Recent Advances in Molecular Tools and Pre-Breeding Activities in White Lupin (Lupinus albus)
Abstract
1. Introduction
2. PGRs in Food Legumes and Plant Breeding Perspective
- Legume Generation (2023–2028) aims to boost the breeding of major food and feed legume crops in Europe to address the protein gap and support biodiversity, such as soybean, lupin, pea, lentil (blue, white and Andean), common bean and clover. The project involves 32 partners from 16 countries. It is funded by the European Union (EU) and UK Research and Innovation, with a total budget of EUR 8.6 million [55].
- BELIS (2023-2028) focuses on enhancing the competitiveness of the legume breeding industry in the EU and associated countries. The project aims to develop cost-effective breeding methodologies, improve regulatory frameworks for variety registration, and facilitate innovation transfer among breeders, seed industries, and agricultural stakeholders. By promoting sustainable practices and increasing the availability of high-quality legume varieties (14 species), BELIS seeks to support the production of food and feed while delivering ecosystem services, working both on forage crops (lucerne, red clover, white clover, annual clovers, sainfoin, birdsfoot trefoil and vetches) and grain crops (pea, fava bean, soybean, white lupin, lentil, chickpea and common bean) [56].
- EUROLEGUME (2014–2017) focused on enhancing legume cultivation in Europe through sustainable cropping practices, selection, characterisation and the identification of genetic resources for fava beans, peas, and cowpeas, with the goal of improving their production, developing novel food products and optimising agricultural management for different agro-climatic conditions [57].
- ABSTRESS (2012–2016) aimed to improve legume crop resistance to combined abiotic and biotic stresses, delivering new plant varieties with greater drought and disease tolerance. This initiative involved multiple national and international partners and sought to enhance the sustainability of European agriculture by reducing reliance on imported protein sources. The project primarily focused on legume species, including pea (Pisum sativum), and Medicago truncatula, a small low-growing clover-like legume native to the Mediterranean region. The last is a fast-growing plant commonly used as a model for legume crops [58].
- EXTRULEG (2013–2017) developed extruded products from legumes (i.e., pea, common bean, carob) and cereals (i.e., rice), through fortification using dietary fiber, and it focused on the nutritional impacts and childhood obesity prevention. The project explored innovative food processing methods to enhance the nutritional value of legume-based snacks [59].
- CropExplore (2018–2022) explored alternative raw materials for the food sector, focusing on nutritional qualities and functionality of various legumes. The project sought to create a knowledge matrix of known and lesser-known crops/raw materials [59].
- LupinChallenge (2012–2015) characterised beta-conglutin seed proteins in lupin, to assess their nutritional value and impact on human health, including allergenicity. Overall, this project sought to improve lupin as a safe and nutritious plant-based protein source [60].
- LegumES project (2024–2027) develops multi-actor driven approaches for monitoring the benefits of legume crops, focusing on beans, chickpea and lentil), legume-based crop rotations and wild legumes in semi-natural ecosystems [61].
- EVA Legumes Network (2024–2027) aims to increase the use of crop genetic diversity in legume breeding and promote legume diversity for sustainable plant-based protein production in Europe [62]. The project covers seven legume crops: beans, chickpeas, fava beans, lentils, lupins, peas, and orphan legumes.
- LegValue project (2017–2021) focused on enhancing the economic viability and sustainability of legume production, addressing the protein gap in European agriculture by improving the quality and marketability of several legume species (i.e., fava beans, lucerne, lupins, peas and soybeans) and promoting their cultivation and consumption. The project involved collaboration among researchers, farmers, and industry stakeholders to develop innovative practices and products [63].
- Legumes Translated (2018–2022) supported the production and use of grain legumes, helping farmers benefit from relevant research, particularly those funded by the European Union [64].
- LEGUMINOSE (2022–2026) promotes legume-based intercropping as a climate-smart farming practice, providing science-based and economically viable systems for European farmers. The project relies on different species and mixture according to the country where the research is conducted (e.g., in Poland, a common intercropping mixture involves cereals, such as oats, barley, wheat, and sometimes maize, paired with legumes such as peas, lupins, fava beans, and sometimes soybeans). The project will (1) investigate the benefits of intercropping beyond the well-studied effects on nitrogen dynamics, (2) identify obstacles and issues related to the adoption of the intercropping practice, and (3) provide farmers across the EU with accessible, actionable and science-based information for a profitable and sustainable agricultural transformation [65].
- DemoNetErBo (2016–2021) established a knowledge transfer network for field peas and fava bean cultivation and utilisation, connecting conventional and organic farmers. The project aimed to build value chains, from breeding to consumer usage, meeting the increasing demand for non-GMO protein crops [66].
- ProVeg (2017-still ongoing) is a global initiative promoting plant-based and cultivated foods, reducing global animal product consumption by 50%, by 2040. ProVeg engages with various stakeholders to foster awareness and facilitate the transition to sustainable diets, emphasising the benefits of healthy plant-based alternatives [6].
- BRESOV (2018–2022) enhanced organic vegetable production by improving breeding and availability of organic seeds for broccoli, snap beans, and tomatoes. The project aimed to explore genetic diversity and increase the crops’ resilience to biotic and abiotic stresses, adapting them to organic farming practices. By collaborating with researchers, breeders, farmers, and stakeholders across Europe, Africa, and Asia, BRESOV promoted sustainable agriculture to meet the growing demand for high-quality organic vegetables [67].
- BEAN ADAPT (2015–2018) contributed to improving resilience and adaptability of common bean (Phaseolus vulgaris) varieties to climate change and variable growing conditions. The project led to a publication in which the complex evolutionary history of common bean has been further dissected, shedding light on the mechanisms behind the successful introduction, dissemination and adaptation of common bean in Europe [68]. Collaborating with farmers, researchers, and industry stakeholders, BEAN ADAPT seeks to improve food security and sustainability in bean production systems across different regions [69].
3. Lupin: A Large Genus of Interesting and Underutilised Species
4. White Lupin
5. Incorporating Landrace Traits in Breeding Programs: Benefits and Limitations
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Grafton, R.Q.; Daugbjerg, C.; Qureshi, M.E. Towards Food Security by 2050. Food Secur. 2015, 7, 179–183. [Google Scholar] [CrossRef]
- FAO. The Future of Food and Agriculture—Alternative Pathways to 2050; Summary Version; FAO: Rome, Italy, 2018; p. 60. [Google Scholar]
- Hopf, E. The Global Significance of Genetic Resources for Food Security. J. Biodivers. Bioprospect. Dev. 2023, 9. [Google Scholar]
- Calicioglu, O.; Flammini, A.; Bracco, S.; Bellù, L.; Sims, R. The Future Challenges of Food and Agriculture: An Integrated Analysis of Trends and Solutions. Sustainability 2019, 11, 222. [Google Scholar] [CrossRef]
- Gerten, D.; Heck, V.; Jägermeyr, J.; Bodirsky, B.L.; Fetzer, I.; Jalava, M.; Kummu, M.; Lucht, W.; Rockström, J.; Schaphoff, S.; et al. Feeding Ten Billion People Is Possible within Four Terrestrial Planetary Boundaries. Nat. Sustain. 2020, 3, 200–208. [Google Scholar] [CrossRef]
- ProVeg. Available online: https://proveg.org/ (accessed on 8 October 2024).
- Green, A.; Blattmann, C.; Chen, C.; Mathys, A. The Role of Alternative Proteins and Future Foods in Sustainable and Contextually-Adapted Flexitarian Diets. Trends Food Sci. Technol. 2022, 124, 250–258. [Google Scholar] [CrossRef]
- Foyer, C.H.; Lam, H.M.; Nguyen, H.T.; Siddique, K.H.M.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M.; et al. Neglecting Legumes Has Compromised Human Health and Sustainable Food Production. Nat. Plants 2016, 2, 16112. [Google Scholar] [CrossRef] [PubMed]
- Bellucci, E.; Mario Aguilar, O.; Alseekh, S.; Bett, K.; Brezeanu, C.; Cook, D.; De la Rosa, L.; Delledonne, M.; Dostatny, D.F.; Ferreira, J.J.; et al. The INCREASE Project: Intelligent Collections of Food-Legume Genetic Resources for European Agrofood Systems. Plant J. 2021, 108, 646–660. [Google Scholar] [CrossRef]
- Ray, D.K.; Ramankutty, N.; Mueller, N.D.; West, P.C.; Foley, J.A. Recent Patterns of Crop Yield Growth and Stagnation. Nat. Commun. 2012, 3, 1293. [Google Scholar] [CrossRef]
- Semba, R.D.; Ramsing, R.; Rahman, N.; Kraemer, K.; Bloem, M.W. Legumes as a Sustainable Source of Protein in Human Diets. Glob. Food Sec. 2021, 28, 100520. [Google Scholar] [CrossRef]
- FAO. The Second Report on the State of the World’s Plant Genetic for Food and Agriculture; FAO: Rome, Italy, 2010. [Google Scholar]
- FAO. The State of the World’s Biodiversity for Food and Agriculture; Bélanger, J., Pilling, D., Eds.; FAO Commission on Genetic Resources for Food and Agriculture Assessments: Rome, Italy, 2019; p. 572. [Google Scholar]
- Sahu, P.K.; Sao, R.; Khute, I.K.; Baghel, S.; Patel, R.R.S.; Thada, A.; Parte, D.; Devi, Y.L.; Nair, S.; Kumar, V.; et al. Plant Genetic Resources: Conservation, Evaluation and Utilization in Plant Breeding. In Advanced Crop Improvement, Volume 2: Case Studies of Economically Important Crops; Springer: Cham, Switzerland, 2023; pp. 1–45. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Chauhan, B.S. Genetic Diversity, Conservation, and Utilization of Plant Genetic Resources. Genes 2023, 14, 174. [Google Scholar] [CrossRef]
- Vaccino, P.; Antonetti, M.; Balconi, C.; Brandolini, A.; Cappellozza, S.; Caputo, A.R.; Carboni, A.; Caruso, M.; Copetta, A.; de Dato, G.; et al. Plant Genetic Resources for Food and Agriculture: The Role and Contribution of CREA (Italy) within the National Program RGV-FAO. Agronomy 2024, 14, 1263. [Google Scholar] [CrossRef]
- Toro, M.A.; Caballero, A. Characterization and Conservation of Genetic Diversity in Subdivided Populations. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1367. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A.; McInnes, K.H. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar] [CrossRef]
- Korir, N.K.; Han, J.; Shangguan, L.; Wang, C.; Kayesh, E.; Zhang, Y.; Fang, J. Plant Variety and Cultivar Identification: Advances and Prospects. Crit. Rev. Biotechnol. 2013, 33, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Berg, T. Landraces and Folk Varieties: A Conceptual Reappraisal of Terminology. Euphytica 2009, 166, 423–430. [Google Scholar] [CrossRef]
- Reif, J.C.; Zhang, A.P.; Dreisigacker, A.S.; Warburton, M.L.; Van Ginkel, A.M.; Hoisington, A.D.; Bohn, M.; Melchinger, A.A.E. Wheat Genetic Diversity Trends during Domestication and Breeding. Theor. Appl. Genet. 2015, 110, 859–864. [Google Scholar] [CrossRef]
- Cavanagh, C.R.; Chao, S.; Wang, S.; Huang, B.E.; Stephen, S.; Kiani, S.; Forrest, K.; Saintenac, C.; Brown-Guedira, G.L.; Akhunova, A.; et al. Genome-Wide Comparative Diversity Uncovers Multiple Targets of Selection for Improvement in Hexaploid Wheat Landraces and Cultivars. Proc. Natl. Acad. Sci. USA 2013, 110, 8057–8062. [Google Scholar] [CrossRef]
- Tirnaz, S.; Zandberg, J.; Thomas, W.J.W.; Marsh, J.; Edwards, D.; Batley, J. Application of Crop Wild Relatives in Modern Breeding: An Overview of Resources, Experimental and Computational Methodologies. Front. Plant Sci. 2022, 13, 1008904. [Google Scholar] [CrossRef]
- Kapazoglou, A.; Gerakari, M.; Lazaridi, E.; Kleftogianni, K.; Sarri, E.; Tani, E.; Bebeli, P.J. Crop Wild Relatives: A Valuable Source of Tolerance to Various Abiotic Stresses. Plants 2023, 12, 328. [Google Scholar] [CrossRef]
- Ceccarelli, S. Specific Adaptation and Breeding for Marginal Conditions. Euphytica 1994, 77, 205–219. [Google Scholar] [CrossRef]
- Plucknett, D.L.; Smith, N.J.H.; Williams, J.T.; Anishetty, N.M. Crop Germplasm Conservation and Developing Countries. Science 1983, 220, 163–169. [Google Scholar] [CrossRef]
- FAO. International Treaty on Plant Genetic Resources for Food and Agriculture. Available online: https://openknowledge.fao.org/server/api/core/bitstreams/a9d0de2a-8e98-4f75-98a8-673078841030/content (accessed on 9 November 2024).
- Ramanatha Rao, V.; Hodgkin, T. Genetic Diversity and Conservation and Utilization of Plant Genetic Resources. Plant Cell Tissue Organ. Cult. 2002, 68, 1–19. [Google Scholar] [CrossRef]
- FAO; CGIAR. The Biodiversity Plan—Crop Trust. Available online: https://www.croptrust.org/about/the-biodiversity-plan/ (accessed on 7 November 2024).
- Bohra, A.; Kilian, B.; Sivasankar, S.; Caccamo, M.; Mba, C.; McCouch, S.R.; Varshney, R.K. Reap the Crop Wild Relatives for Breeding Future Crops. Trends Biotechnol. 2022, 40, 412–431. [Google Scholar] [CrossRef] [PubMed]
- CGIAR. Genebank Platform—CGIAR. Available online: https://www.cgiar.org/research/program-platform/genebank-platform/ (accessed on 12 November 2024).
- CGIAR. Genebank Platform. Available online: https://www.genebanks.org/genebanks/ (accessed on 10 November 2024).
- Gepts, P. Plant Genetic Resources Conservation and Utilization: The Accomplishments and Future of a Societal Insurance Policy. Crop Sci. 2006, 46, 2278–2292. [Google Scholar] [CrossRef]
- Priyanka, V.; Kumar, R.; Dhaliwal, I.; Kaushik, P. Germplasm Conservation: Instrumental in Agricultural Biodiversity—A Review. Sustainability 2021, 13, 6743. [Google Scholar] [CrossRef]
- Hammer, K.; Gladis, T.; Diederichsen, A. In Situ and On-Farm Management of Plant Genetic Resources. Eur. J. Agron. 2003, 19, 509–517. [Google Scholar] [CrossRef]
- Rajpurohit, D.; Jhang, T. In Situ and Ex Situ Conservation of Plant Genetic Resources and Traditional Knowledge. In Plant Genetic Resources and Traditional Knowledge for Food Security; Springer: Singapore, 2015; pp. 137–162. [Google Scholar] [CrossRef]
- FAO. Conservation and Sustainable Use Under the International Treaty. Available online: https://www.fao.org/4/i2579e/i2579e.pdf (accessed on 10 November 2024).
- Cohen, J.I.; Williams, J.T.; Plucknett, D.L.; Shands, H. Ex Situ Conservation of Plant Genetic Resources: Global Development and Environmental Concerns. Science 1991, 253, 866–872. [Google Scholar] [CrossRef]
- Eastwood, R.J.; Cody, S.; Westengen, O.; Von Bothmer, R. Conservation Roles of the Millennium Seed Bank and the Svalbard Global Seed Vault. In Crop Wild Relatives and Climate Change; Wiley-Blackwell: Hoboken, NJ, USA, 2015; pp. 173–186. [Google Scholar] [CrossRef]
- Zeven, A.C. Landraces: A Review of Definitions and Classifications. Euphytica 1998, 104, 127–139. [Google Scholar] [CrossRef]
- Harlan, J. Crops and Man, 2nd ed.; American Society of Agronomy-Crop Science Society: Madison, WI, USA, 1992. [Google Scholar]
- Bitocchi, E.; Nanni, L.; Rossi, M.; Rau, D.; Bellucci, E.; Giardini, A.; Buonamici, A.; Vendramin, G.G.; Papa, R. Introgression from Modern Hybrid Varieties into Landrace Populations of Maize (Zea mays ssp. mays L.) in Central Italy. Mol. Ecol. 2009, 18, 603–621. [Google Scholar] [CrossRef]
- Villa, T.C.C.; Maxted, N.; Scholten, M.; Ford-Lloyd, B. Defining and Identifying Crop Landraces. Plant Genet. Resour. 2005, 3, 373–384. [Google Scholar] [CrossRef]
- Casañas, F.; Simó, J.; Casals, J.; Prohens, J. Toward an Evolved Concept of Landrace. Front. Plant Sci. 2017, 8, 245305. [Google Scholar] [CrossRef]
- Motley, T.J.; Zerega, N.; Cross, H. Darwin’s Harvest: New Approaches to the Origins, Evolution, and Conservation of Crops; Columbia University Press: New York, NY, USA, 2006. [Google Scholar] [CrossRef]
- Jones, H.; Lister, D.L.; Bower, M.A.; Leigh, F.J.; Smith, L.M.; Jones, M.K. Approaches and Constraints of Using Existing Landrace and Extant Plant Material to Understand Agricultural Spread in Prehistory. Plant Genet. Resour. 2008, 6, 98–112. [Google Scholar] [CrossRef]
- Bhullar, N.K.; Street, K.; Mackay, M.; Yahiaoui, N.; Keller, B. Unlocking Wheat Genetic Resources for the Molecular Identification of Previously Undescribed Functional Alleles at the Pm3 Resistance Locus. Proc. Natl. Acad. Sci. USA 2009, 106, 9519–9524. [Google Scholar] [CrossRef] [PubMed]
- Stephen Baenziger, P.; Depauw, R.M. Wheat Breeding: Procedures and Strategies. In Wheat Science and Trade; Wiley: Hoboken, NJ, USA, 2009; pp. 273–308. [Google Scholar] [CrossRef]
- Clark, S. Plant Guide for White Lupine (Lupinus albus L.); United States Department of Agriculture: Washington, DC, USA, 2014.
- Annicchiarico, P.; de Buck, A.J.; Vlachostergios, D.N.; Heupink, D.; Koskosidis, A.; Nazzicari, N.; Crosta, M. White Lupin Adaptation to Moderately Calcareous Soils: Phenotypic Variation and Genome-Enabled Prediction. Plants 2023, 12, 1139. [Google Scholar] [CrossRef]
- Kezeya Sepngang, B.; Muel, F.; Smadja, T.; Stauss, W. Report on Legume Markets in the EU Deliverable D3.1 of the EU-Project LegValue; Fachhochschule Südwestfalen: Soest, Germany, 2020; ISBN 978-3-94095-689-7. [Google Scholar]
- Pilorgé, E.; Muel, F. What Vegetable Oils and Proteins for 2030? Would the Protein Fraction Be the Future of Oil and Protein Crops? OCL 2016, 23, D402. [Google Scholar] [CrossRef]
- Watson, C.A.; Reckling, M.; Preissel, S.; Bachinger, J.; Bergkvist, G.; Kuhlman, T.; Lindström, K.; Nemecek, T.; Topp, C.F.E.; Vanhatalo, A.; et al. Grain Legume Production and Use in European Agricultural Systems. Adv. Agron. 2017, 144, 235–303. [Google Scholar] [CrossRef]
- European Commission. Market Developments and Policy Evaluation Aspects of the Plant Protein Sector in the EU; Agrosynergie EEIG for the European Commission: Brussels, Belgium, 2018. [CrossRef]
- Legume Generation Project. Available online: https://www.legumegeneration.eu/ (accessed on 12 November 2024).
- Belis Project. Available online: http://www.belisproject.eu/ (accessed on 12 November 2024).
- Eurolegume Project. Available online: https://eurolegume.utad.pt/ (accessed on 12 November 2024).
- ABSTRESS Project. Available online: https://eu-cap-network.ec.europa.eu/projects/abstress-improving-resistance-legume-crops-combined-abiotic-and-biotic-stress_en (accessed on 13 November 2024).
- Projects—The International Legume Society. Available online: https://www.legumesociety.org/post_full_width/projects-all/ (accessed on 13 November 2024).
- LupinChallenge Project. Available online: https://cordis.europa.eu/project/id/301550 (accessed on 13 November 2024).
- Legumes. Available online: https://legumesproject.eu/ (accessed on 13 November 2024).
- ECPGR: Legumes. Available online: https://www.ecpgr.org/eva/eva-networks/legumes (accessed on 13 November 2024).
- LEGVALUE—Legumehub.Eu. Available online: https://www.legumehub.eu/legvalue/ (accessed on 13 November 2024).
- Legumes Translated. Available online: https://www.legumestranslated.eu/ (accessed on 13 November 2024).
- Sustainable Agriculture Through Legume-Cereal Intercropping. Available online: https://www.leguminose.eu/ (accessed on 13 November 2024).
- Legunet. Available online: https://www.legunet.de/ (accessed on 13 November 2024).
- Bresov. Shaping the Future of Organic Breeding & Farming. Available online: https://bresov.eu/ (accessed on 12 November 2024).
- Bellucci, E.; Benazzo, A.; Xu, C.; Bitocchi, E.; Rodriguez, M.; Alseekh, S.; Di Vittori, V.; Gioia, T.; Neumann, K.; Cortinovis, G.; et al. Selection and Adaptive Introgression Guided the Complex Evolutionary History of the European Common Bean. Nat. Commun. 2023, 14, 1908. [Google Scholar] [CrossRef]
- ERA-CAPS. BEAN ADAPT-Evolution in a Changing Environment: The Genetic Architecture of Adaptation Outside Centers of Domestication of Phaseolus vulgaris and P. Coccineus. Available online: https://www.eracaps.org/joint-calls/era-caps-funded-projects/era-caps-second-call-2014/evolution-changing-environment (accessed on 12 November 2024).
- Cortinovis, G.; Oppermann, M.; Neumann, K.; Graner, A.; Gioia, T.; Marsella, M.; Alseekh, S.; Fernie, A.R.; Papa, R.; Bellucci, E.; et al. Towards the Development, Maintenance, and Standardized Phenotypic Characterization of Single-Seed-Descent Genetic Resources for Common Bean. Curr. Protoc. 2021, 1, e133. [Google Scholar] [CrossRef]
- Bulut, M.; Wendenburg, R.; Bitocchi, E.; Bellucci, E.; Kroc, M.; Gioia, T.; Susek, K.; Papa, R.; Fernie, A.R.; Alseekh, S. A Comprehensive Metabolomics and Lipidomics Atlas for the Legumes Common Bean, Chickpea, Lentil and Lupin. Plant J. 2023, 116, 1152–1171. [Google Scholar] [CrossRef]
- Kroc, M.; Tomaszewska, M.; Czepiel, K.; Bitocchi, E.; Oppermann, M.; Neumann, K.; Guasch, L.; Bellucci, E.; Alseekh, S.; Graner, A.; et al. Towards Development, Maintenance, and Standardized Phenotypic Characterization of Single-Seed-Descent Genetic Resources for Lupins. Curr. Protoc. 2021, 1, e191. [Google Scholar] [CrossRef]
- Rocchetti, L.; Gioia, T.; Logozzo, G.; Brezeanu, C.; Pereira, L.G.; De la Rosa, L.; Marzario, S.; Pieri, A.; Fernie, A.R.; Alseekh, S.; et al. Towards the Development, Maintenance and Standardized Phenotypic Characterization of Single-Seed-Descent Genetic Resources for Chickpea. Curr. Protoc. 2022, 2, e371. [Google Scholar] [CrossRef]
- Guerra-García, A.; Gioia, T.; von Wettberg, E.; Logozzo, G.; Papa, R.; Bitocchi, E.; Bett, K.E. Intelligent Characterization of Lentil Genetic Resources: Evolutionary History, Genetic Diversity of Germplasm, and the Need for Well-Represented Collections. Curr. Protoc. 2021, 1, e134. [Google Scholar] [CrossRef]
- The International Legume Society—Official Website of ILS. Available online: https://www.legumesociety.org/ (accessed on 13 November 2024).
- Consuela Lorela Cristiana, D.; Man, S.; Chiş, S.; Pop, A.; Muste, S.; Păucean, A. Lupin—A Review of the Chemical Composition, Health Benefits and Its Uses in Bakery and Pastry Products. Hop Med. Plants 2022, 30, 98–125. [Google Scholar]
- Gladstones, J.S.; Atkins, C.A.; Hamblin, J. Distribution, Origin, Taxonomy, History and Importance. In Lupins as Crop Plant; CAB International: Wallingford, UK, 1998; p. 465. [Google Scholar]
- Kurlovich, B.S. Lupins: Geography, Classification, Genetic Resources and Breeding; Intan: St. Petersburg, Russia, 2002. [Google Scholar]
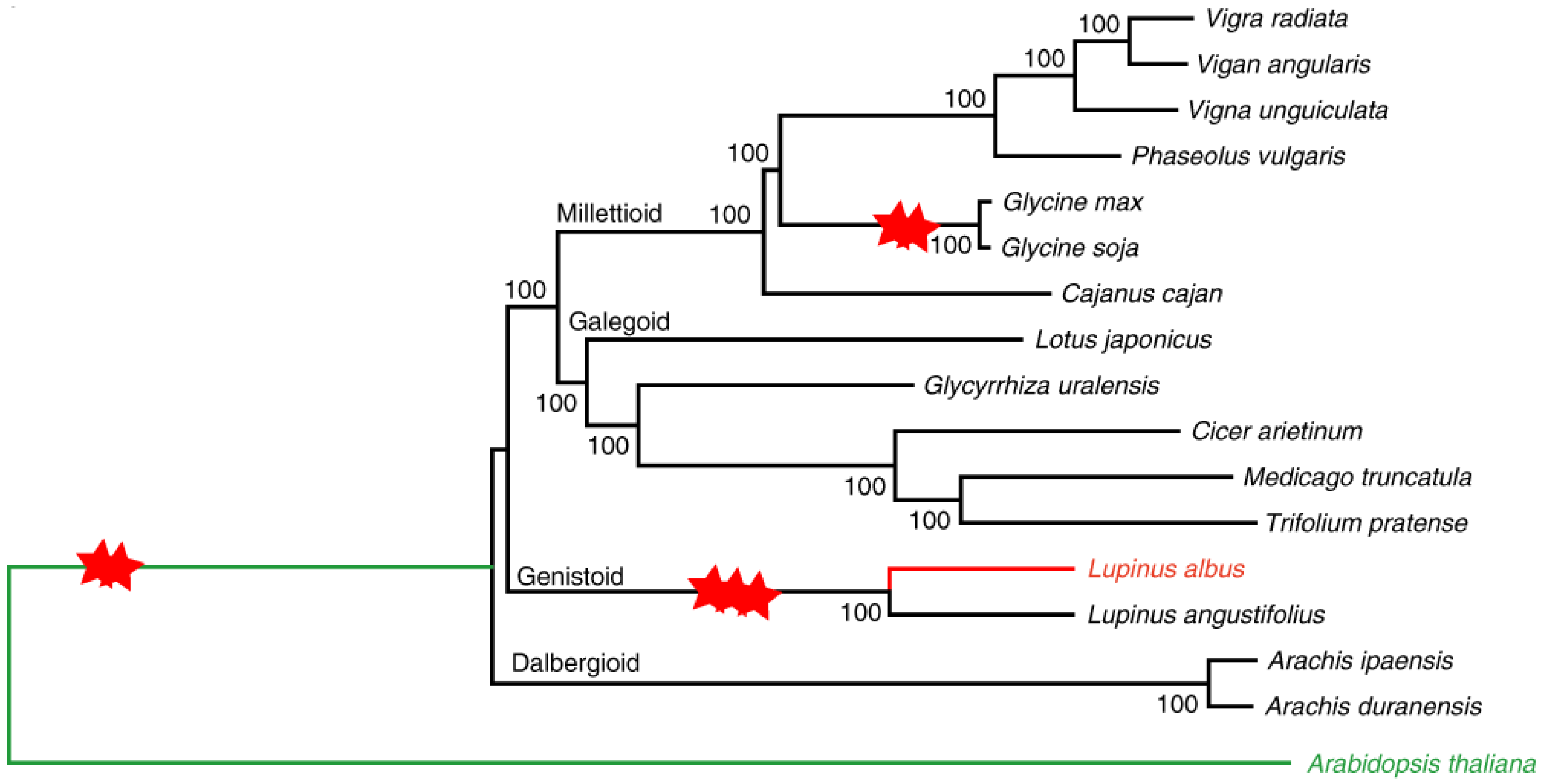
- Drummond, C.S.; Eastwood, R.J.; Miotto, S.T.S.; Hughes, C.E. Multiple Continental Radiations and Correlates of Diversification in Lupinus (Leguminosae): Testing for Key Innovation with Incomplete Taxon Sampling. Syst. Biol. 2012, 61, 443–460. [Google Scholar] [CrossRef]
- Susek, K.; Bielski, W.; Czyż, K.B.; Hasterok, R.; Jackson, S.A.; Wolko, B.; Naganowska, B. Impact of Chromosomal Rearrangements on the Interpretation of Lupin Karyotype Evolution. Genes 2019, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.L.; De Angelis, G.; Nelson, M.N. How Have Narrow-Leafed Lupin Genomic Resources Enhanced Our Understanding of Lupin Domestication? In The Lupin Genome; Springer: Cham, Switzerland, 2020; pp. 95–108. [Google Scholar] [CrossRef]
- Atchison, G.W.; Nevado, B.; Eastwood, R.J.; Contreras-Ortiz, N.; Reynel, C.; Madriñán, S.; Filatov, D.A.; Hughes, C.E. Lost Crops of the Incas: Origins of Domestication of the Andean Pulse Crop Tarwi, Lupinus Mutabilis. Am. J. Bot. 2016, 103, 1592–1606. [Google Scholar] [CrossRef]
- Garg, G.; Kamphuis, L.G.; Bayer, P.E.; Kaur, P.; Dudchenko, O.; Taylor, C.M.; Frick, K.M.; Foley, R.C.; Gao, L.L.; Aiden, E.L.; et al. A Pan-genome and Chromosome-length Reference Genome of Narrow-leafed Lupin (Lupinus angustifolius) Reveals Genomic Diversity and Insights into Key Industry and Biological Traits. Plant J. 2022, 111, 1252. [Google Scholar] [CrossRef]
- Iqbal, M.M.; Huynh, M.; Udall, J.A.; Kilian, A.; Adhikari, K.N.; Berger, J.D.; Erskine, W.; Nelson, M.N. The First Genetic Map for Yellow Lupin Enables Genetic Dissection of Adaptation Traits in an Orphan Grain Legume Crop. BMC Genet. 2019, 20, 68. [Google Scholar] [CrossRef]
- Kohajdová, Z.; Karovičová, J.; Schmidt, Š. Lupin Composition and Possible Use in Bakery—A Review. Czech J. Food Sci. 2011, 29, 203–211. [Google Scholar] [CrossRef]
- Sharasia, P.L.; Garg, M.R.; Bhanderi, B.M. Pulses and Their By-Products as Animal Feed; FAO: Rome, Italy, 2018. [Google Scholar]
- Adamidou, S.; Nengas, I.; Grigorakis, K.; Nikolopoulou, D.; Jauncey, K. Chemical Composition and Antinutritional Factors of Field Peas (Pisum sativum), Chickpeas (Cicer arietinum), and Faba Beans (Vicia faba) as Affected by Extrusion Preconditioning and Drying Temperatures. Cereal Chem. 2011, 88, 80–86. [Google Scholar] [CrossRef]
- Badjona, A.; Bradshaw, R.; Millman, C.; Howarth, M.; Dubey, B. Faba Beans Protein as an Unconventional Protein Source for the Food Industry: Processing Influence on Nutritional, Techno-Functionality, and Bioactivity. Food Rev. Int. 2024, 40, 1999–2023. [Google Scholar] [CrossRef]
- Gulisano, A.; Alves, S.; Martins, J.N.; Trindade, L.M. Genetics and Breeding of Lupinus Mutabilis: An Emerging Protein Crop. Front. Plant Sci. 2019, 10, 485490. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Larenas, F.E.; Linnemann, A.R.; Nout, M.J.R.; Koziol, M.; van Boekel, M.A.J.S. Lupinus Mutabilis: Composition, Uses, Toxicology, and Debittering. Crit. Rev. Food Sci. Nutr. 2016, 56, 1454–1487. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Ramos, F.; Sanches Silva, A. Lupin (Lupinus albus L.) Seeds: Balancing the Good and the Bad and Addressing Future Challenges. Molecules 2022, 27, 8557. [Google Scholar] [CrossRef] [PubMed]
- Celmeli, T.; Sari, H.; Canci, H.; Sari, D.; Adak, A.; Eker, T.; Toker, C. The Nutritional Content of Common Bean (Phaseolus vulgaris L.) Landraces in Comparison to Modern Varieties. Agronomy 2018, 8, 166. [Google Scholar] [CrossRef]
- Rybiński, W.; Święcicki, W.; Bocianowski, J.; Börner, A.; Starzycka-Korbas, E.; Starzycki, M. Variability of Fat Content and Fatty Acids Profiles in Seeds of a Polish White Lupin (Lupinus albus L.) Collection. Genet. Resour. Crop Evol. 2018, 65, 417–431. [Google Scholar] [CrossRef]
- Arnoldi, A.; Boschin, G.; Zanoni, C.; Lammi, C. The Health Benefits of Sweet Lupin Seed Flours and Isolated Proteins. J. Funct. Foods 2015, 18, 550–563. [Google Scholar] [CrossRef]
- Boschin, G.; Arnoldi, A. Legumes Are Valuable Sources of Tocopherols. Food Chem. 2011, 127, 1199–1203. [Google Scholar] [CrossRef]
- Fontanari, G.G.; Batistuti, J.P.; Da Cruz, R.J.; Saldiva, P.H.N.; Arêas, J.A.G. Cholesterol-Lowering Effect of Whole Lupin (Lupinus albus) Seed and Its Protein Isolate. Food Chem. 2012, 132, 1521–1526. [Google Scholar] [CrossRef]
- Wink, M.; Botschen, F.; Gosmann, C.; Schäfer, H.; Waterman, P.G. Chemotaxonomy Seen from a Phylogenetic Perspective and Evolution of Secondary Metabolism. In Biochemistry of Plant Secondary Metabolism, 2nd ed.; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2010; Volume 40, pp. 364–433. [Google Scholar] [CrossRef]
- Frick, K.M.; Kamphuis, L.G.; Siddique, K.H.M.; Singh, K.B.; Foley, R.C. Quinolizidine Alkaloid Biosynthesis in Lupins and Prospects for Grain Quality Improvement. Front. Plant Sci. 2017, 8, 245678. [Google Scholar] [CrossRef]
- Unkovich, M.; Baldock, J.; Forbes, M. Variability in Harvest Index of Grain Crops and Potential Significance for Carbon Accounting: Examples from Australian Agriculture. Adv. Agron. 2010, 105, 173–219. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, Q.; Yuan, W.; Xu, F.; Muhammad Aslam, M.; Miao, R.; Li, Y.; Wang, Q.; Li, X.; Zhang, X.; et al. The Genome Evolution and Low-Phosphorus Adaptation in White Lupin. Nat. Commun. 2020, 11, 1069. [Google Scholar] [CrossRef] [PubMed]
- Pueyo, J.J.; Quiñones, M.A.; Coba de la Peña, T.; Fedorova, E.E.; Lucas, M.M. Nitrogen and Phosphorus Interplay in Lupin Root Nodules and Cluster Roots. Front. Plant Sci. 2021, 12, 644218. [Google Scholar] [CrossRef] [PubMed]
- Le Thanh, T.; Hufnagel, B.; Soriano, A.; Divol, F.; Brottier, L.; Casset, C.; Péret, B.; Doumas, P.; Marquès, L. Dynamic Development of White Lupin Rootlets Along a Cluster Root. Front. Plant Sci. 2021, 12, 738172. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Bucciarelli, B.; Liu, J.; Zinn, K.; Miller, S.; Patton-Vogt, J.; Allan, D.; Shen, J.; Vance, C.P. White Lupin Cluster Root Acclimation to Phosphorus Deficiency and Root Hair Development Involve Unique Glycerophosphodiester Phosphodiesterases. Plant Physiol. 2011, 156, 1131–1148. [Google Scholar] [CrossRef]
- Xia, T.; Zhu, X.; Zhan, Y.; Liu, B.; Zhou, X.; Zhang, Q.; Xu, W. The White Lupin Trehalase Gene LaTRE1 Regulates Cluster Root Formation and Function under Phosphorus Deficiency. Plant Physiol. 2024, 196, 2184–2198. [Google Scholar] [CrossRef]
- Quiñones, M.A.; Lucas, M.M.; Pueyo, J.J. Adaptive Mechanisms Make Lupin a Choice Crop for Acidic Soils Affected by Aluminum Toxicity. Front. Plant Sci. 2022, 12, 810692. [Google Scholar] [CrossRef]
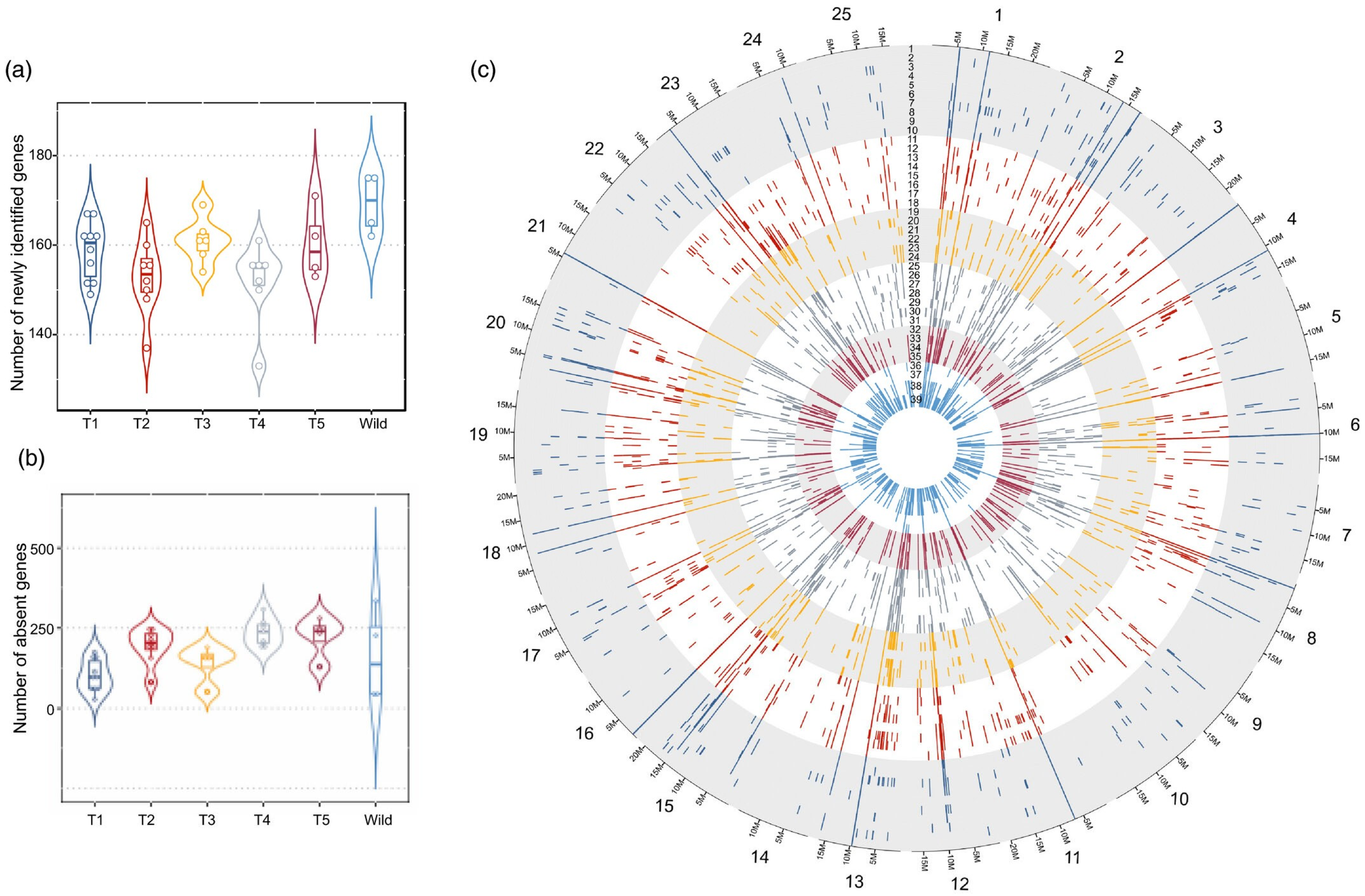
- Hufnagel, B.; Soriano, A.; Taylor, J.; Divol, F.; Kroc, M.; Sanders, H.; Yeheyis, L.; Nelson, M.; Péret, B. Pangenome of White Lupin Provides Insights into the Diversity of the Species. Plant Biotechnol. J. 2021, 19, 2532–2543. [Google Scholar] [CrossRef]
- Hufnagel, B.; Marques, A.; Soriano, A.; Marquès, L.; Divol, F.; Doumas, P.; Sallet, E.; Mancinotti, D.; Carrere, S.; Marande, W.; et al. High-Quality Genome Sequence of White Lupin Provides Insight into Soil Exploration and Seed Quality. Nat. Commun. 2020, 11, 492. [Google Scholar] [CrossRef]
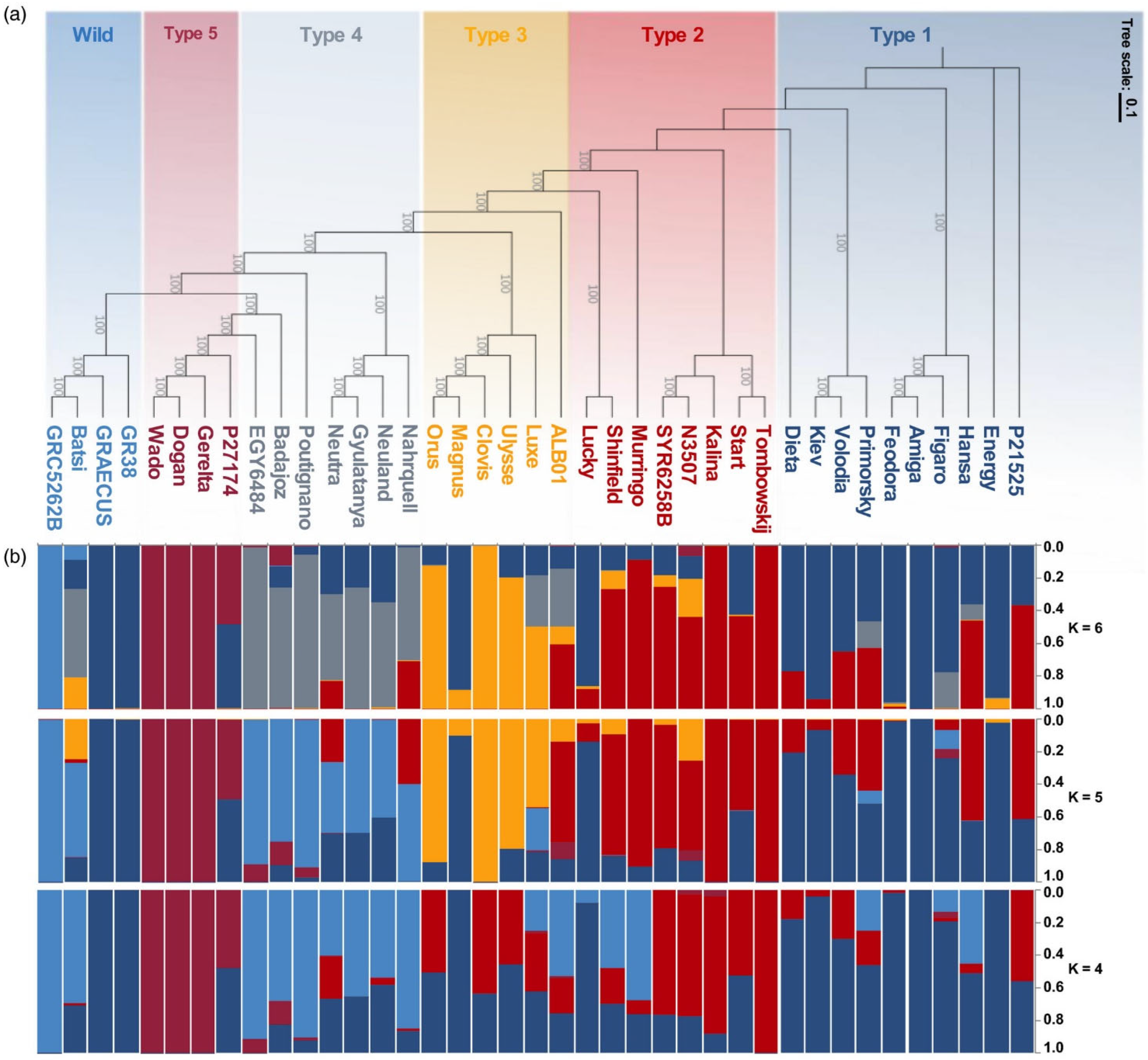
- Zafeiriou, I.; Polidoros, A.N.; Baira, E.; Kasiotis, K.M.; Machera, K.; Mylona, P.V. Mediterranean White Lupin Landraces as a Valuable Genetic Reserve for Breeding. Plants 2021, 10, 2403. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Romani, M.; Pecetti, L. White Lupin (Lupinus albus) Variation for Adaptation to Severe Drought Stress. Plant Breed. 2018, 137, 782–789. [Google Scholar] [CrossRef]
- Schwertfirm, G.; Schneider, M.; Haase, F.; Riedel, C.; Lazzaro, M.; Ruge-Wehling, B.; Schweizer, G. Genome-Wide Association Study Revealed Significant SNPs for Anthracnose Resistance, Seed Alkaloids and Protein Content in White Lupin. Theor. Appl. Genet. 2024, 137, 155. [Google Scholar] [CrossRef] [PubMed]
- Lucas, M.M.; Stoddard, F.L.; Annicchiarico, P.; Frías, J.; Martínez-Villaluenga, C.; Sussmann, D.; Duranti, M.; Seger, A.; Zander, P.M.; Pueyo, J.J. The Future of Lupin as a Protein Crop in Europe. Front. Plant Sci. 2015, 6, 705. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. Available online: https://www.fao.org/faostat/en/#data/QCL/visualize (accessed on 24 October 2024).
- Caramona, A.; Martins, A.M.; Seixas, J.; Marto, J. The Use, Reuse and Valorization of Lupin and Its Industry by-Products for Dermocosmetics Applications. Sustain. Chem. Pharm. 2024, 38, 101477. [Google Scholar] [CrossRef]
- DBRM Lupin Market Size, Share, Industry Trends & Forecast 2029. Available online: https://www.databridgemarketresearch.com/reports/global-lupin-market (accessed on 24 October 2024).
- Wolko, B.; Clements, J.C.; Naganowska, B.; Nelson, M.N.; Yang, H. Lupinus. In Wild Crop Relatives: Genomic and Breeding Resources; Springer: Berlin/Heidelberg, Germany, 2010; pp. 153–206. [Google Scholar] [CrossRef]
- Tanwar, U.K.; Tomaszewska, M.; Czepiel, K.; Neji, M.; Jamil, H.; Rocchetti, L.; Pieri, A.; Bitocchi, E.; Bellucci, E.; Pipan, B.; et al. Genetic and Phenotypic Characterization of Global Lupinus albus Genetic Resources for the Development of a CORE Collection. bioRxiv 2024. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome Sequence of the Palaeopolyploid Soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Susek, K.; Franco, E.; Tomaszewska, M.; Kroc, M.; Jamil, H.; Tanwar, U.; Nelson, M.N.; Papa, R.; Delledonne, M.; Jackson, S.A. The Unexplored Diversity of Wild Lupins Provides Rich Genomic Resources and Insights into Lupin Evolution. bioRxiv 2024. [Google Scholar] [CrossRef]
- Cortinovis, G.; Vincenzi, L.; Anderson, R.; Marturano, G.; Marsh, J.I.; Bayer, P.E.; Rocchetti, L.; Frascarelli, G.; Lanzavecchia, G.; Pieri, A.; et al. Adaptive Gene Loss in the Common Bean Pan-Genome during Range Expansion and Domestication. Nat. Commun. 2024, 15, 6698. [Google Scholar] [CrossRef]
- Raman, R.; Cowley, R.B.; Raman, H.; Luckett, D.J. Analyses Using SSR and DArT Molecular Markers Reveal That Ethiopian Accessions of White Lupin (Lupinus albus L.) Represent a Unique Genepool. Open J. Genet. 2014, 2014, 87–98. [Google Scholar] [CrossRef]
- Kinghorn, A.D.; Hussain, R.A.; Robbins, E.F.; Balandrin, M.F.; Stirton, C.H.; Evans, S.V. Alkaloid Distribution in Seeds of Ormosia, Pericopsis and Haplormosia. Phytochemistry 1988, 27, 439–444. [Google Scholar] [CrossRef]
- Wink, M.; Mohamed, G.I.A. Evolution of Chemical Defense Traits in the Leguminosae: Mapping of Distribution Patterns of Secondary Metabolites on a Molecular Phylogeny Inferred from Nucleotide Sequences of the RbcL Gene. Biochem. Syst. Ecol. 2003, 31, 897–917. [Google Scholar] [CrossRef]
- Mancinotti, D.; Czepiel, K.; Taylor, J.L.; Galehshahi, H.G.; Møller, L.A.; Jensen, M.K.; Motawia, M.S.; Hufnagel, B.; Soriano, A.; Yeheyis, L.; et al. The Causal Mutation Leading to Sweetness in Modern White Lupin Cultivars. Sci. Adv. 2023, 9, eadg8866. [Google Scholar] [CrossRef] [PubMed]
- Kroc, M.; Rybiński, W.; Wilczura, P.; Kamel, K.; Kaczmarek, Z.; Barzyk, P.; Święcicki, W. Quantitative and Qualitative Analysis of Alkaloids Composition in the Seeds of a White Lupin (Lupinus albus L.) Collection. Genet. Resour. Crop Evol. 2017, 64, 1853–1860. [Google Scholar] [CrossRef]
- Valente, I.M.; Sousa, C.; Almeida, M.; Miranda, C.; Pinheiro, V.; Garcia-Santos, S.; Ferreira, L.M.M.; Guedes, C.M.; Maia, M.R.G.; Cabrita, A.R.J.; et al. Insights from the Yield, Protein Production, and Detailed Alkaloid Composition of White (Lupinus albus), Narrow-Leafed (Lupinus angustifolius), and Yellow (Lupinus luteus) Lupin Cultivars in the Mediterranean Region. Front. Plant Sci. 2023, 14, 1231777. [Google Scholar] [CrossRef] [PubMed]
- Gresta, F.; Oteri, M.; Scordia, D.; Costale, A.; Armone, R.; Meineri, G.; Chiofalo, B. White Lupin (Lupinus albus L.), an Alternative Legume for Animal Feeding in the Mediterranean Area. Agriculture 2023, 13, 434. [Google Scholar] [CrossRef]
- Schrenk, D.; Bodin, L.; Chipman, J.K.; del Mazo, J.; Grasl-Kraupp, B.; Hogstrand, C.; Hoogenboom, L.; Leblanc, J.C.; Nebbia, C.S.; Nielsen, E.; et al. Scientific Opinion on the Risks for Animal and Human Health Related to the Presence of Quinolizidine Alkaloids in Feed and Food, in Particular in Lupins and Lupin-Derived Products. EFSA J. 2019, 17, e05860. [Google Scholar] [CrossRef]
- Mancinotti, D.; Frick, K.M.; Geu-Flores, F. Biosynthesis of Quinolizidine Alkaloids in Lupins: Mechanistic Considerations and Prospects for Pathway Elucidation. Nat. Prod. Rep. 2022, 39, 1423–1437. [Google Scholar] [CrossRef]
- Harrison, J.E.M.; Williams, W. Genetical Control of Alkaloids in Lupinus albus. Euphytica 1982, 31, 357–364. [Google Scholar] [CrossRef]
- Osorio, C.E.; Till, B.J. A Bitter-Sweet Story: Unraveling the Genes Involved in Quinolizidine Alkaloid Synthesis in Lupinus albus. Front. Plant Sci. 2022, 12, 795091. [Google Scholar] [CrossRef]
- Lin, R.; Renshaw, D.; Luckett, D.; Clements, J.; Yan, G.; Adhikari, K.; Buirchell, B.; Sweetingham, M.; Yang, H. Development of a Sequence-Specific PCR Marker Linked to the Gene “Pauper” Conferring Low-Alkaloids in White Lupin (Lupinus albus L.) for Marker Assisted Selection. Mol. Breed. 2009, 23, 153–161. [Google Scholar] [CrossRef]
- Rychel, S.; Książkiewicz, M. Development of Gene-Based Molecular Markers Tagging Low Alkaloid Pauper Locus in White Lupin (Lupinus albus L.). J. Appl. Genet. 2019, 60, 269–281. [Google Scholar] [CrossRef]
- Rodés-Bachs, C.; Van der Fels-Klerx, H.J. Impact of Environmental Factors on the Presence of Quinolizidine Alkaloids in Lupins: A Review. Food Addit. Contam. Part A 2023, 40, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Jansen, G.; Jürgens, H.-U.; Schliephake, E.; Ordon, F. Effect of the Soil PH on the Alkaloid Content of Lupinus angustifolius. Int. J. Agron. 2012, 2012, 269878. [Google Scholar] [CrossRef]
- Valente, I.M.; Monteiro, A.; Sousa, C.; Miranda, C.; Maia, M.R.G.; Castro, C.; Cabrita, A.R.J.; Trindade, H.; Fonseca, A.J.M. Agronomic, Nutritional Traits, and Alkaloids of Lupinus albus, Lupinus angustifolius and Lupinus luteus Genotypes: Effect of Sowing Dates and Locations. ACS Agric. Sci. Technol. 2024, 4, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Annicchiarico, P.; Harzic, N.; Huyghe, C.; Carroni, A.M. Ecological Classification of White Lupin Landrace Genetic Resources. Euphytica 2011, 180, 17–25. [Google Scholar] [CrossRef]
- Brand, J.D.; Tang, C.; Rathjen, A.J. Screening Rough-Seeded Lupins (Lupinus pilosus Murr. and Lupinus atlanticus Glads.) for Tolerance to Calcareous Soils. Plant Soil 2002, 245, 261–275. [Google Scholar] [CrossRef]
- Tang, C.; Robson, A.D.; Longnecker, N.E.; Buirchell, B.J. The Growth of Lupinus Species on Alkaline Soils. Aust. J. Agric. Res. 1995, 46, 255–268. [Google Scholar] [CrossRef]
- Tang, C.; Buirchell, B.J.; Longnecker, N.E.; Robson, A.D. Variation in the Growth of Lupin Species and Genotypes on Alkaline Soil. Plant Soil 1993, 155–156, 513–516. [Google Scholar] [CrossRef]
- Annicchiarico, P.; Harzic, N.; Carroni, A.M. Adaptation, Diversity, and Exploitation of Global White Lupin (Lupinus albus L.) Landrace Genetic Resources. Field Crops Res. 2010, 119, 114–124. [Google Scholar] [CrossRef]
- Arief, O.M.; Pang, J.; Shaltout, K.H.; Lambers, H. Performance of Two Lupinus albus L. Cultivars in Response to Three Soil PH Levels. Exp. Agric. 2020, 56, 321–330. [Google Scholar] [CrossRef]
- Christiansen, J.L.; Raza, S.; Jørnsgård, B.; Mahmoud, S.A.; Ortiz, R. Potential of Landrace Germplasm for Genetic Enhancement of White Lupin in Egypt. Genet. Resour. Crop Evol. 2000, 47, 425–430. [Google Scholar] [CrossRef]
- Raza, S.; Abdel-Wahab, A.; Jørnsgård, B.; Christiansen, J.L. Calcium Tolerance and Ion Uptake of Egyptian Lupin Landraces on Calcareous Soils. Available online: https://www.bioline.org.br/request?cs01021 (accessed on 6 January 2025).
- Kerley, S.J.; Norgaard, C.; Leach, J.E.; Christiansen, J.L.; Huyghe, C.; Römer, P. The Development of Potential Screens Based on Shoot Calcium and Iron Concentrations for the Evaluation of Tolerance in Egyptian Genotypes of White Lupin (Lupinus albus L.) to Limed Soils. Ann. Bot. 2002, 89, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Tang, C. Comparative Performance of Lupinus albus Genotypes in Response to Soil Alkalinity. Aust. J. Agric. Res. 1999, 50, 1435–1442. [Google Scholar] [CrossRef]
- Koseoglou, E. Selection of White Lupine Genotypes for Yield and Tolerance to Alkaline Soils. In Proceedings of the General EUCARPIA Congress “Plant Breeding for Future Generations”, Zurich, Switzerland, 29 August–1 September 2016. [Google Scholar]
- Annicchiarico, P.; Alami, I.T. Enhancing White Lupin (Lupinus albus L.) Adaptation to Calcareous Soils through Selection of Lime-Tolerant Plant Germplasm and Bradyrhizobium Strains. Plant Soil 2012, 350, 131–144. [Google Scholar] [CrossRef]
- Dinkelaker, B.; Römheld, V.; Marschner, H. Citric Acid Excretion and Precipitation of Calcium Citrate in the Rhizosphere of White Lupin (Lupinus albus L.). Plant Cell Environ. 1989, 12, 285–292. [Google Scholar] [CrossRef]
- Tang, C.; Thomson, B.D. Effects of Solution Ph and Bicarbonate on the Growth and Nodulation of a Range of Grain Legume Species. Plant Soil 1996, 186, 321–330. [Google Scholar] [CrossRef]
- Kerley, S.J. The Effect of Soil Liming on Shoot Development, Root Growth, and Cluster Root Activity of White Lupin. Biol. Fertil. Soils 2000, 32, 94–101. [Google Scholar] [CrossRef]
- Tang, C.; Zheng, S.J.; Qiao, Y.F.; Wang, G.H.; Han, X.Z. Interactions between High PH and Iron Supply on Nodulation and Iron Nutrition of Lupinus albus L. Genotypes Differing in Sensitivity to Iron Deficiency. Plant Soil 2006, 279, 153–162. [Google Scholar] [CrossRef]
- Kerley, S.J.; Huyghe, C. Comparison of Acid and Alkaline Soil and Liquid Culture Growth Systems for Studies of Shoot and Root Characteristics of White Lupin (Lupinus albus L.) Genotypes. Plant Soil 2001, 236, 275–286. [Google Scholar] [CrossRef]
- Vasconcelos, M.; Eckert, H.; Arahana, V.; Graef, G.; Grusak, M.A.; Clemente, T. Molecular and Phenotypic Characterization of Transgenic Soybean Expressing the Arabidopsis Ferric Chelate Reductase Gene, FRO2. Planta 2006, 224, 1116–1128. [Google Scholar] [CrossRef]
- Tian, Q.; Zhang, X.; Yang, A.; Wang, T.; Zhang, W.H. CIPK23 Is Involved in Iron Acquisition of Arabidopsis by Affecting Ferric Chelate Reductase Activity. Plant Sci. 2016, 246, 70–79. [Google Scholar] [CrossRef]
- Pestana, M.; David, M.; de Varennes, A.; Abadía, J.; Faria, E.A. Responses of “Newhall” Orange Trees to Iron Deficiency in Hydroponics: Effects on Leaf Chlorophyll, Photosynthetic Efficiency, and Root Ferric Chelate Reductase Activity. J. Plant Nutr. 2001, 24, 1609–1620. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, S.; Li, F.; Sun, M.; Liang, Z.; Sun, Z.; Yu, F.; Li, H. The Low Ferric Chelate Reductase Activity and High Apoplastic PH in Leaves Cause Iron Deficiency Chlorosis in ‘Huangguan’ Pears Grafted onto Quince A Grown in Calcareous Soil. Sci. Hortic. 2023, 310, 111754. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Kim, S.; Tsukamoto, T.; Oki, H.; Kobayashi, T.; Watanabe, S.; Matsuhashi, S.; Takahashi, M.; Nakanishi, H.; Mori, S.; et al. Mutational Reconstructed Ferric Chelate Reductase Confers Enhanced Tolerance in Rice to Iron Deficiency in Calcareous Soil. Proc. Natl. Acad. Sci. USA 2007, 104, 7373–7378. [Google Scholar] [CrossRef] [PubMed]
- Mori, S. Iron Acquisition by Plants. Curr. Opin. Plant Biol. 1999, 2, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, S. Landraces: Importance and Use in Breeding and Environmentally Friendly Agronomic Systems. In Agrobiodiversity Conservation: Securing the Diversity of Crop Wild Relatives and Landraces; CABI Publishing: Wallingford, UK, 2011; pp. 103–117. ISBN 978-1-84593-851-2. [Google Scholar]
- Dwivedi, S.L.; Ceccarelli, S.; Blair, M.W.; Upadhyaya, H.D.; Are, A.K.; Ortiz, R. Landrace Germplasm for Improving Yield and Abiotic Stress Adaptation. In Agrobiodiversity Conservation: Securing the Diversity of Crop Wild Relatives and Landraces; CABI Publishing: Wallingford, UK, 2016; pp. 103–117. ISBN 978-1-84593-851-2. [Google Scholar]
- Ceccarelli, S. Adaptation to Low/High Input Cultivation. Euphytica 1997, 92, 203–214. [Google Scholar] [CrossRef]
- Ceccarelli, S. Positive Interpretation of Genotype by Environment Interaction in Relation to Sustainability and Biodiversity. In Plant Adaptation and Crop Improvement; CAB International: Wallingford, UK, 1996. [Google Scholar]
- Sharma, S.; Upadhyaya, H.D.; Varshney, R.K.; Gowda, C.L.L. Pre-Breeding for Diversification of Primary Gene Pool and Genetic Enhancement of Grain Legumes. Front. Plant Sci. 2013, 4, 309. [Google Scholar] [CrossRef]
- Pratap, A.; Das, A.; Kumar, S.; Gupta, S. Current Perspectives on Introgression Breeding in Food Legumes. Front. Plant Sci. 2021, 11, 589189. [Google Scholar] [CrossRef]
- Abraham, E.M.; Ganopoulos, I.; Madesis, P.; Mavromatis, A.; Mylona, P.; Nianiou-Obeidat, I.; Parissi, Z.; Polidoros, A.; Tani, E.; Vlachostergios, D. The Use of Lupin as a Source of Protein in Animal Feeding: Genomic Tools and Breeding Approaches. Int. J. Mol. Sci. 2019, 20, 851. [Google Scholar] [CrossRef]
- Mavromatis, A.; Nianiou-Obeidat, I.; Polidoros, A.; Parissi, Z.; Tani, E.; Irakli, M.; Aliferis, K.A.; Zafeiriou, I.; Mylona, P.V.; Sarri, E.; et al. Characterization of Lupin Cultivars Based on Phenotypical, Molecular and Metabolomic Analyses. Agronomy 2023, 13, 370. [Google Scholar] [CrossRef]
- Lazaridi, E.; Kapazoglou, A.; Gerakari, M.; Kleftogianni, K.; Passa, K.; Sarri, E.; Papasotiropoulos, V.; Tani, E.; Bebeli, P.J. Crop Landraces and Indigenous Varieties: A Valuable Source of Genes for Plant Breeding. Plants 2024, 13, 758. [Google Scholar] [CrossRef]
- Marone, D.; Russo, M.A.; Mores, A.; Ficco, D.B.M.; Laidò, G.; Mastrangelo, A.M.; Borrelli, G.M. Importance of Landraces in Cereal Breeding for Stress Tolerance. Plants 2021, 10, 1267. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Z.; Bao, Z.; Li, H.; Lyu, Y.; Zan, Y.; Wu, Y.; Cheng, L.; Fang, Y.; Wu, K.; et al. Graph Pangenome Captures Missing Heritability and Empowers Tomato Breeding. Nature 2022, 606, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, N.W. Introgression and incorporation. Strategies for the use of crop genetic resources. Biol. Rev. 1993, 68, 539–562. [Google Scholar] [CrossRef]
- Cheng, S.; Feng, C.; Wingen, L.U.; Cheng, H.; Riche, A.B.; Jiang, M.; Leverington-Waite, M.; Huang, Z.; Collier, S.; Orford, S.; et al. Harnessing Landrace Diversity Empowers Wheat Breeding. Nature 2024, 632, 823–831. [Google Scholar] [CrossRef]
- Huang, K.; Jahani, M.; Gouzy, J.; Legendre, A.; Carrere, S.; Lázaro-Guevara, J.M.; González Segovia, E.G.; Todesco, M.; Mayjonade, B.; Rodde, N.; et al. The Genomics of Linkage Drag in Inbred Lines of Sunflower. Proc. Natl. Acad. Sci. USA 2023, 120, e2205783119. [Google Scholar] [CrossRef]
- Smýkal, P.; Coyne, C.; Von Wettberg, E.; Hübner, S.; Kantar, M.B. Tapping Diversity from the Wild: From Sampling to Implementation. Front. Plant Sci. 2021, 12, 626565. [Google Scholar] [CrossRef]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L. Past and Future Use of Wild Relatives in Crop Breeding. Crop Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA Molecular Markers in Plant Breeding: Current Status and Recent Advancements in Genomic Selection and Genome Editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef]
- Hospital, F. Size of Donor Chromosome Segments Around Introgressed Loci and Reduction of Linkage Drag in Marker-Assisted Backcross Programs. Genetics 2001, 158, 1363–1379. [Google Scholar] [CrossRef]
- Zhang, F.; Shi, Y.; Ali, J.; Xu, J.; Li, Z. Breeding by Selective Introgression: Theory, Practices, and Lessons Learned from Rice. Crop J. 2021, 9, 646–657. [Google Scholar] [CrossRef]
- Gupta, S.; Buirchell, B.J.; Cowling, W.A. Interspecific Reproductive Barriers and Genomic Similarity among the Rough-Seeded Lupinus Species. Plant Breed. 1996, 115, 123–127. [Google Scholar] [CrossRef]


| Species | PROTEIN % | OIL % | References |
|---|---|---|---|
| White lupin | 38–44 | 9–13 | [89,91] |
| Andean lupin | 41–51 | 14–24 | [89] |
| Narrow leaf lupin | 30–35 | 5–10 | [89] |
| Yellow lupin | 30–40 | 14–24 | [89] |
| Soybean | 36.9 | 18.1 | [89,91] |
| Chickpea | 23.6 | 6.4 | [87,91] |
| Pea | 21.9 | 2.3 | [87,91] |
| Lentil | 20.6 | 2.15 | [91] |
| Common bean | 21.3 | 1.6 | [92] |
| Fava bean | 25 | 1.3 | [87,91] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tosoroni, A.; Di Vittori, V.; Nanni, L.; Musari, E.; Papalini, S.; Bitocchi, E.; Bellucci, E.; Pieri, A.; Ghitarrini, S.; Susek, K.; et al. Recent Advances in Molecular Tools and Pre-Breeding Activities in White Lupin (Lupinus albus). Plants 2025, 14, 914. https://doi.org/10.3390/plants14060914
Tosoroni A, Di Vittori V, Nanni L, Musari E, Papalini S, Bitocchi E, Bellucci E, Pieri A, Ghitarrini S, Susek K, et al. Recent Advances in Molecular Tools and Pre-Breeding Activities in White Lupin (Lupinus albus). Plants. 2025; 14(6):914. https://doi.org/10.3390/plants14060914
Chicago/Turabian StyleTosoroni, Andrea, Valerio Di Vittori, Laura Nanni, Evan Musari, Simone Papalini, Elena Bitocchi, Elisa Bellucci, Alice Pieri, Sofia Ghitarrini, Karolina Susek, and et al. 2025. "Recent Advances in Molecular Tools and Pre-Breeding Activities in White Lupin (Lupinus albus)" Plants 14, no. 6: 914. https://doi.org/10.3390/plants14060914
APA StyleTosoroni, A., Di Vittori, V., Nanni, L., Musari, E., Papalini, S., Bitocchi, E., Bellucci, E., Pieri, A., Ghitarrini, S., Susek, K., & Papa, R. (2025). Recent Advances in Molecular Tools and Pre-Breeding Activities in White Lupin (Lupinus albus). Plants, 14(6), 914. https://doi.org/10.3390/plants14060914








