Anthocyanins and Anthocyanin Biosynthesis Gene Expression in Passiflora Flower Corona Filaments
Abstract
1. Introduction
2. Results
2.1. Identification and Quantification of Anthocyanins in Different Passiflora Species
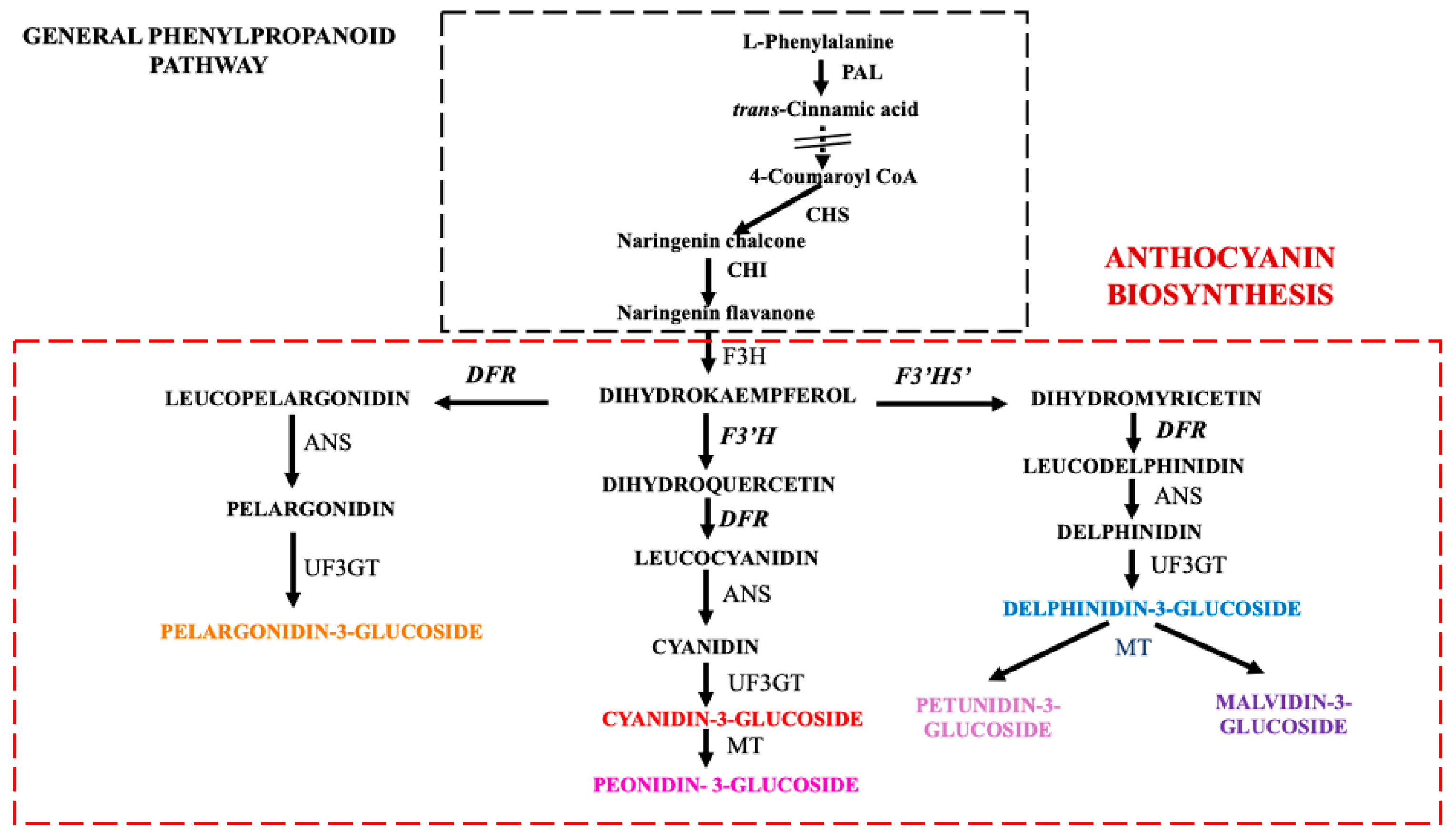
2.2. Expression Profiles of the Anthocyanin Biosynthesis-Related Genes
3. Discussion
3.1. Anthocyanin Presence in Corona Filaments
3.2. Expression Analysis of Anthocyanin Pathway Genes and Transcription Factors
4. Materials and Methods
4.1. Plant Materials
4.2. HPLC/DAD/TOF Analysis
4.3. Identification of Passiflora Anthocyanin Biosynthesis Genes
4.4. RNA Extraction and Gene Expression Analysis by qRT-PCR
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ocampo Perez, J.A.; Coppens d’Eeckenbrugge, G.; Restrepo, M.T.; Jarvis, A.; Salazar, M.H.; Caetano, C.M. Diversity of Colombian Passifloraceae: Biogeography and an updated list for conservation. Biota Colomb. 2007, 8, 1–45. [Google Scholar]
- Malacrida, C.R.; Jorge, N. Yellow passion fruit seed oil (Passiflora edulis f. flavicarpa): Physical and chemical characteristics. Braz. Arc. Biol. Tech. 2012, 55, 127–134. [Google Scholar] [CrossRef]
- Ulmer, T.; MacDougal, J.M. Passionflowers of the World; Timber Press: Portland, OR, USA, 2004. [Google Scholar]
- Thomson, J.D.; Wilson, P. Explaining evolutionary shifts between bee and hummingbird pollination: Convergence, divergence, and directionality. Int. J. Plant Sci. 2008, 169, 23–38. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Brugliera, F.; Kalc, G.; Senior, M.; Dyson, B.; Nakamura, N.; Katsumoto, Y.; Chandler, S. Flower color modification by 586 engineering of the flavonoid biosynthetic pathway: Practical perspectives. Biosci. Biotechnol. Biochem. 2010, 74, 1760–1769. [Google Scholar] [CrossRef]
- Alan, H.; Ingo, A.; Cathie, M. Natural blues: Structure meets function in anthocyanins. Plants 2021, 10, 726. [Google Scholar] [CrossRef] [PubMed]
- Giuseppe, M.; Carla, G.; Andrea, E.; Graziella, S.; Cinzia, M.B. Anthocyanins: Biosynthesis, distribution, ecological role, and use of biostimulants to increase their content in plant foods. Agriculture 2021, 11, 212. [Google Scholar] [CrossRef]
- Kazuma, K.; Noda, N.; Suzuki, M. Flavonoid Composition Related to Petal Color in Different Lines of Clitoria ternatea. Phytochemistry 2003, 64, 1133–1139. [Google Scholar] [CrossRef]
- Sasaki, N.; Nakayama, T. Achievements and perspectives in biochemistry concerning anthocyanin modification for blue flower coloration. Plant Cell Physiol. 2014, 56, 28–40. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, Y.; Long, T.; Wang, S.; Yang, J. Regulation mechanism of plant pigments biosynthesis: Anthocyanins, carotenoids, and betalains. Metabolites 2022, 12, 871. [Google Scholar] [CrossRef]
- Pires, T.C.S.P.; Dias, M.I.; Barros, L.; Barreira, J.C.M.; Santos-Buelga, C.; Ferreira, I.C.F.R. Incorporation of natural colorants obtained from edible flowers in yogurts. LWT 2018, 97, 668–675. [Google Scholar] [CrossRef]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [PubMed]
- Teng, H.; Fang, T.; Lin, Q.; Song, H.; Liu, B.; Chen, L. Red raspberry and its anthocyanins: Bioactivity beyond antioxidant capacity. Trends Food Sci. Technol. 2017, 66, 153–165. [Google Scholar] [CrossRef]
- Horbowicz, M.; Kosson, R.; Grzesiuk, A.; Dębski, H. Anthocyanins of Fruits and Vegetables—Their Occurrence, Analysis and Role in Human Nutrition. J. Fruit Ornam. Plant Res. 2008, 68, 5–22. [Google Scholar] [CrossRef]
- Wang, L.S.; Stoner, G.D. Anthocyanins and their role in cancer prevention. Cancer Lett. 2008, 269, 281–290. [Google Scholar] [CrossRef]
- Chen, J.; Xu, B.; Sun, J.; Jiang, X.; Bai, W. Anthocyanin supplement as a dietary strategy in cancer prevention and management: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2022, 62, 7242–7254. [Google Scholar] [CrossRef]
- Aizza, L.; Sawaya, A.; Dornelas, M.C. Identification of anthocyanins in the corona of two species of Passiflora and their hybrid by ultra-high-performance chromatography with electrospray ionization tandem mass spectrometry (UHPLC-ESI-MS/MS). Biochem. Syst. Ecol. 2019, 85, 60–67. [Google Scholar] [CrossRef]
- Teixeira, M.; Tao, W.; Fernandes, A.; Faria, A.; Ferreira, I.M.P.L.V.O.; He, J.; de Freitas, V.; Mateus, N.; Oliveira, H. Anthocyanin-rich edible flowers, current understanding of a potential new trend in dietary patterns. Trends Food Sci. Technol. 2023, 138, 708–725. [Google Scholar] [CrossRef]
- Yang, J.; Chen, R.; Wang, C.; Li, C.; Ye, W.; Zhang, Z.; Wang, S. A widely targeted metabolite modificomics strategy for modified metabolites identification in tomato. J. Integ. Plant Biol. 2024, 66, 810–823. [Google Scholar] [CrossRef]
- Quattrocchio, F.; Wing, J.; van der Woude, K.; Souer, E.; de Vetten, N.; Mol, J.; Koes, R. Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell 1999, 8, 1433–1444. [Google Scholar] [CrossRef]
- Helariutta, Y.; Elomaa, P.; Kotilainen, M.; Seppanen, P.; Teeri, T.H. Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of DFR expression in the corollas of Gerbera hybrida var. Regina (Compositae). Plant Mol. Biol. 1993, 2, 183–193. [Google Scholar]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef]
- Yue, M.; Jiang, L.; Zhang, N.; Zhang, L.; Liu, Y.; Lin, Y.; Zhang, Y.; Luo, Y.; Zhang, Y.; Wang, Y. Regulation of flavonoids in strawberry fruits by FaMYB5/FaMYB10 dominated MYB-bHLH-WD40 ternary complexes. Front. Plant Sci. 2023, 14, 1145670. [Google Scholar] [CrossRef]
- Huq, E.; Quail, P.-H. A phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2022, 15, 2441–2450. [Google Scholar] [CrossRef]
- Lee, S.; Lee, S.-h.; Yang, K.-Y.; Kim, Y.-M.; Park, S.-Y.; Kim, S.-Y.; Soh, M.-S. Overexpression of PRE1 and itshomologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 591–600. [Google Scholar] [CrossRef]
- Duek, P.-D.; Fankhauser, C. bHLH class transcription factors take Centre stage in phytochrome signaling. Trends Plant Sci. 2005, 10, 51–54. [Google Scholar] [CrossRef]
- Hu, D.-G.; Sun, C.-H.; Zhang, Q.-Y.; An, J.-P.; You, C.-X.; Hao, Y.-J. Glucose Sensor MdHXK1 Phosphorylates and Stabilizes MdbHLH3 to Promote Anthocyanin Biosynthesis in Apple. PLoS Genet. 2016, 25, e1006273. [Google Scholar] [CrossRef]
- Zhang, H.; Jin, P.-J.; Tang, L.; Zhao, Y.; Gu, X.-C.; Gao, G.; Luo, J.-C. PlantTFDB 2.0: Update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011, 9, D1114–D1117. [Google Scholar] [CrossRef]
- Jin, P.J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J.C. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef]
- Liu, H.-T.; Yu, X.-H.; Li, K.-W.; Klejnot, J.; Yang, H.-Y.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, J.; Liang, C.; Liu, F.; Hou, X.; Zou, X. Genome-wide identification and characterization of the bHLH transcription factor family in Pepper (Capsicum annuum L.). Front. Genet. 2020, 11, 570156. [Google Scholar] [CrossRef]
- Varaud, E.; Brioudes, F.; Szecsi, J.; Leroux, J.; Brown, S.; Perrot-Rechenmann, C. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETAL. Plant Cell 2011, 23, 973–983. [Google Scholar] [CrossRef]
- Kidoy, L.; Nygard, A.M.; Andersen, O.M.; Pedersen, A.T.; Aksnes, D.W.; Kiremire, B.T. Anthocyanins in fruits of Passiflora edulis and Passiflora suberosa. J. Food Compos. Anal. 1997, 10, 49–54. [Google Scholar] [CrossRef]
- Aizza, L.C.; Dornelas, M.C. A genomic approach to study anthocyanin synthesis and flower pigmentation in Passionflowers. J. Nucleic Acids. 2011, 2011, 371517. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, D.; Ma, F.; Yang, L.; Wu, B.; Xing, W.; Sun, P.; Chen, D.; Xu, B.; Song, S. Identification of key genes involved in flavonoid and terpenoid biosynthesis and the pathway of triterpenoid biosynthesis in Passiflora edulis. J. Integ. Agric. 2023, 22, 1412–1423. [Google Scholar] [CrossRef]
- Shi, M.; Ali, M.M.; Sun, K.; Gull, S.; Hu, X.; Kayima, V.; Cai, S.; Hou, Y.; Chen, F. Changes in fruit anthocyanins, their biosynthesis-related enzymes and related genes during fruit development of purple and yellow passion fruits. Fruit Res. 2023, 3, 17. [Google Scholar] [CrossRef]
- Zheng, Y.-Y.; Chen, L.-H.; Fan, B.-L.; Xu, Z.; Wang, Q.; Zhao, B.-Y.; Gao, M.; Yuan, M.-H.; Qamar, M.T.u.; Jiang, Y.; et al. Integrative multiomics profiling of passion fruit reveals the genetic basis for fruit color and aroma. Plant Physiol. 2024, 194, 2491–2510. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Xiong, B.-X.; Huang, R.-B.; Ni, Y.; Li, X. Integrated metabolomics and proteomics analysis of anthocyanin biosynthesis regulations in passion fruit (Passiflora edulis) pericarp. Plant Physiol. Biochem. 2025, 220, 109441. [Google Scholar] [CrossRef]
- Bochi, V.C.; Godoy, H.T.; Giusti, M.M. Anthocyanin and other phenolic compounds in Ceylon gooseberry (Dovyalis hebecarpa) fruits. Food Chem. 2015, 176, 234–243. [Google Scholar] [CrossRef][Green Version]
- Lou, Q.; Wang, L.; Liu, H.; Liu, Y. Anthocyanin profiles in flowers of grape Hyacinth. Molecules 2017, 22, 688. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Lai, L.; Wang, F.; Sun, W.; Zhang, L.; Li, X.; Wang, L.; Jiang, L.; Zheng, Y. Characterisation of flower colouration in 30 Rhododendron species via anthocyanin and flavonol identification and quantitative traits. Plant Biol. 2018, 20, 121–129. [Google Scholar] [CrossRef]
- Katja, K.; Declan, J.L.; Nick, W.A.; Nelli, M.; Tony, M.; Andrew, C.A.; Bilal, M.A.; Hely, H.; Richard, V.E.; Laura, J. MYBA and MYBPA transcription factors co-regulate anthocyanin biosynthesis in blue-coloured berries. New Phytol. 2021, 232, 1350–1367. [Google Scholar] [CrossRef]
- Campbell, S.M.; Pearson, B.; Marble, S.C. Butterfly Pea (Clitoria ternatea) Flower Extract (BPFE) and Its Use as a PH-Dependent Natural Colorant. EDIS 2019, 2019, 467–472. [Google Scholar] [CrossRef]
- Mori, S.; Asano, S.; Kobayashi, H.; Nakano, M. Analyses of anthocyanidins and anthocyanins in flowers of Muscari spp. Bull. Fac. Agr. Niigata Univ. 2002, 55, 13–18. [Google Scholar]
- Sakata, Y.; Aoki, N.; Tsunematsu, S.; Nishikouri, H.; Johjima, T. Petal coloration and pigmentation of tree peony bred and selected in Daikon Island (Shimane Prefecture). J. Jpn. Soc. Hortic. Sci. 1995, 64, 351–357. [Google Scholar] [CrossRef]
- Fournier-Level, A.; Hugueney, P.; Verrieès, A.; Ageorges, A. Genetic mechanisms underlying the methylation level of anthocyanins in grape (Vitis vinifera L.). BMC Plant Biol. 2011, 11, 179. [Google Scholar] [CrossRef]
- Meyer, P.; Heidmann, I.; Forkmann, G.; Saedler, H. A new petunia flower color generated by transformation of a mutant with a 588-maize gene. Nature 1987, 330, 677–678. [Google Scholar] [CrossRef]
- Mekapogu, M.; Song, H.-Y.; Lim, S.-H.; Jung, J.-A. Genetic engineering and genome editing advances to enhance the floral attributes in ornamental plants: An update. Plants 2023, 12, 3983. [Google Scholar] [CrossRef]
- Boase, M.R.; Lewis, D.H.; Davies, K.M.; Marshall, G.B.; Patel, D.; Schwinn, K.E.; Deroles, S.C. Isolation and antisense suppression 595 of flavonoid 3′5′-hydroxylase modifies flower pigments and color in cyclamen. BMC Plant Biol. 2010, 10, 107. [Google Scholar] [CrossRef]
- Brugliera, F.; Tao, G.Q.; Tems, U.; Kalc, G.; Mouradova, E.; Price, K.; Stevenson, K.; Nakamura, N.; Stacey, I.; Katsumoto, Y.; et al. Violet/blue chrysanthemums- Metabolic engineering of the anthocyanin biosynthetic pathway results in novel petal 610 colors. Plant Cell Physiol. 2013, 54, 1696–1710. [Google Scholar] [CrossRef] [PubMed]
- Seitz, C.; Vitten, M.; Steinbach, P.; Hartl, S.; Hirsche, J.; Rathje, W.; Treutter, D.; Forkmann, G. Redirection of anthocyanin synthesis 597 in Osteospermum hybrida by a two-enzyme manipulation strategy. Phytochem 2007, 68, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuka, T.; Nishihara, M.; Mishiba, K.; Yamamura, S. Heterologous expression of two gentian cytochrome P450 genes can modulate the intensity of flower pigmentation in transgenic tobacco plants. Mol. Breed. 2006, 17, 91–99. [Google Scholar] [CrossRef]
- Tanaka, Y.; Ohmiya, A. Seeing is believing Engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotechnol. 2008, 19, 190–197. [Google Scholar] [CrossRef]
- Gopaulchan, D.; Lennon, A.M.; Umaharan, P. Expression analysis of the anthocyanin genes in pink spathes of anthurium with different color intensities. J. Amer. Soc. Hort. Sci. 2015, 140, 480–489. [Google Scholar] [CrossRef]
- Brugliera, F.; Barri-Rewell, G.; Holton, T.A.; Mason, J.G. Isolation and characterization of a flavonoid 3#-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J. 1999, 19, 441–451. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 36–242. [Google Scholar] [CrossRef]
- Fraser, L.G.; Seal, A.G.; Montefiori, M.; McGhie, T.K.; Tsang, G.K.; Datson, P.M.; Hilario, E.; Marsh, H.E.; Dunn, J.K.; Hellens, R.P.; et al. An R2R3 MYB transcription factor determines red petal colour in an Actnidia (kiwifruit) hybrid population. BMC Genom. 2013, 14, 28. [Google Scholar] [CrossRef]
- Peng, Y.; Lin-Wang, K.; Cooney, J.-M.; Wang, T.; Espley, R.-V.; Allan, A.-C. Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). Horticul. Res. 2019, 6, 3. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, H.; Wang, Y.; Guan, S.; Wang, F.; Tang, J.; Zhang, R.; Xie, L.; Lu, Y. Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. J. Exp. Bot. 2015, 66, 3775–3789. [Google Scholar] [CrossRef]
- Blando, F.; Scardino, A.P.; De Bellis, L.; Nicoletti, I.; Giovinazzo, G. Characterization of in vitro anthocyanin-producing sour cherry (Prunus cerasus L.) callus cultures. Food Res. Int. 2005, 38, 937–942. [Google Scholar] [CrossRef]
- Liang, J.; Fang, Y.; An, C.; Yao, Y.; Wang, X.; Zhang, W.; Liu, R.; Wang, L.; Aslam, M.; Cheng, Y.; et al. Genome-wide identification and expression analysis of the bHLH gene family in passion fruit (Passiflora edulis) and its response to abiotic stress. Int. J. Biol. Macromol. 2023, 225, 389–403. [Google Scholar] [CrossRef]
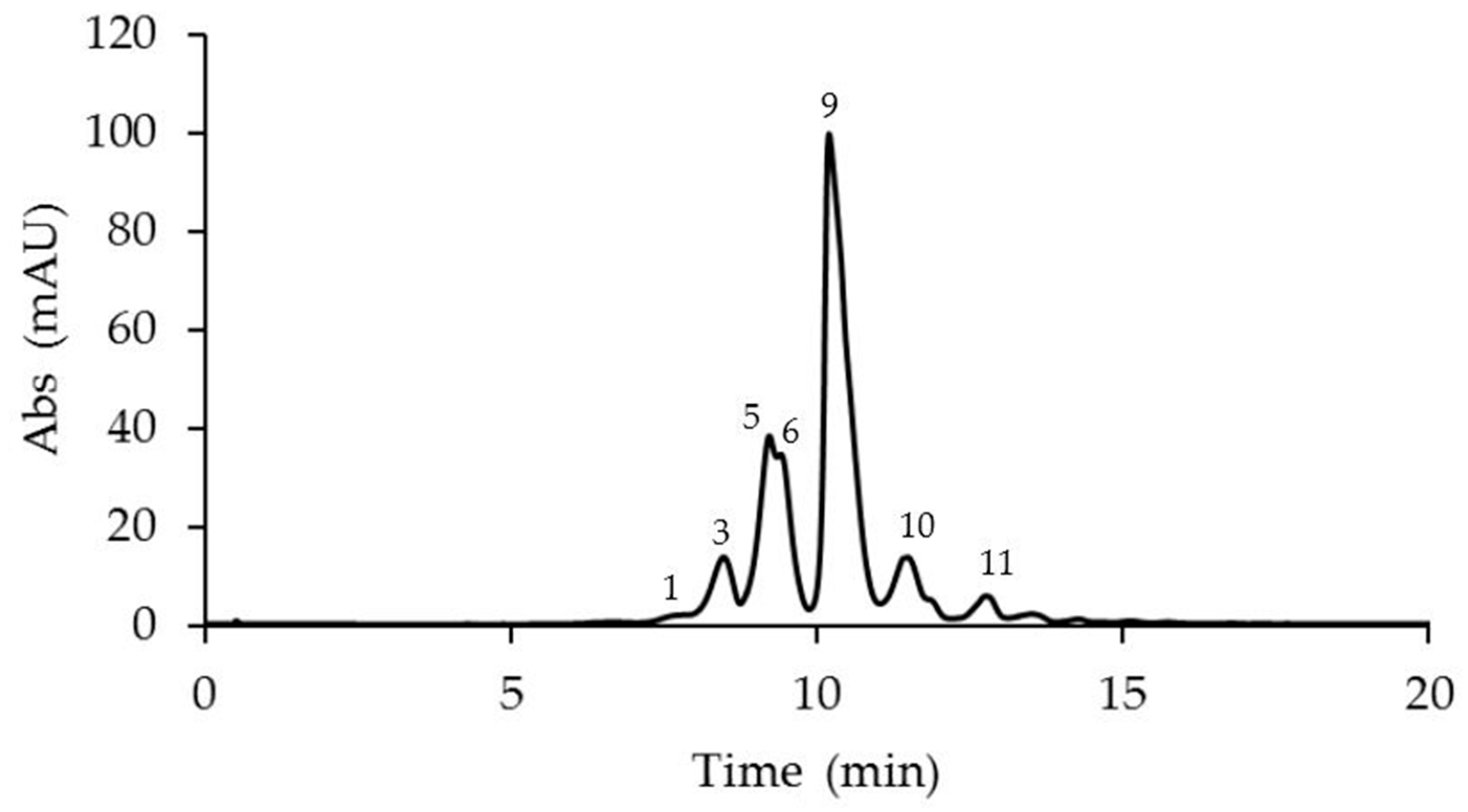
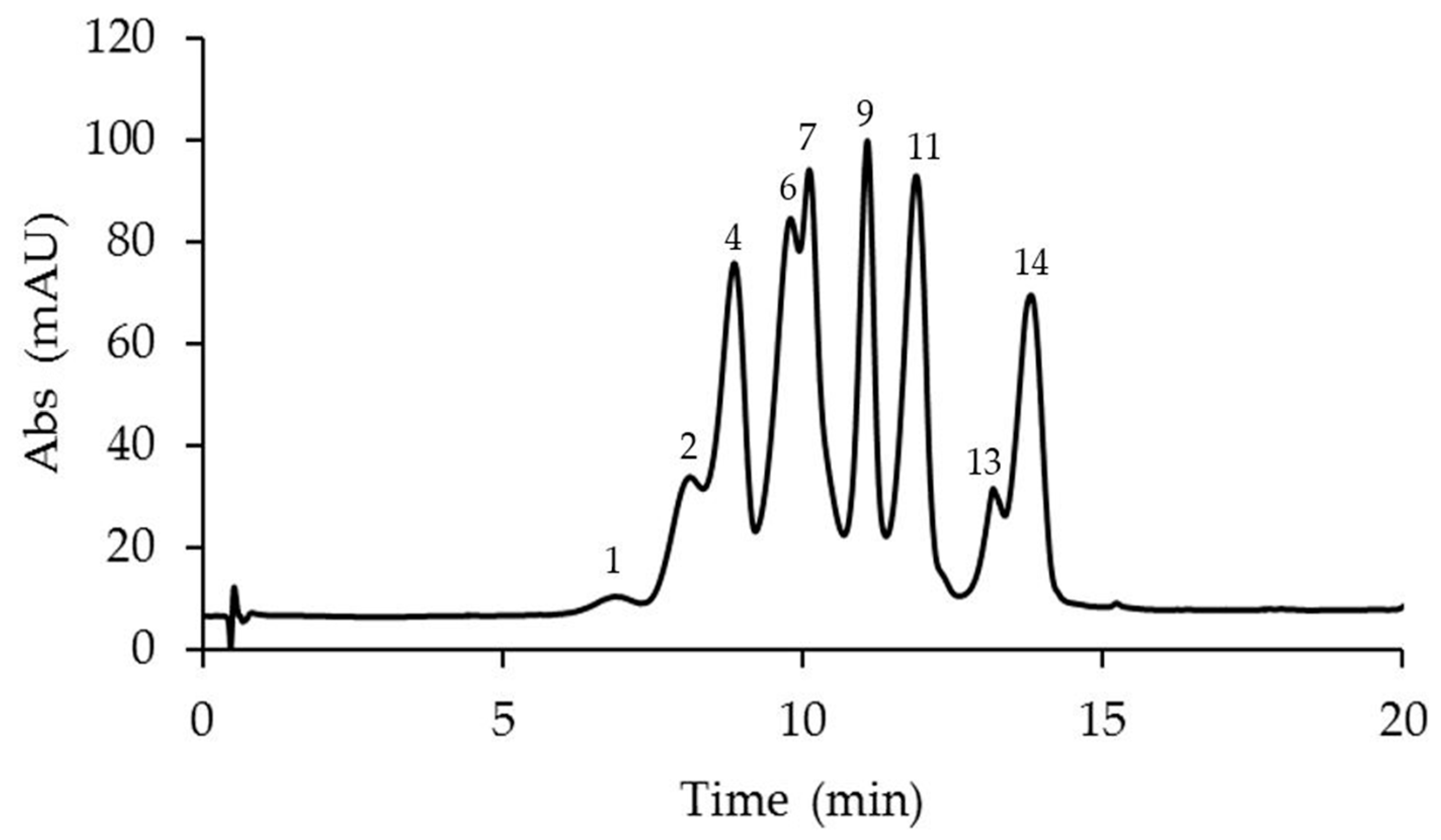
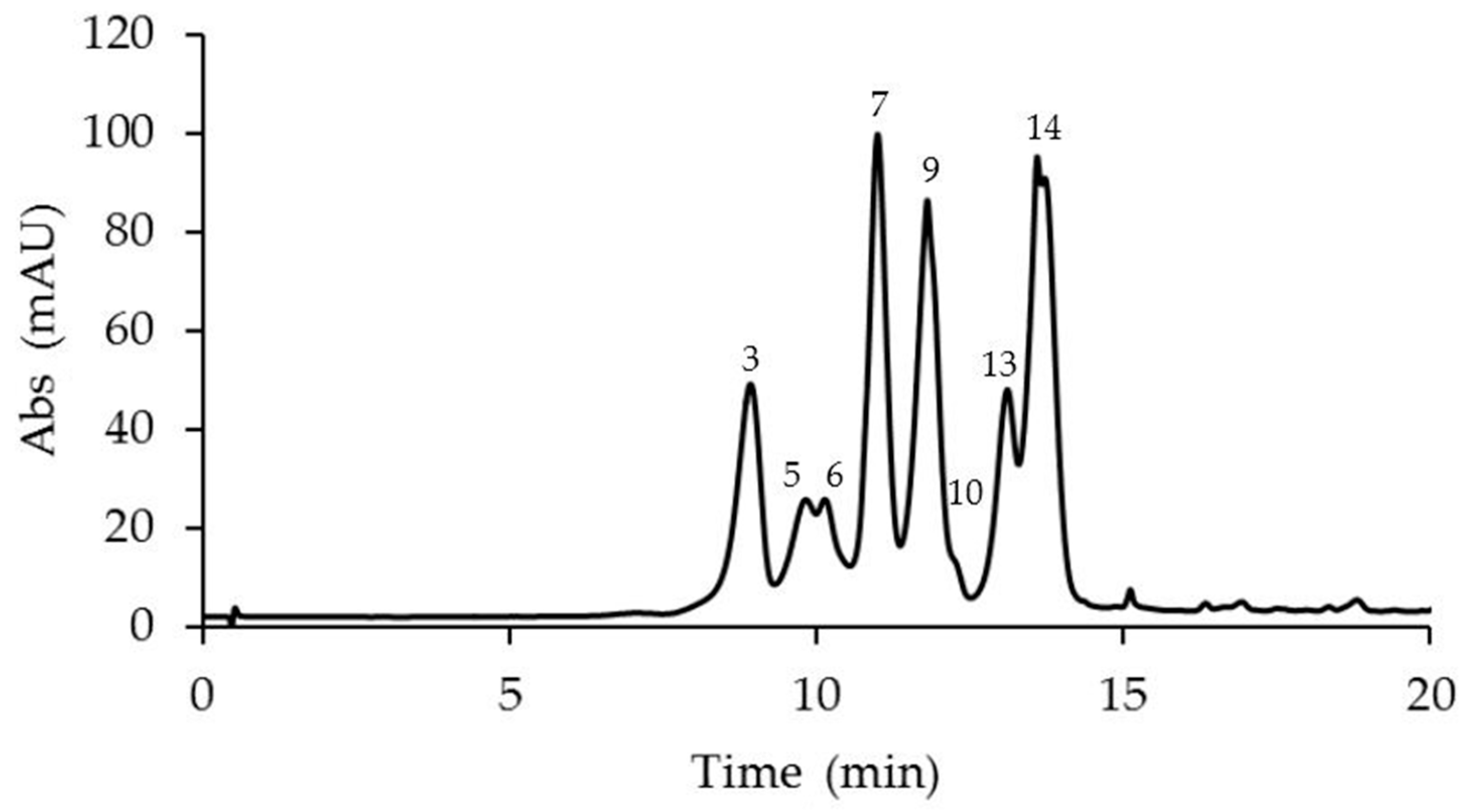
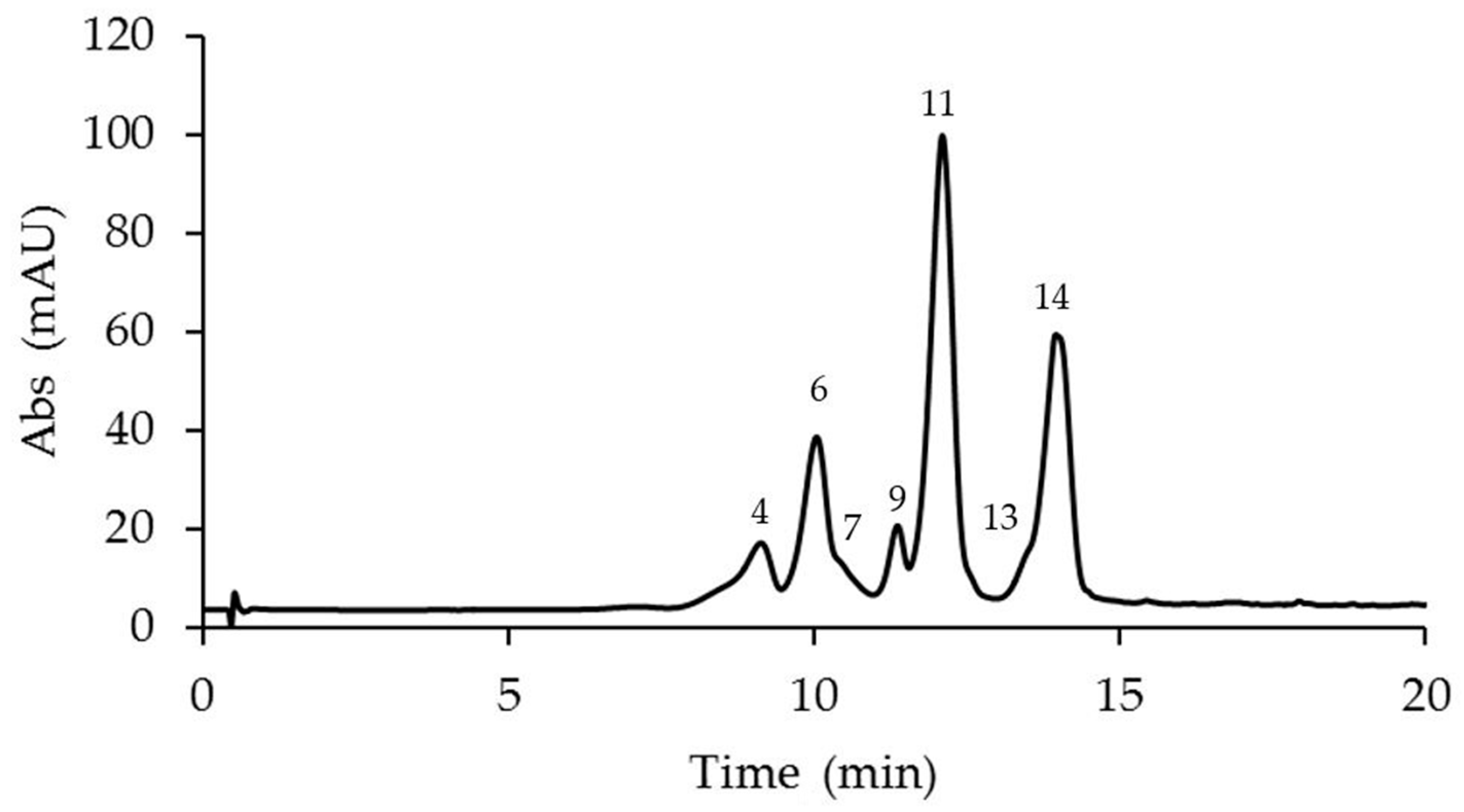


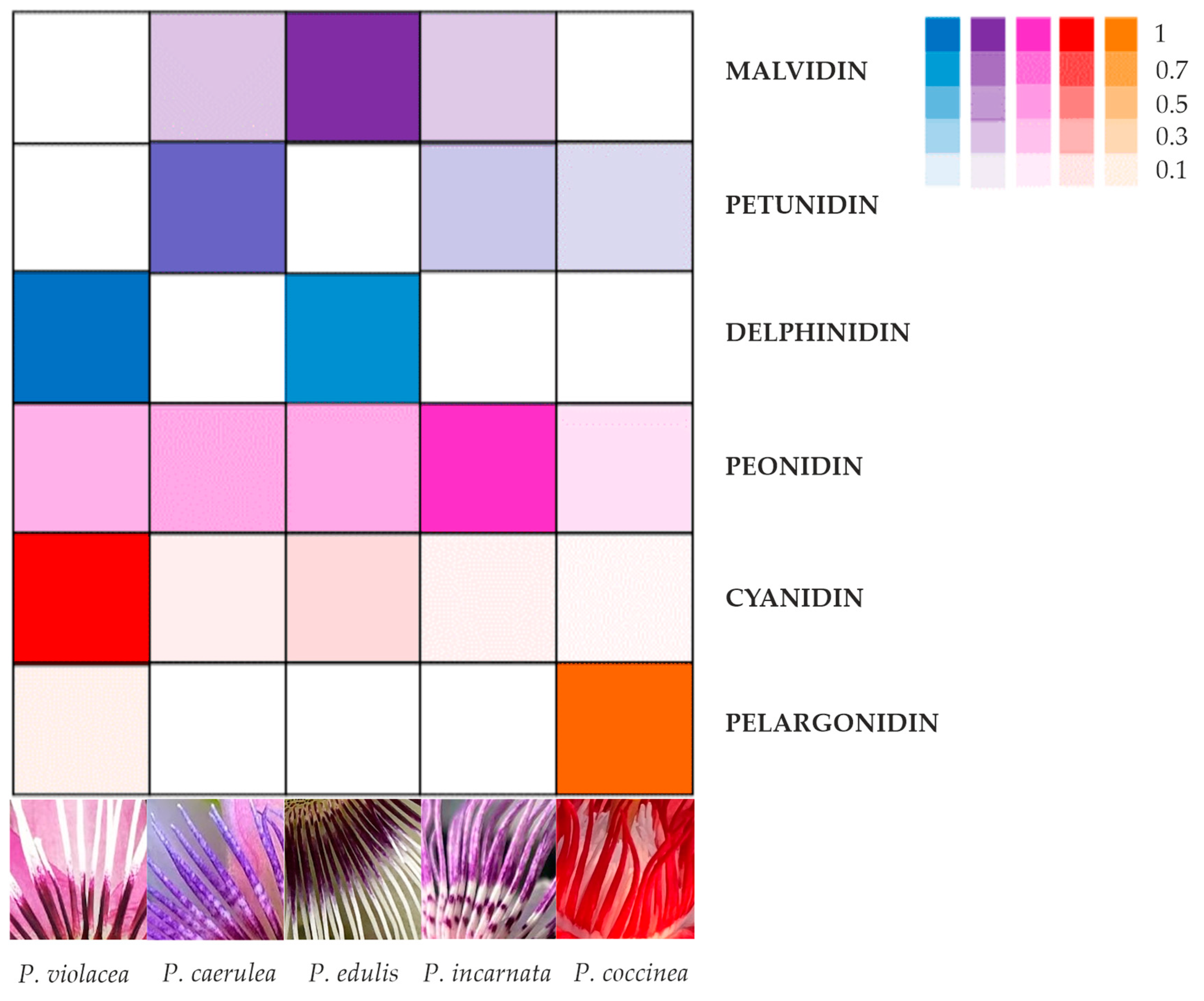
| N. | Compound | M-H+ (Exp.) | M-H+ (Calc.) | Error Δ ppm | Anthocyanins Amount in Passiflora Species (μg/g Fresh Weight) ** | ||||
|---|---|---|---|---|---|---|---|---|---|
| P. violacea | P. caerulea | P. edulis | P. incarnata | P. coccinea | |||||
| 1 | Cyanidin 3-O-sophoroside | 611.1612 | 611.1607 | −0.60 | 20.76 ± 0.61 a | 2.54 ± 0.03 b | <LOD | <LOD | <LOD |
| 2 | Petunidin 3-O-rutinoside | 625.1746 | 625.1763 | 2.77 | <LOD | 15.28 ± 0.21 a | <LOD | <LOD | 4.42 ± 0.34 b |
| 3 | Delphinidin 3-O-glucoside | 465.1030 | 465.1028 | −0.57 | 90.24 ± 0.03 a | <LOD | 71.02 ± 0.25 b | <LOD | <LOD |
| 4 | Petunidin 3,5-O-diglucoside | 641.1717 | 641.1712 | −0.13 | <LOD | 43.07 ± 0.25 a | <LOD | 21.49 ± 0.36 b | <LOD |
| 5 | * Cyanidin 3-O-glucoside | 449.1088 | 449.1078 | −2.17 | 168.5 ± 0.52 a | <LOD | 28.17 ± 0.99 b | <LOD | 29.13 ± 0.36 b |
| 6 | * Cyanidin 3,5-O-diglucoside | 611.1610 | 611.1607 | −0.51 | 176.15 ± 0.38 a | 37.78 ± 0.86 b | 29.56 ± 0.92 b | 38.46 ± 0.21 b | <LOD |
| 7 | Malvidin 3,5-O-diglucoside | 655.1867 | 655.1869 | 0.69 | <LOD | 40.91 ± 0.33 b | 117.11 ± 0.87 a | 9.27 ± 0.84 c | <LOD |
| 8 | Pelargonidin 3-O-glucoside | 433.1131 | 433.1129 | −0.52 | <LOD | <LOD | <LOD | <LOD | 528.55 ± 0.45 a |
| 9 | Cyanidin 3-O-rutinoside | 595.1666 | 595.1657 | −1.46 | 758.12 ± 0.55 a | 35.43 ± 0.58 c | 110.38 ± 0.69 b | 12.5 ± 0.31 d | <LOD |
| 10 | Pelargonidin 3,5-O-diglucoside | 595.1662 | 595.1657 | −1.38 | 136.39 ± 0.21b | <LOD | 8.50 ± 0.20 c | <LOD | 778.84 ± 0.35 a |
| 11 | Peonidin 3,5-O-diglucoside | 625.1777 | 625.1763 | −1.15 | 46.05 ± 0.54 b | 43.17 ± 0.29 b | <LOD | 110.38 ± 0.82 a | <LOD |
| 12 | Pelargonidin 3-O-rutinoside | 579.1722 | 579.1708 | −2.41 | <LOD | <LOD | <LOD | <LOD | 206.87 ± 0.38 a |
| 13 | Peonidin 3-O-rutinoside | 609.1814 | 609.1814 | 0 | <LOD | 8.07 ± 0.32 c | 51.34 ± 0.38 a | 10.14 ± 0.41 bc | 19.42 ± 0.30 b |
| 14 | Malvidin 3-O-rutinoside | 639.1922 | 639.1920 | −0.42 | <LOD | 32.54 ± 0.49 c | 135.72 ± 0.81 a | 60.76 ± 0.59 b | <LOD |
| Compound | Equation of Curve | R2 | Linear Range μg/ml | LOD μg/ml | LOQ μg/ml | RSD% Conc. Intra-Day (n = 5) | RSD% Conc. Inter-Day (n = 5) |
|---|---|---|---|---|---|---|---|
| Cy3gluc | Y = 377,129x – 94,124 | 0.999 | 1–80 | 0.3 | 1 | 0.21% | 0.85% |
| Cy3,5digluc | Y = 381,475x – 85,623 | 0.998 | 1–80 | 0.3 | 1 | 0.23% | 0.93% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nutricati, E.; Sabella, E.; Negro, C.; Min Allah, S.; Luvisi, A.; De Bellis, L.; Accogli, R.A. Anthocyanins and Anthocyanin Biosynthesis Gene Expression in Passiflora Flower Corona Filaments. Plants 2025, 14, 1050. https://doi.org/10.3390/plants14071050
Nutricati E, Sabella E, Negro C, Min Allah S, Luvisi A, De Bellis L, Accogli RA. Anthocyanins and Anthocyanin Biosynthesis Gene Expression in Passiflora Flower Corona Filaments. Plants. 2025; 14(7):1050. https://doi.org/10.3390/plants14071050
Chicago/Turabian StyleNutricati, Eliana, Erika Sabella, Carmine Negro, Samar Min Allah, Andrea Luvisi, Luigi De Bellis, and Rita Annunziata Accogli. 2025. "Anthocyanins and Anthocyanin Biosynthesis Gene Expression in Passiflora Flower Corona Filaments" Plants 14, no. 7: 1050. https://doi.org/10.3390/plants14071050
APA StyleNutricati, E., Sabella, E., Negro, C., Min Allah, S., Luvisi, A., De Bellis, L., & Accogli, R. A. (2025). Anthocyanins and Anthocyanin Biosynthesis Gene Expression in Passiflora Flower Corona Filaments. Plants, 14(7), 1050. https://doi.org/10.3390/plants14071050










