wide Identification and Characterization of Cucumber BPC Transcription Factors and Their Responses to Abiotic Stresses and Exogenous Phytohormones. Int. J. Mol. Sci. 2019, 20, 5048. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Chawla, H.S.; Obermeier, C.; Dreyer, F.; Abbadi, A.; Snowdon, R. Chromosome-Scale Assembly of Winter Oilseed Rape Brassica napus. Front. Plant Sci. 2020, 11, 496. [Google Scholar] [CrossRef]
- Guruprasad, K.; Reddy, B.V.B.; Pandit, M. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Gray, W. Hormonal Regulation of Plant Growth and Development. PLoS Biol. 2004, 2, e311. [Google Scholar] [CrossRef]
- Wu, S.; Gallagher, K.L. Transcription factors on the move. Curr. Opin. Plant Biol. 2012, 15, 645–651. [Google Scholar] [CrossRef]
- Cavell, A.C.; Lydiate, D.J.; Parkin, I.A.P.; Dean, C.; Trick, M. Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 1998, 41, 62–69. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef]
- Shaul, O. How introns enhance gene expression. Int. J. Biochem. Cell Biol. 2017, 91 Pt B, 145–155. [Google Scholar]
- Chen, J.; Wang, B.; Chung, J.S.; Chai, H.; Liu, C.; Ruan, Y.; Shi, H. The role of promoter cis-element, mRNA capping, and ROS in the repression and salt-inducible expression of AtSOT12 in Arabidopsis. Front. Plant Sci. 2015, 6, 974. [Google Scholar] [CrossRef]
- Lescot, M.; Gert, G.; Marchal, K.; Moreau, Y.; Déhais, P.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Hecker, A.; Brand, L.H.; Peter, S.; Simoncello, N.; Kilian, J.; Harter, K.; Gaudin, V.; Wanke, D. The Arabidopsis GAGA-Binding Factor BASIC PENTACYSTEINE6 Recruits the POLYCOMB-REPRESSIVE COMPLEX1 Component LIKE HETEROCHROMATIN PROTEIN1 to GAGA DNA Motifs. Plant Physiol. 2015, 168, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [PubMed]

| Gene ID | Gene Name | Location | Length | Molecular Weight (Da) | pI | Instability Index | Aliphatic Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| BnaA06G0096700ZS | BnaA06.BPC2 | A06:5788186..5791223 | 570 | 65,234.18 | 8.66 | 55.78 | 68.6 | −0.791 | nuclear |
| BnaA08G0270900ZS | BnaA08.BPC2 | A08:26026857..26027678 | 273 | 30,490.56 | 9.51 | 57.73 | 52.53 | −0.84 | nuclear |
| BnaA09G0631800ZS | BnaA09.BPC2 | A09:61219562..61220401 | 279 | 31,216.44 | 9.65 | 53.63 | 50.36 | −0.853 | nuclear |
| BnaC05G0118800ZS | BnaC05.BPC2 | C05:7417299..7420376 | 569 | 65,338.51 | 8.92 | 57.30 | 71.97 | −0.764 | nuclear |
| BnaC08G0230300ZS | BnaC08.BPC2-2 | C08:32814996..32815811 | 271 | 30,205.25 | 9.51 | 54.96 | 51.48 | −0.845 | nuclear |
| BnaC08G0489500ZS | BnaC08.BPC2 | C08:50925950..50926789 | 279 | 31,200.44 | 9.65 | 52.14 | 51.04 | −0.837 | nuclear |
| BnaA02G0171100ZS | BnaA02.BPC3 | A02:10179794..10182350 | 308 | 34,783.84 | 9.58 | 49.55 | 63.64 | −0.605 | nuclear |
| BnaC02G0219700ZS | BnaC02.BPC3 | C02:19442545..19474170 | 370 | 41,534.44 | 9.55 | 52.48 | 65.59 | −0.561 | nuclear |
| BnaA04G0137800ZS | BnaA04.BPC4 | A04:15262407..15263357 | 286 | 31,885.16 | 9.17 | 34.87 | 70.70 | −0.699 | nuclear |
| BnaC04G0428300ZS | BnaC04.BPC4 | C04:55284453..55285403 | 284 | 31,618.83 | 9.05 | 37.51 | 66.73 | −0.708 | nuclear |
| BnaC08G0454400ZS | BnaC08.BPC4 | C08:48985111..48985935 | 134 | 14,968.89 | 10.46 | 26.50 | 82.31 | −0.753 | nuclear |
| BnaA01G0002800ZS | BnaA01.BPC5 | A01:296800..297762 | 287 | 32,212.38 | 9.44 | 55.38 | 65.68 | −0.819 | nuclear |
| BnaA06G0450000ZS | BnaA06.BPC5 | A06:48524214..48525002 | 262 | 29,681.82 | 9.27 | 57.63 | 66.37 | −0.833 | nuclear |
| BnaA08G0198900ZS | BnaA08.BPC5 | A08:22511939..22512885 | 284 | 31,838.19 | 9.29 | 54.63 | 63.63 | −0.866 | nuclear |
| BnaC01G0002300ZS | BnaC01.BPC5 | C01:161333..162266 | 282 | 31,760.9 | 9.46 | 52.04 | 66.13 | −0.828 | nuclear |
| BnaC03G0671500ZS | BnaC03.BPC5 | C03:64845945..64846875 | 279 | 31,413.76 | 9.40 | 55.93 | 62.33 | −0.861 | nuclear |
| BnaC07G0547100ZS | BnaC07.BPC5 | C07:60724181..60724990 | 269 | 30,443.6 | 9.26 | 58.41 | 64.65 | −0.863 | nuclear |
| BnaA04G0017300ZS | BnaA04.BPC6 | A04:1121999..1122989 | 305 | 34,288.76 | 9.35 | 59.04 | 58.00 | −0.922 | nuclear |
| BnaA07G0211100ZS | BnaA07.BPC6 | A07:22160943..22163249 | 341 | 38,028.13 | 9.28 | 52.71 | 57.01 | −0.859 | nuclear |
| BnaA09G0548200ZS | BnaA09.BPC6 | A09:56780945..56782210 | 316 | 35,778.63 | 9.47 | 50.79 | 58.45 | −0.876 | nuclear |
| BnaC04G0278000ZS | BnaC04.BPC6 | C04:38436311..38437344 | 315 | 35,646.12 | 9.25 | 51.23 | 55.52 | −0.979 | nuclear |
| BnaC06G0222600ZS | BnaC06.BPC6 | C06:33449718..33450832 | 340 | 38,057.17 | 9.20 | 52.30 | 57.44 | −0.869 | nuclear |
| BnaC08G0394500ZS | BnaC08.BPC6 | C08:45482887..45484108 | 300 | 33,903.27 | 9.51 | 57.66 | 53.77 | −1.010 | nuclear |
| BnaA03G0169700ZS | BnaA03.BPC7 | A03:8638747..8639847 | 270 | 29,757.7 | 9.92 | 52.33 | 72.93 | −0.375 | nuclear |
| BnaC03G0196600ZS | BnaC03.BPC7 | C03:11259402..11260091 | 229 | 25,501.36 | 9.75 | 58.04 | 60.00 | −0.601 | nuclear |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, H.; Jiang, Y.; Liu, C.; Zhang, Y.; Chen, M.; Liu, Z. Genome-Wide Identification and Characterization of Basic Pentacysteine Transcription Factors in Brassica napus. Plants 2025, 14, 1136. https://doi.org/10.3390/plants14071136
Hu H, Jiang Y, Liu C, Zhang Y, Chen M, Liu Z. Genome-Wide Identification and Characterization of Basic Pentacysteine Transcription Factors in Brassica napus. Plants. 2025; 14(7):1136. https://doi.org/10.3390/plants14071136
Chicago/Turabian StyleHu, Huan, Yuqin Jiang, Chiyuan Liu, Ying Zhang, Mingxun Chen, and Zijin Liu. 2025. "Genome-Wide Identification and Characterization of Basic Pentacysteine Transcription Factors in Brassica napus" Plants 14, no. 7: 1136. https://doi.org/10.3390/plants14071136
APA StyleHu, H., Jiang, Y., Liu, C., Zhang, Y., Chen, M., & Liu, Z. (2025). Genome-Wide Identification and Characterization of Basic Pentacysteine Transcription Factors in Brassica napus. Plants, 14(7), 1136. https://doi.org/10.3390/plants14071136








