Characterization of OsCAF1 Protein Function in Rice Response to Thermal Stress
Abstract
1. Introduction
2. Results
2.1. High Temperature Induces Re-Localization of OsCAF1 Proteins in Rice Cells
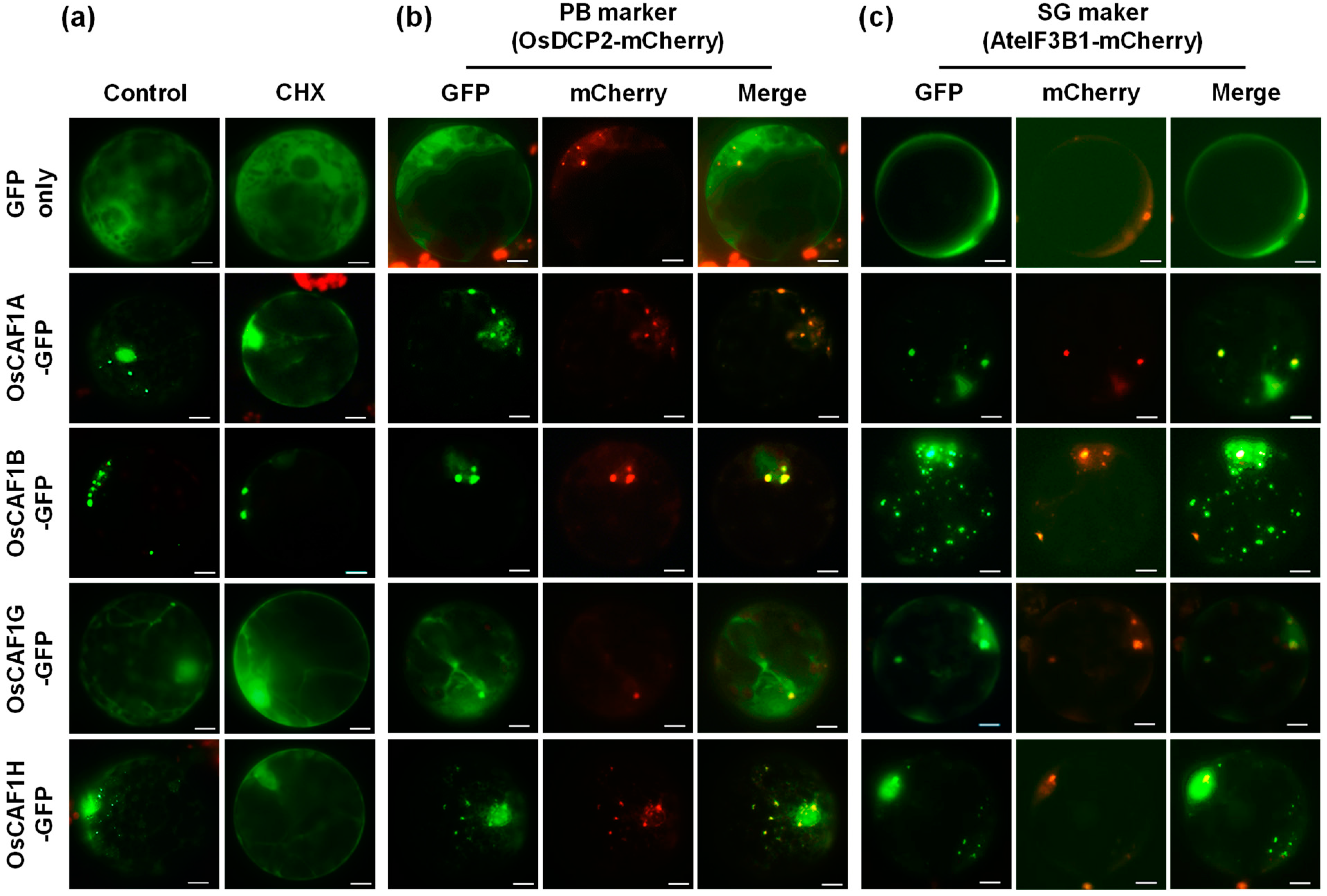
2.2. OsCAF1 Proteins Localize to Processing Bodies and Stress Granules Under High-Temperature Conditions
2.3. OseIF4AIIb and OsRH8 Interacts with OsCAF1 Proteins
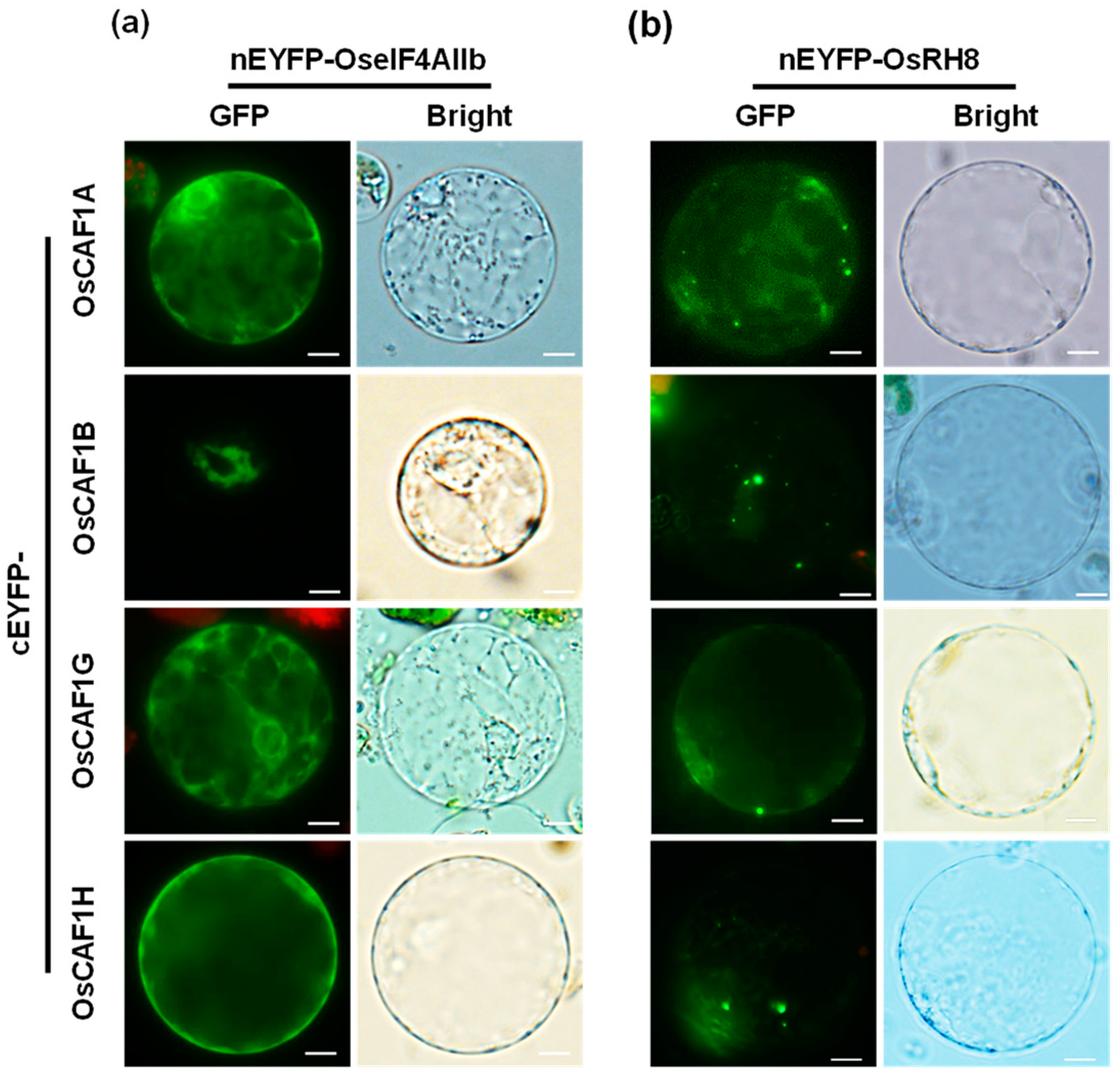
2.4. Subcellular Localization of OseIF4AIIb and OsRH8
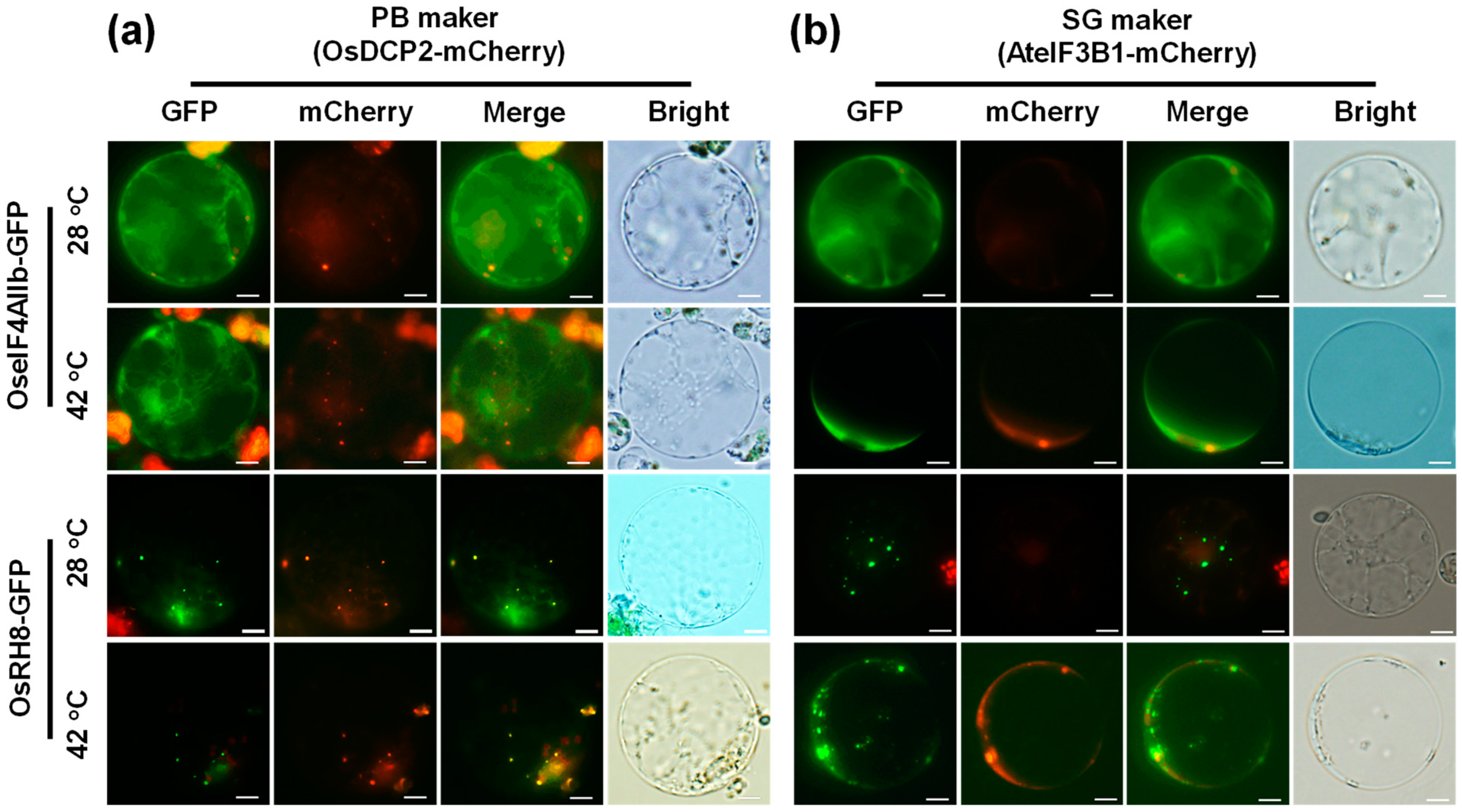
2.5. High Temperature-Mediated Localization of P-Bodies of the OsCAF1s Is Reduced by OseIF4AIIb and Promoted by OsRH8
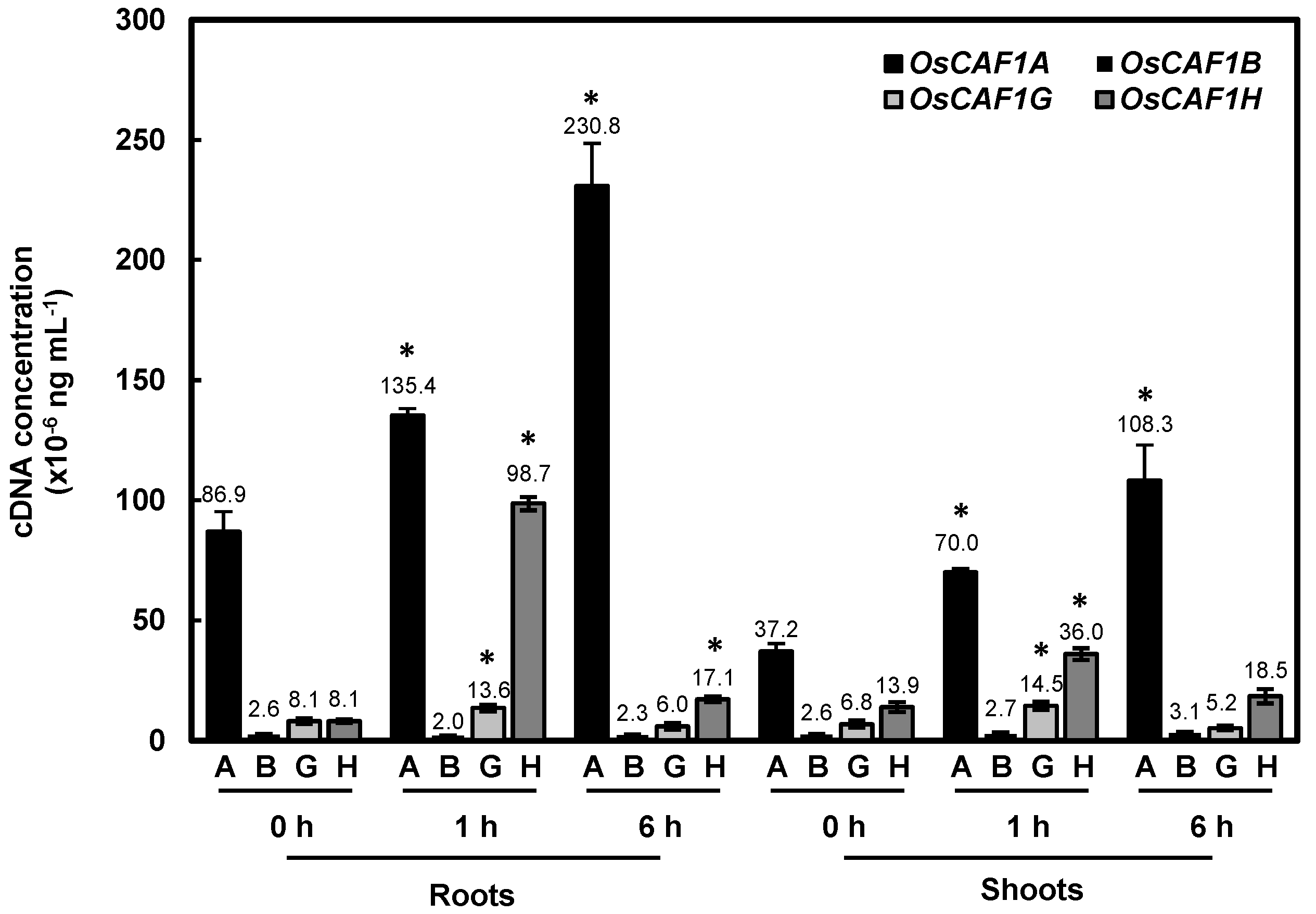
2.6. OsCAF1A mRNA Is Highly Expressed in Rice Seedlings and Further Induced Under High-Temperature Conditions
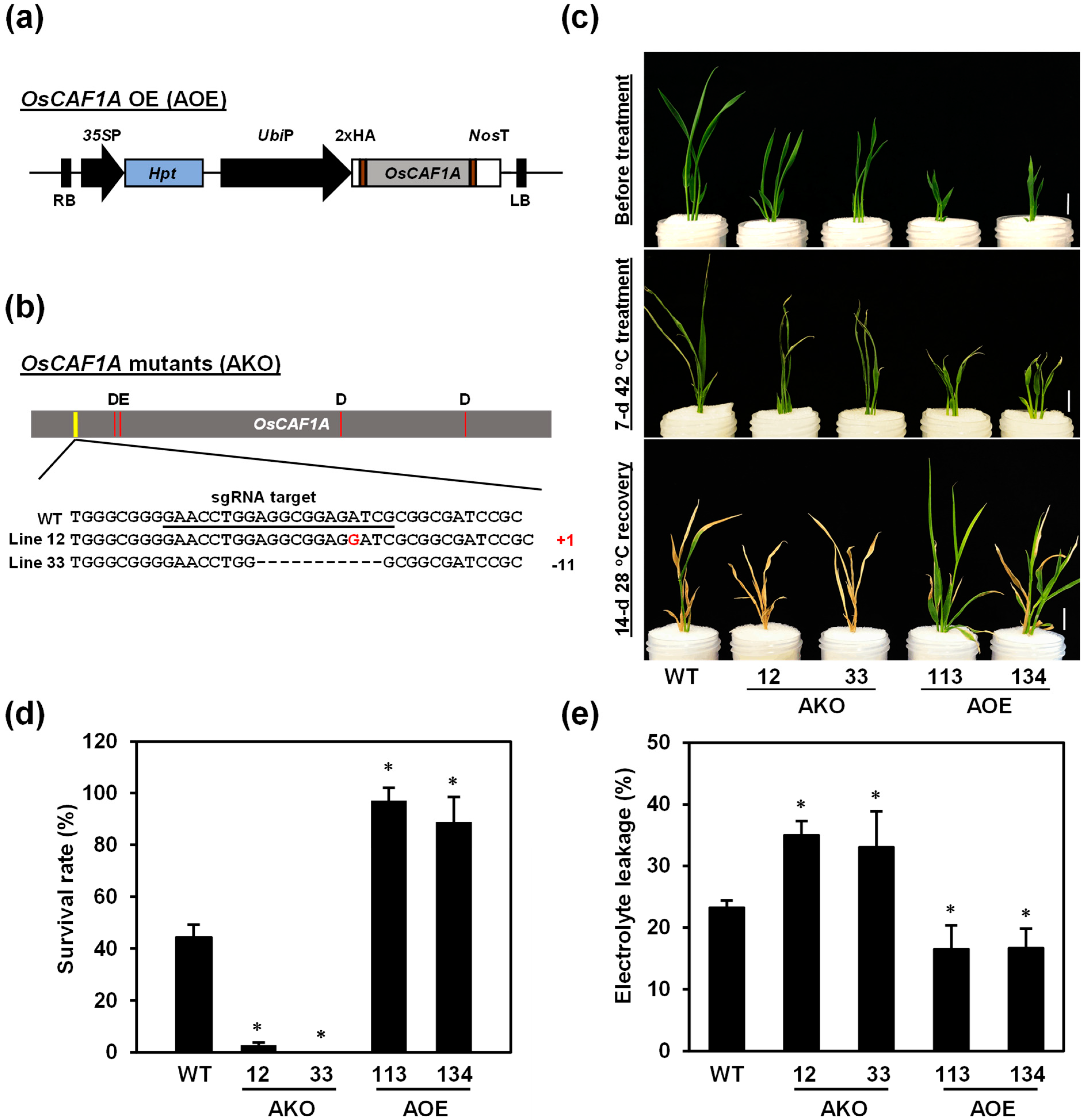
2.7. OsCAF1A Is Required for Rice Seedling Response to High Temperature
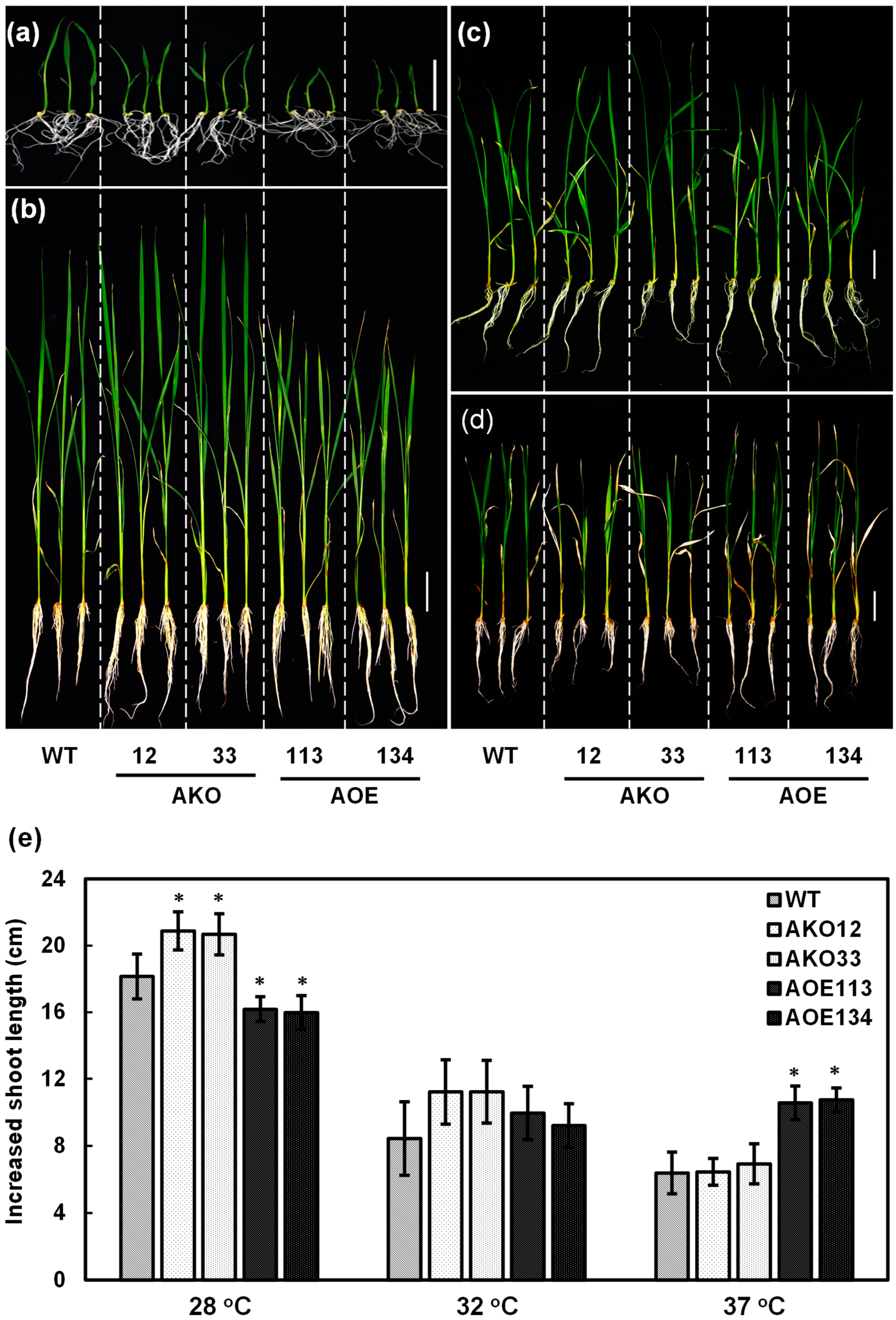
2.8. OsCAF1A Promotes Rice Seedling Growth Under Sublethal High-Temperature Treatment
3. Discussion
3.1. OsCAF1 Proteins as Components of PBs in Rice Under High-Temperature Conditions
3.2. Mechanistic Insights into the Regulation of OsCAF1 Protein Re-Localization by OseIF4AIIb and OsRH8
3.3. OsCAF1A Confers Heat Stress Tolerance but Compromises Growth in Rice
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid
4.3. Plasmid Construction
4.4. RT-PCR and RT-qPCR Analyses
4.5. Standard Curves and Absolute Quantification
4.6. Plant Transformation
4.7. Phenotypic Analysis of Heat Stress-Treated Plants
4.8. Subcellular Localization Analysis and Bimolecular Fluorescence Complementation (BiFC) Assay
4.9. Co-Immunoprecipitation and Mass Spectrometry (IP-MS) Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BiFC | Bimolecular fluorescence complementation |
| CCR4-NOT | Carbon Catabolite Repression 4-Negative On TATA-less |
| CHX | Cycloheximide |
| co-IP | co-immunoprecipitation |
| CRISPR/Cas9 | Clustered regularly interspaced palindromic repeats |
| DMSO | Dimethyl sulfoxide |
| LC-MS/MS | Liquid chromatography-tandem mass spectrometry |
| PB | Processing body |
| SG | Stress granule |
| TNG67 | Tainung 67 |
References
- Eckardt, N.A.; Ainsworth, E.A.; Bahuguna, R.N.; Broadley, M.R.; Busch, W.; Carpita, N.C.; Castrillo, G.; Chory, J.; DeHaan, L.R.; Duarte, C.M. Climate change challenges, plant science solutions. Plant Cell 2023, 35, 24–66. [Google Scholar] [CrossRef] [PubMed]
- Jägermeyr, J.; Müller, C.; Ruane, A.C.; Elliott, J.; Balkovic, J.; Castillo, O.; Faye, B.; Foster, I.; Folberth, C.; Franke, J.A. Climate impacts on global agriculture emerge earlier in new generation of climate and crop models. Nat. Food 2021, 2, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Mahat, J.; Shrestha, J.; Madhav, K.; Paudel, K. Influence of high-temperature stress on rice growth and development. A review. Heliyon 2022, 8, e12651. [Google Scholar] [CrossRef]
- Xu, Y.; Chu, C.; Yao, S. The impact of high-temperature stress on rice: Challenges and solutions. Crop J. 2021, 9, 963–976. [Google Scholar]
- Liu, J.; Hasanuzzaman, M.; Wen, H.; Zhang, J.; Peng, T.; Sun, H.; Zhao, Q. High temperature and drought stress cause abscisic acid and reactive oxygen species accumulation and suppress seed germination growth in rice. Protoplasma 2019, 256, 1217–1227. [Google Scholar]
- Djanaguiraman, M.; Perumal, R.; Jagadish, S.; Ciampitti, I.; Welti, R.; Prasad, P. Sensitivity of sorghum pollen and pistil to high-temperature stress. Plant Cell Environ. 2018, 41, 1065–1082. [Google Scholar] [PubMed]
- Endo, M.; Tsuchiya, T.; Hamada, K.; Kawamura, S.; Yano, K.; Ohshima, M.; Higashitani, A.; Watanabe, M.; Kawagishi-Kobayashi, M. High temperatures cause male sterility in rice plants with transcriptional alterations during pollen development. Plant Cell Physiol. 2009, 50, 1911–1922. [Google Scholar] [CrossRef]
- Wu, C.; Cui, K.; Wang, W.; Li, Q.; Fahad, S.; Hu, Q.; Huang, J.; Nie, L.; Peng, S. Heat-induced phytohormone changes are associated with disrupted early reproductive development and reduced yield in rice. Sci. Rep. 2016, 6, 34978. [Google Scholar] [CrossRef]
- Li, Z.; Li, Z.; Ji, Y.; Wang, C.; Wang, S.; Shi, Y.; Le, J.; Zhang, M. The heat shock factor 20-HSF4-cellulose synthase A2 module regulates heat stress tolerance in maize. Plant Cell 2024, 36, 2652–2667. [Google Scholar]
- Chang, Y.; Fang, Y.; Liu, J.; Ye, T.; Li, X.; Tu, H.; Ye, Y.; Wang, Y.; Xiong, L. Stress-induced nuclear translocation of ONAC023 improves drought and heat tolerance through multiple processes in rice. Nat. Commun. 2024, 15, 5877. [Google Scholar] [CrossRef]
- Liu, X.H.; Lyu, Y.S.; Yang, W.; Yang, Z.T.; Lu, S.J.; Liu, J.X. A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol. J. 2020, 18, 1317–1329. [Google Scholar]
- Zhang, M.; Zhao, R.; Huang, K.; Huang, S.; Wang, H.; Wei, Z.; Li, Z.; Bian, M.; Jiang, W.; Wu, T. The OsWRKY63–OsWRKY76–OsDREB1B module regulates chilling tolerance in rice. Plant J. 2022, 112, 383–398. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Fu, D.; Wang, X.; Zeng, R.; Zhang, X.; Tian, J.; Zhang, S.; Yang, X.; Tian, F.; Lai, J. The transcription factor bZIP68 negatively regulates cold tolerance in maize. Plant Cell 2022, 34, 2833–2851. [Google Scholar]
- Chantarachot, T.; Sorenson, R.S.; Hummel, M.; Ke, H.; Kettenburg, A.T.; Chen, D.; Aiyetiwa, K.; Dehesh, K.; Eulgem, T.; Sieburth, L.E. DHH1/DDX6-like RNA helicases maintain ephemeral half-lives of stress-response mRNAs. Nat. Plants 2020, 6, 675–685. [Google Scholar] [PubMed]
- Fang, J.-C.; Liu, H.-Y.; Tsai, Y.-C.; Chou, W.-L.; Chang, C.-C.; Lu, C.-A. A CCR4 association factor 1, OsCAF1B, participates in the αAmy3 mRNA poly (A) tail shortening and plays a role in germination and seedling growth. Plant Cell Physiol. 2020, 61, 554–564. [Google Scholar] [PubMed]
- Fang, J.-C.; Tsai, Y.-C.; Chou, W.-L.; Liu, H.-Y.; Chang, C.-C.; Wu, S.-J.; Lu, C.-A. A CCR4-associated factor 1, OsCAF1B, confers tolerance of low-temperature stress to rice seedlings. Plant Mol. Biol. 2021, 105, 177–192. [Google Scholar]
- Tiwari, R.; Rajam, M.V. RNA-and miRNA-interference to enhance abiotic stress tolerance in plants. J. Plant Biochem. Biotechnol. 2022, 31, 689–704. [Google Scholar]
- Collart, M.A.; Audebert, L.; Bushell, M. Roles of the CCR4-Not complex in translation and dynamics of co-translation events. Wiley Interdiscip. Rev. RNA 2024, 15, e1827. [Google Scholar] [CrossRef]
- Mostafa, D.; Takahashi, A.; Yanagiya, A.; Yamaguchi, T.; Abe, T.; Kureha, T.; Kuba, K.; Kanegae, Y.; Furuta, Y.; Yamamoto, T. Essential functions of the CNOT7/8 catalytic subunits of the CCR4-NOT complex in mRNA regulation and cell viability. RNA Biol. 2020, 17, 403–416. [Google Scholar] [CrossRef]
- Soeda, S.; Oyama, M.; Kozuka-Hata, H.; Yamamoto, T. The CCR4–NOT complex suppresses untimely translational activation of maternal mRNAs. Development 2023, 150, dev201773. [Google Scholar] [CrossRef]
- Tucker, M.; Valencia-Sanchez, M.A.; Staples, R.R.; Chen, J.; Denis, C.L.; Parker, R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 2001, 104, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Arae, T.; Morita, K.; Imahori, R.; Suzuki, Y.; Yasuda, S.; Sato, T.; Yamaguchi, J.; Chiba, Y. Identification of Arabidopsis CCR4-NOT complexes with pumilio RNA-binding proteins, APUM5 and APUM2. Plant Cell Physiol. 2019, 60, 2015–2025. [Google Scholar]
- Chou, W.-L.; Huang, L.-F.; Fang, J.-C.; Yeh, C.-H.; Hong, C.-Y.; Wu, S.-J.; Lu, C.-A. Divergence of the expression and subcellular localization of CCR4-associated factor 1 (CAF1) deadenylase proteins in Oryza sativa. Plant Mol. Biol. 2014, 85, 443–458. [Google Scholar] [PubMed]
- Walley, J.W.; Kelley, D.R.; Nestorova, G.; Hirschberg, D.L.; Dehesh, K. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol. 2010, 152, 866–875. [Google Scholar] [PubMed]
- Wang, P.; Zhou, J.; Sun, W.; Li, H.; Rehman, S.; Xu, C.; Li, D.; Zhuge, Q. Poplar CCR4-associated factor PtCAF1I is necessary for poplar development and defense response. Int. J. Biol. Macromol. 2023, 242, 125090. [Google Scholar]
- Chantarachot, T.; Bailey-Serres, J. Polysomes, stress granules, and processing bodies: A dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol. 2018, 176, 254–269. [Google Scholar]
- Ivanov, P.; Kedersha, N.; Anderson, P. Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol. 2019, 11, a032813. [Google Scholar]
- Chen, C.-Y.A.; Shyu, A.-B. Deadenylation and P-bodies. Ten Years Prog. GW/P Body Res. 2012, 768, 183–195. [Google Scholar]
- Song, X.-H.; Liao, X.-Y.; Zheng, X.-Y.; Liu, J.-Q.; Zhang, Z.-W.; Zhang, L.-N.; Yan, Y.-B. Human Ccr4 and Caf1 deadenylases regulate proliferation and tumorigenicity of human gastric cancer cells via modulating cell cycle progression. Cancers 2021, 13, 834. [Google Scholar] [CrossRef]
- Lu, C.-A.; Huang, C.-K.; Huang, W.-S.; Huang, T.-S.; Liu, H.-Y.; Chen, Y.-F. DEAD-box RNA helicase 42 plays a critical role in pre-mRNA splicing under cold stress. Plant Physiol. 2020, 182, 255–271. [Google Scholar]
- Li, X.; Li, C.; Zhu, J.; Zhong, S.; Zhu, H.; Zhang, X. Functions and mechanisms of RNA helicases in plants. J. Exp. Bot. 2023, 74, 2295–2310. [Google Scholar] [CrossRef] [PubMed]
- Wilczynska, A.; Gillen, S.L.; Schmidt, T.; Meijer, H.A.; Jukes-Jones, R.; Langlais, C.; Kopra, K.; Lu, W.-T.; Godfrey, J.D.; Hawley, B.R. eIF4A2 drives repression of translation at initiation by Ccr4-Not through purine-rich motifs in the 5′ UTR. Genome Biol. 2019, 20, 262. [Google Scholar] [CrossRef] [PubMed]
- Meijer, H.A.; Schmidt, T.; Gillen, S.L.; Langlais, C.; Jukes-Jones, R.; de Moor, C.H.; Cain, K.; Wilczynska, A.; Bushell, M. DEAD-box helicase eIF4A2 inhibits CNOT7 deadenylation activity. Nucleic Acids Res. 2019, 47, 8224–8238. [Google Scholar] [CrossRef]
- Teixeira, D.; Parker, R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 2274–2287. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Ezzeddine, N.; Chen, C.-Y.A.; Zhu, W.; He, X.; Shyu, A.-B. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 2008, 182, 89–101. [Google Scholar] [CrossRef]
- Shi, J.X.; Li, J.S.; Hu, R.; Zhao, X.C.; Liang, C.C.; Li, X.M.; Wang, H.; Shi, Y.; Su, X. CNOT1 is involved in TTP-mediated ICAM-1 and IL-8 mRNA decay. Mol. Med. Rep. 2018, 18, 2321–2327. [Google Scholar] [CrossRef]
- Gutierrez-Beltran, E.; Moschou, P.N.; Smertenko, A.P.; Bozhkov, P.V. Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 2015, 27, 926–943. [Google Scholar] [CrossRef]
- Tsuda, K.; Suzuki, T.; Mimura, M.; Nonomura, K.-I. Comparison of constitutive promoter activities and development of maize ubiquitin promoter-and Gateway-based binary vectors for rice. Plant Biotechnol. 2022, 39, 139–146. [Google Scholar] [CrossRef]
- Suzuki, Y.; Arae, T.; Green, P.J.; Yamaguchi, J.; Chiba, Y. AtCCR4a and AtCCR4b are involved in determining the poly (A) length of granule-bound starch synthase 1 transcript and modulating sucrose and starch metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 863–874. [Google Scholar] [CrossRef]
- Yilmazer, I.; Abt, M.R.; Liang, Y.; Seung, D.; Zeeman, S.C.; Sharma, M. Determining Protein-Protein Interaction with GFP-Trap Beads. In Fluorescent Proteins: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2022; pp. 317–323. [Google Scholar]
- Liu, H.; Sadygov, R.G.; Yates, J.R. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004, 76, 4193–4201. [Google Scholar] [CrossRef]
- Meijer, H.; Kong, Y.; Lu, W.; Wilczynska, A.; Spriggs, R.; Robinson, S.; Godfrey, J.; Willis, A.; Bushell, M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 2013, 340, 82–85. [Google Scholar]
- Coller, J.M.; Tucker, M.; Sheth, U.; Valencia-Sanchez, M.A.; Parker, R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 2001, 7, 1717–1727. [Google Scholar] [PubMed]
- Hata, H.; Mitsui, H.; Liu, H.; Bai, Y.; Denis, C.L.; Shimizu, Y.; Sakai, A. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 1998, 148, 571–579. [Google Scholar] [PubMed]
- Lu, W.-T.; Wilczynska, A.; Smith, E.; Bushell, M. The diverse roles of the eIF4A family: You are the company you keep. Biochem. Soc. Trans. 2014, 42, 166–172. [Google Scholar] [PubMed]
- Nyikó, T.; Auber, A.; Bucher, E. Functional and molecular characterization of the conserved Arabidopsis PUMILIO protein, APUM9. Plant Mol. Biol. 2019, 100, 199–214. [Google Scholar]
- Shim, J.S.; Park, S.-H.; Lee, D.-K.; Kim, Y.S.; Park, S.-C.; Redillas, M.C.F.R.; Seo, J.S.; Kim, J.-K. The rice GLYCINE-RICH PROTEIN 3 confers drought tolerance by regulating mRNA stability of ROS scavenging-related genes. Rice 2021, 14, 31. [Google Scholar] [CrossRef]
- Dubourg-Felonneau, G.; Abbasi, A.; Akiva, E.; Lee, L. Improving protein subcellular localization prediction with structural prediction & graph neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Maldonado-Bonilla, L.D. Composition and function of P bodies in Arabidopsis thaliana. Front. Plant Sci. 2014, 5, 201. [Google Scholar]
- Zhou, G.; Niu, R.; Zhou, Y.; Luo, M.; Peng, Y.; Wang, H.; Wang, Z.; Xu, G. Proximity editing to identify RNAs in phase-separated RNA binding protein condensates. Cell Discov. 2021, 7, 72. [Google Scholar]
- Nguyen, T.M.; Lu, C.-A.; Huang, L.-F. Applications of CRISPR/Cas9 in a rice protein expression system via an intron-targeted insertion approach. Plant Sci. 2022, 315, 111132. [Google Scholar]
- Chou, W.-L.; Chung, Y.-L.; Fang, J.-C.; Lu, C.-A. Novel interaction between CCR4 and CAF1 in rice CCR4–NOT deadenylase complex. Plant Mol. Biol. 2017, 93, 79–96. [Google Scholar] [PubMed]
- Huang, C.-K.; Sie, Y.-S.; Chen, Y.-F.; Huang, T.-S.; Lu, C.-A. Two highly similar DEAD box proteins, OsRH2 and OsRH34, homologous to eukaryotic initiation factor 4AIII, play roles of the exon junction complex in regulating growth and development in rice. BMC Plant Biol. 2016, 16, 84. [Google Scholar]
- Yokosho, K.; Yamaji, N.; Ma, J.F. Buckwheat FeNramp5 mediates high manganese uptake in roots. Plant Cell Physiol. 2021, 62, 600–609. [Google Scholar] [PubMed]


| Accession of Gene | Gene Name | MW [kDa] | Normalized PSMs | Gene Description | |
|---|---|---|---|---|---|
| OsCAF1A -GFP | GFP Only | ||||
| XP_015614670.1 | OsCCR4a | 67.2 | 65.1 | - | Carbon catabolite repressor protein 4 homolog 1 |
| XP_025879986.1 | OsCCR4b | 67.5 | 44.0 | - | Carbon catabolite repressor protein 4 homolog 1 |
| XP_015614203.1 | OsNOT1 | 267.6 | 118.2 | - | CCR4-NOT transcription complex subunit 1 isoform X1 |
| XP_015627126.1 | OsNOT2a | 66.3 | 53.2 | - | Probable NOT transcription complex subunit VIP2 |
| XP_015630454.1 | OsNOT2 | 60.9 | 63.2 | - | Probable NOT transcription complex subunit VIP2 isoform X3 |
| XP_015630453.1 | OsNOT2b | 66.5 | 63.0 | - | NOT2/NOT3/NOT5 family protein |
| XP_015629837.1 | OsNOT9c | 35.6 | 30.2 | - | CCR4-NOT transcription complex subunit 9 |
| XP_025883045.1 | OsNOT9 | 35.2 | 19.2 | - | CCR4-NOT transcription complex subunit 9 isoform X2 |
| Accession of Gene | Accession Number of Proteins | MW [kDa] | Normalized PSMs | Gene Description | |
|---|---|---|---|---|---|
| OsCAF1A -GFP | GFP Only | ||||
| XP_015626460.1 | Q6Z2Z4 | 47.1 | 62.3 | - | Eukaryotic initiation factor 4AIIb |
| XP_015627069.1 | Q6H7S | 58.1 | 39.4 | - | DEAD-box ATP-dependent RNA helicase 8-like |
| XP_015650041.1 | P41095 | 34.4 | 34.8 | - | 60S acidic ribosomal protein P0-like |
| XP_015632031.1 | Q10RW9 | 61.0 | 33.0 | - | Chaperonin CPN60-1, mitochondrial |
| XP_015614255.1 | Q7XE16 | 89.8 | 29.3 | - | Cell division control protein 48 homolog E |
| XP_015632250.1 | C7IZI5 | 44.5 | 24.7 | - | 60S ribosomal protein L4 |
| XP_015630377.1 | A3AIN7 | 28.0 | 22.9 | - | 40S ribosomal protein S6 |
| XP_025881412.1 | B7F8T1 | 27.3 | 21.0 | - | Uncharacterized protein LOC9269814 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, V.-B.; Lu, C.-A. Characterization of OsCAF1 Protein Function in Rice Response to Thermal Stress. Plants 2025, 14, 1036. https://doi.org/10.3390/plants14071036
Nguyen V-B, Lu C-A. Characterization of OsCAF1 Protein Function in Rice Response to Thermal Stress. Plants. 2025; 14(7):1036. https://doi.org/10.3390/plants14071036
Chicago/Turabian StyleNguyen, Vu-Bao, and Chung-An Lu. 2025. "Characterization of OsCAF1 Protein Function in Rice Response to Thermal Stress" Plants 14, no. 7: 1036. https://doi.org/10.3390/plants14071036
APA StyleNguyen, V.-B., & Lu, C.-A. (2025). Characterization of OsCAF1 Protein Function in Rice Response to Thermal Stress. Plants, 14(7), 1036. https://doi.org/10.3390/plants14071036




