Comparative Analysis of Floral Transcriptomes in Gossypium hirsutum (Malvaceae)
Abstract
1. Introduction
2. Results
2.1. Overall Transcript Expression
2.2. Functional Analysis of Differentially Expressed Transcripts
2.3. Differentially Enriched Transcription Factors
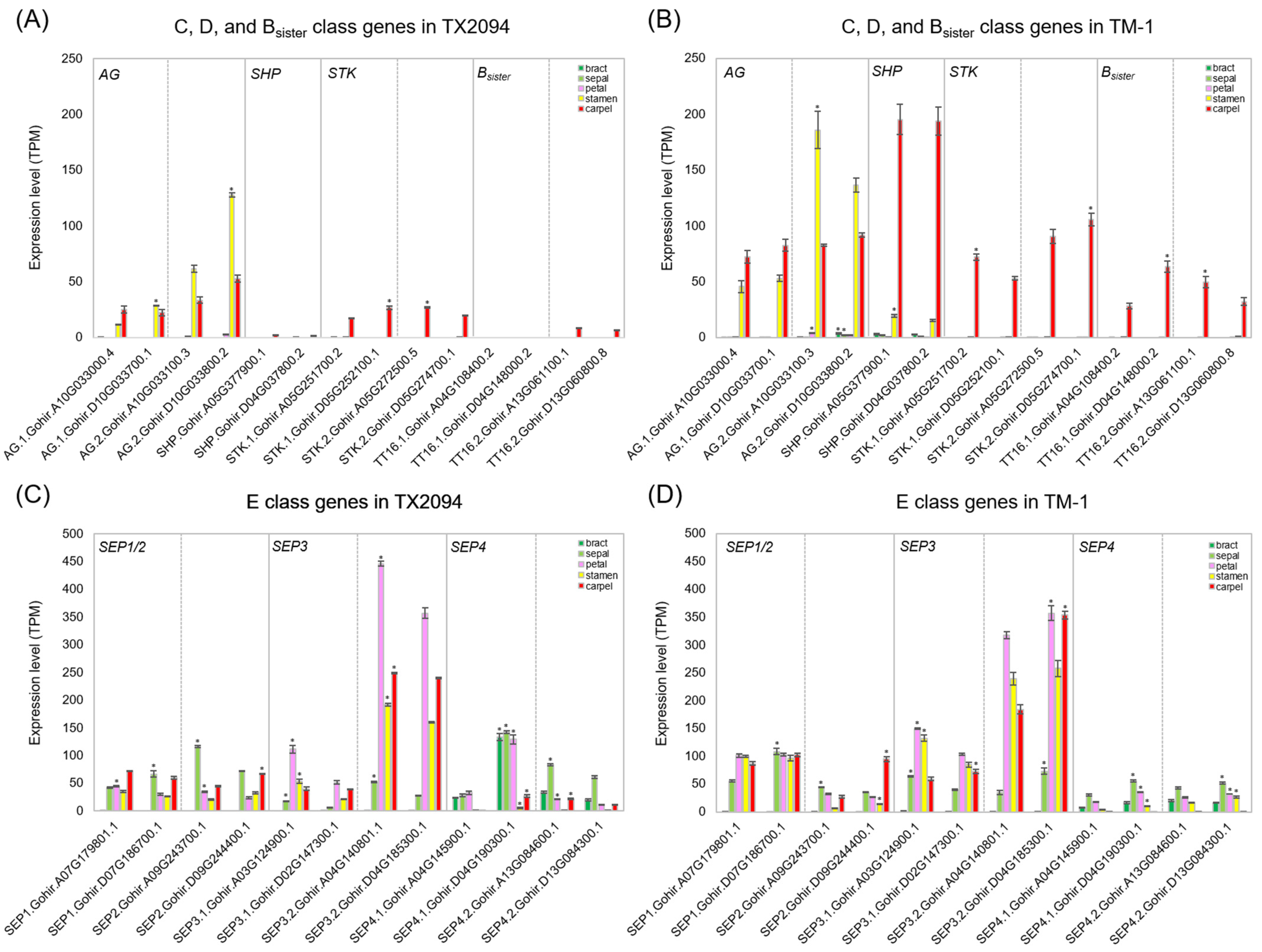
2.3.1. MIKC-Type MADS-Box Gene Expression in Floral Organs
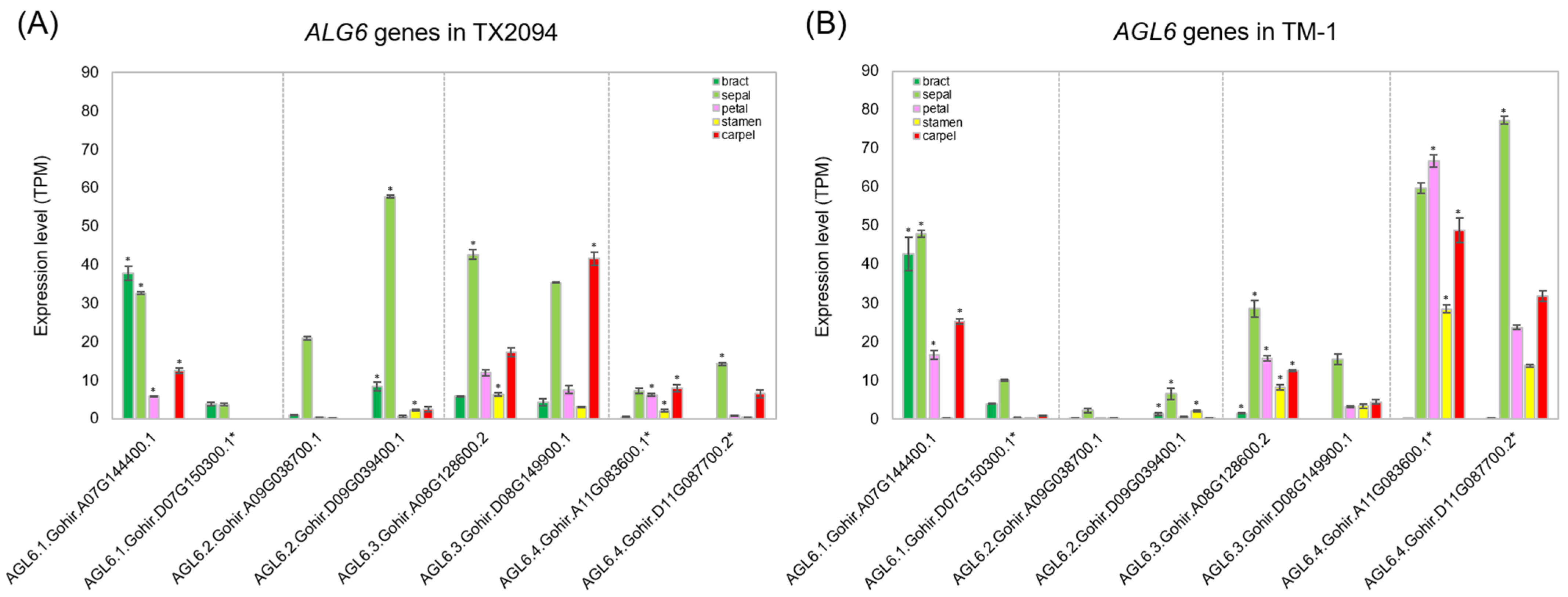
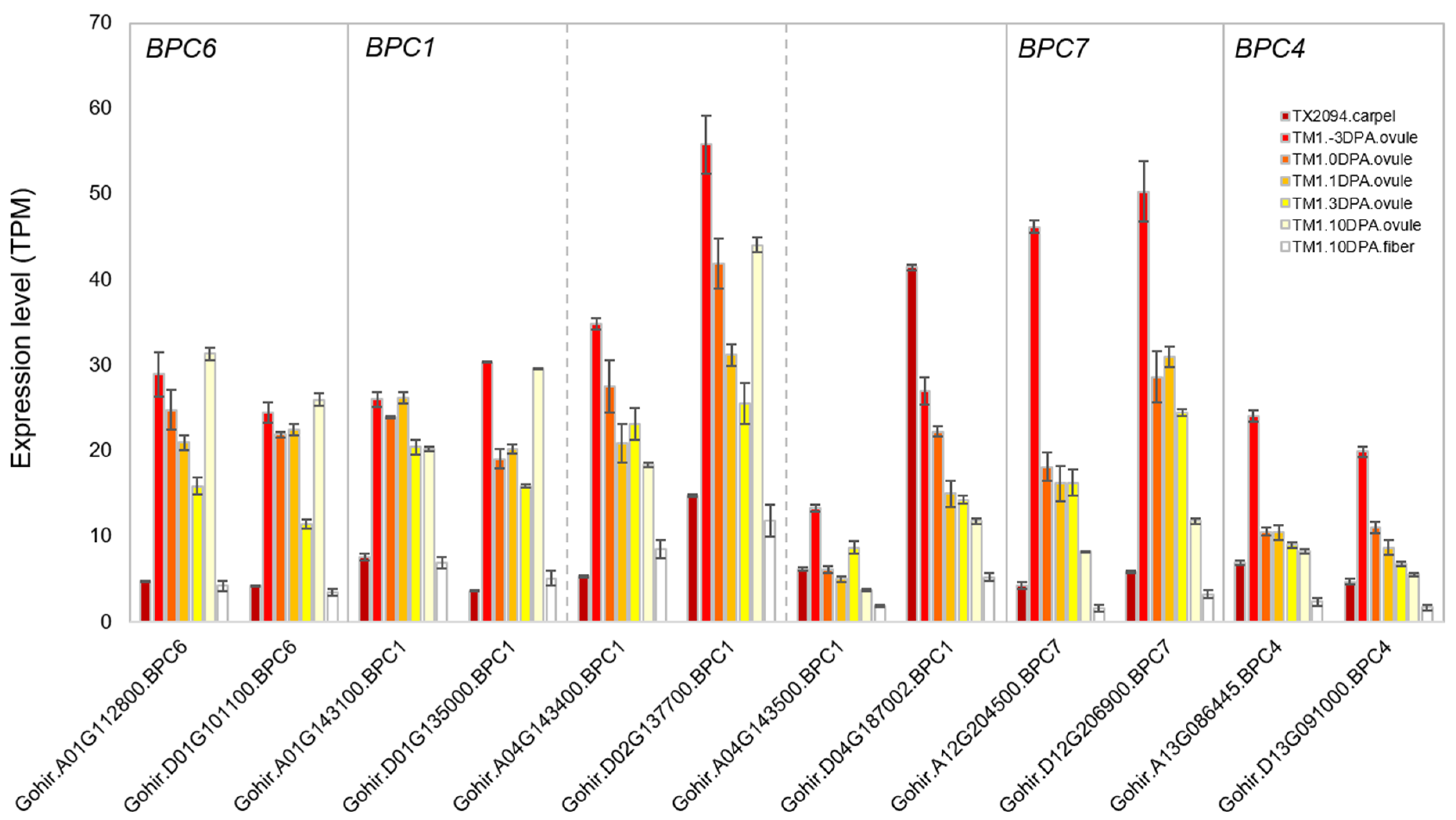
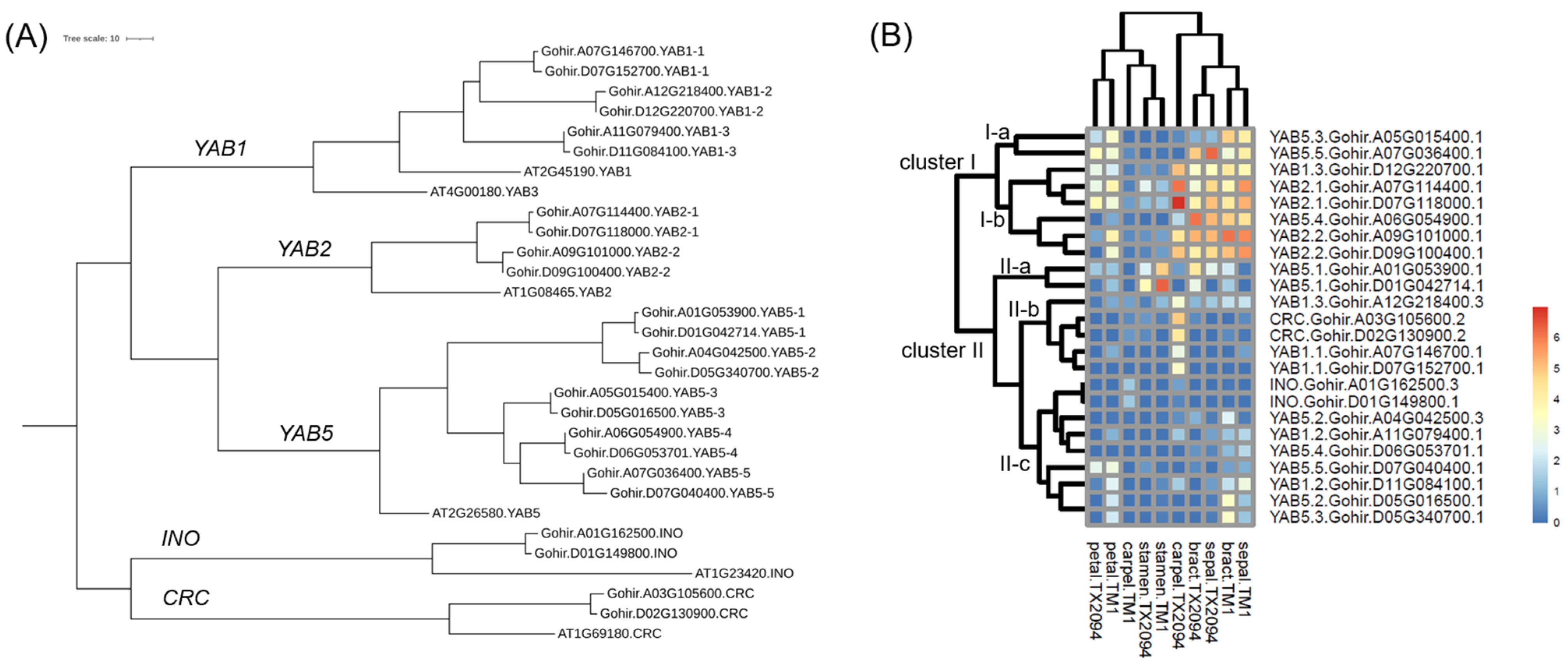
2.3.2. Other Transcription Factors
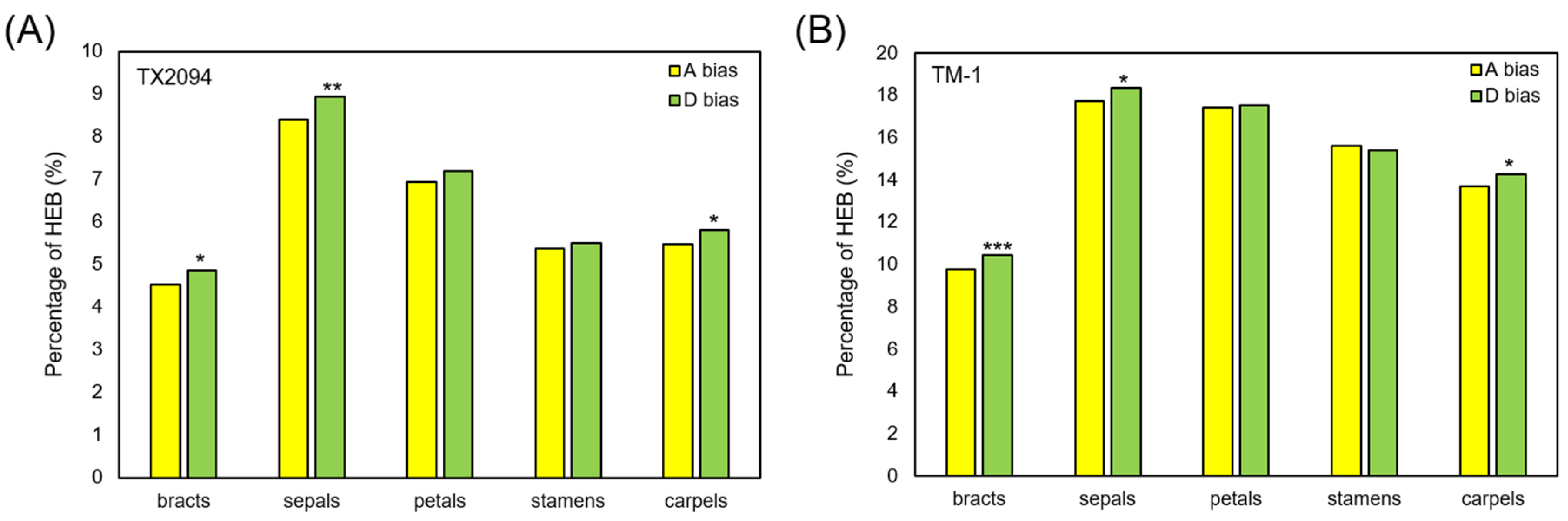
2.4. Homoeolog Expression Bias (HEB) in Floral Organs
2.4.1. Overall Comparison of HEB Between TX2094 and TM-1
2.4.2. HEB in MIKC-MADS Box Genes
2.5. Validation of RNA-Seq Results Using Quantitative Real-Time PCR (qRT-PCR)
3. Discussion
3.1. The ABCDE Model in Gossypium hirsutum
3.2. Transcriptome Profiles in Each Floral Organ
3.3. HEB of MADS-Box Genes in Wild and Domesticated Cotton
4. Materials and Methods
4.1. Plant Materials and Library Construction
4.2. Analysis of RNA-Seq Data
4.3. Validation of RNA-seq Results Using qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Becker, A.; Theissen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Bowman, J.L.; Moyroud, E. Reflections on the ABC model of flower development. Plant Cell 2024, 36, 1334–1357. [Google Scholar] [CrossRef] [PubMed]
- Castelan-Munoz, N.; Herrera, J.; Cajero-Sanchez, W.; Arrizubieta, M.; Trejo, C.; Garcia-Ponce, B.; Sanchez, M.P.; Alvarez-Buylla, E.R.; Garay-Arroyo, A. MADS-Box Genes Are Key Components of Genetic Regulatory Networks Involved in Abiotic Stress and Plastic Developmental Responses in Plants. Front. Plant Sci. 2019, 10, 853. [Google Scholar] [CrossRef]
- De Bodt, S.; Raes, J.; Florquin, K.; Rombauts, S.; Rouze, P.; Theissen, G.; Van de Peer, Y. Genomewide structural annotation and evolutionary analysis of the type I MADS-box genes in plants. J. Mol. Evol. 2003, 56, 573–586. [Google Scholar] [CrossRef]
- Gramzow, L.; Theissen, G. A hitchhiker’s guide to the MADS world of plants. Genome Biol. 2010, 11, 214. [Google Scholar] [CrossRef]
- Parenicova, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef]
- Theissen, G.; Kim, J.T.; Saedler, H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 1996, 43, 484–516. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Ribas de Pouplana, L.; Martinez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Honma, T.; Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001, 409, 525–529. [Google Scholar] [CrossRef]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef]
- Veron, A.S.; Kaufmann, K.; Bornberg-Bauer, E. Evidence of interaction network evolution by whole-genome duplications: A case study in MADS-box proteins. Mol. Biol. Evol. 2007, 24, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.Y.; Hu, Y.; Tudor, M.; Ma, H. Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 1997, 12, 999–1010. [Google Scholar] [CrossRef]
- Liu, C.; Xi, W.; Shen, L.; Tan, C.; Yu, H. Regulation of floral patterning by flowering time genes. Dev. Cell 2009, 16, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Gregis, V.; Sessa, A.; Dorca-Fornell, C.; Kater, M.M. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 2009, 60, 626–637. [Google Scholar] [CrossRef]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Meyerowitz, E.M. Genetic control of pattern formation during flower development in Arabidopsis. Symp. Soc. Exp. Biol. 1991, 45, 89–115. [Google Scholar] [PubMed]
- Theissen, G.; Saedler, H. Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef]
- Murai, K. Homeotic Genes and the ABCDE Model for Floral Organ Formation in Wheat. Plants 2013, 2, 379–395. [Google Scholar] [CrossRef]
- Ali, Z.; Raza, Q.; Atif, R.M.; Aslam, U.; Ajmal, M.; Chung, G. Genetic and Molecular Control of Floral Organ Identity in Cereals. Int. J. Mol. Sci. 2019, 20, 2743. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991, 112, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; dePamphilis, C. The ABCs of floral evolution. Cell 2000, 101, 5–8. [Google Scholar] [CrossRef]
- Zahn, L.M.; Feng, B.; Ma, H. Beyond the ABC-Model: Regulation of floral homeotic genes. Adv. Bot. Res. 2006, 44, 163–207. [Google Scholar]
- Silva, C.S.; Puranik, S.; Round, A.; Brennich, M.; Jourdain, A.; Parcy, F.; Hugouvieux, V.; Zubieta, C. Evolution of the plant reproduction master regulators LFY and the MADS transcription factors: The role of protein structure in the evolutionary development of the flower. Front. Plant Sci. 2015, 6, 1193. [Google Scholar] [CrossRef]
- Byrne, M.E.; Groover, A.T.; Fontana, J.R.; Martienssen, R.A. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 2003, 130, 3941–3950. [Google Scholar] [CrossRef] [PubMed]
- Peaucelle, A.; Louvet, R.; Johansen, J.N.; Salsac, F.; Morin, H.; Fournet, F.; Belcram, K.; Gillet, F.; Hofte, H.; Laufs, P.; et al. The transcription factor BELLRINGER modulates phyllotaxis by regulating the expression of a pectin methylesterase in Arabidopsis. Development 2011, 138, 4733–4741. [Google Scholar] [CrossRef]
- Bao, X.; Franks, R.G.; Levin, J.Z.; Liu, Z. Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell 2004, 16, 1478–1489. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Mislata, A.; Bencivenga, S.; Bush, M.; Schiessl, K.; Boden, S.; Sablowski, R. DELLA genes restrict inflorescence meristem function independently of plant height. Nat. Plants 2017, 3, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Qin, L.; Lee, S.; Fu, X.; Richards, D.E.; Cao, D.; Luo, D.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 2004, 131, 1055–1064. [Google Scholar] [CrossRef]
- Wendel, J.F.; Cronn, R.C. Polyploidy and the evolutionary history of cotton. Adv. Agron. 2003, 87, 139–186. [Google Scholar]
- Wendel, J.F.; Grover, C.E. Taxonomy and Evolution of the Cotton Genus, Gossypium. In Cotton, Agronomy Monograph; Fang, D., Percy, R., Eds.; American Society of Agronomy Inc.: Madison, WI, USA, 2015; Volume 24, pp. 25–44. [Google Scholar]
- Hu, G.; Grover, C.E.; Yuan, D.; Dong, Y.; Miller, E.; Conover, J.L.; Wendel, J.F. Evolution and diversity of the cotton genome. In Cotton Precision Breeding; Rahman, M., Zafar, Y., Zhang, T., Eds.; Springer: Cham, Switzerland, 2021; pp. 25–78. [Google Scholar]
- Viot, C.R.; Wendel, J.F. Evolution of the cotton genus Gossypium and its domestication in the americas: A review. Crit. Rev. Plant Sci. 2023, 42, 1–33. [Google Scholar] [CrossRef]
- Gallagher, J.P.; Grover, C.E.; Rex, K.; Moran, M.; Wendel, J.F. A new species of cotton from Wake Atoll, Gossypium stephensii (Malvaceae). Syst. Bot. 2017, 42, 115–123. [Google Scholar] [CrossRef]
- Wendel, J.F.; Brubaker, C.L.; Alvarez, I.; Cronn, R.C.; Stewart, J.M. Genetics and Genomics of Cotton. In Plant Genetics and Genomics: Crops and Models; Paterson, A.H., Ed.; Springer: New York, NY, USA, 2009; pp. 3–22. [Google Scholar]
- Guo, Y.; Zhu, Q.; Zheng, S.; Li, M. Cloning of a MADS box gene (GhMADS3) from cotton and analysis of its homeotic role in transgenic tobacco. J. Genet. Genom. 2007, 34, 527–535. [Google Scholar] [CrossRef]
- Lightfoot, D.J.; Malone, K.M.; Timmis, J.N.; Orford, S.J. Evidence for alternative splicing of MADS-box transcripts in developing cotton fibre cells. Mol. Genet. Genom. 2008, 279, 75–85. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, B.Y.; Li, M.; Li, X.J.; Zhang, Z.T.; Li, Y.; Li, X.B. A MADS-box gene is specifically expressed in fibers of cotton (Gossypium hirsutum) and influences plant growth of transgenic Arabidopsis in a GA-dependent manner. Plant Physiol. Biochem. 2014, 75, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.Q.; Li, B.Y.; Zhang, Z.T.; Zhou, Y.; Jiang, J.; Li, X.B. Expression of a cotton MADS-box gene is regulated in anther development and in response to phytohormone signaling. J. Genet. Genom. 2010, 37, 805–816. [Google Scholar] [CrossRef]
- Jiang, S.-C.; Pang, C.-U.; Song, M.-Z.; Wei, H.-L.; Fan, S.-L.; Yu, S.-X. Analysis of MIKCC−Type MADS-Box Gene Family in Gossypium hirsutum. J. Integr. Agric. 2014, 13, 1239–1249. [Google Scholar] [CrossRef]
- Nardeli, S.M.; Artico, S.; Aoyagi, G.M.; de Moura, S.M.; da Franca Silva, T.; Grossi-de-Sa, M.F.; Romanel, E.; Alves-Ferreira, M. Genome-wide analysis of the MADS-box gene family in polyploid cotton (Gossypium hirsutum) and in its diploid parental species (Gossypium arboreum and Gossypium raimondii). Plant Physiol. Biochem. 2018, 127, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Yu, D.; Yang, Z.; Li, C.; Qanmber, G.; Li, Y.; Li, J.; Liu, Z.; Lu, L.; Wang, L.; et al. Genome-wide identification of the MIKC-Type MADS-box gene family in Gossypium hirsutum L. unravels their roles in flowering. Front. Plant Sci. 2017, 8, 384. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Sreedasyam, A.; Ando, A.; Song, Q.X.; De Santiago, L.M.; Hulse-Kemp, A.M.; Ding, M.Q.; Ye, W.X.; Kirkbride, R.C.; Jenkins, J.; et al. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 2020, 52, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef]
- Griffiths, S.; Dunford, R.P.; Coupland, G.; Laurie, D.A. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003, 131, 1855–1867. [Google Scholar] [CrossRef] [PubMed]
- Hamant, O.; Pautot, V. Plant development: A TALE story. Comptes Rendus Biol. 2010, 333, 371–381. [Google Scholar] [CrossRef]
- Siefers, N.; Dang, K.K.; Kumimoto, R.W.; Bynum, W.E.t.; Tayrose, G.; Holt, B.F., 3rd. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009, 149, 625–641. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, X.; Zhang, J.; Zhao, H.; Tan, S.; Xu, W.; Pan, J.; Yang, F.; Pi, E. ERF subfamily transcription factors and their function in plant responses to abiotic stresses. Front. Plant Sci. 2022, 13, 1042084. [Google Scholar] [CrossRef]
- Muller, M.; Munne-Bosch, S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef]
- Martin-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Sharoni, A.M.; Satoh, K.; Moumeni, A.; Venuprasad, R.; Serraj, R.; Kumar, A.; Leung, H.; Attia, K.; Kikuchi, S. Comprehensive gene expression analysis of the NAC gene family under normal growth conditions, hormone treatment, and drought stress conditions in rice using near-isogenic lines (NILs) generated from crossing Aday Selection (drought tolerant) and IR64. Mol. Genet. Genom. 2012, 287, 389–410. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Wang, H.; Tang, X. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Koyama, H.; Bhati, K.K.; Alok, A. The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 2021, 134, 475–495. [Google Scholar] [CrossRef] [PubMed]
- Litt, A.; Irish, V.F. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: Implications for the evolution of floral development. Genetics 2003, 165, 821–833. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef]
- Fang, Y.; You, J.; Xie, K.; Xie, W.; Xiong, L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 2008, 280, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Theune, M.L.; Bloss, U.; Brand, L.H.; Ladwig, F.; Wanke, D. Phylogenetic analyses and GAGA-motif binding studies of BBR/BPC proteins lend to clues in GAGA-motif recognition and a regulatory role in brassinosteroid signaling. Front. Plant Sci. 2019, 10, 466. [Google Scholar] [CrossRef] [PubMed]
- Petrella, R.; Caselli, F.; Roig-Villanova, I.; Vignati, V.; Chiara, M.; Ezquer, I.; Tadini, L.; Kater, M.M.; Gregis, V. BPC transcription factors and a Polycomb Group protein confine the expression of the ovule identity gene SEEDSTICK in Arabidopsis. Plant J. 2020, 102, 582–599. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Gong, Q.; Wang, L.; Jin, Y.; Xi, J.; Li, Z.; Qin, W.; Yang, Z.; Lu, L.; Chen, Q.; et al. Genome-Wide Study of YABBY Genes in Upland Cotton and Their Expression Patterns under Different Stresses. Front. Genet. 2018, 9, 33. [Google Scholar] [CrossRef]
- Grover, C.E.; Gallagher, J.P.; Szadkowski, E.P.; Yoo, M.J.; Flagel, L.E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 2012, 196, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.P.; Grover, C.E.; Hu, G.; Jareczek, J.J.; Wendel, J.F. Conservation and Divergence in Duplicated Fiber Coexpression Networks Accompanying Domestication of the Polyploid Gossypium hirsutum L. G3 2020, 10, 2879–2892. [Google Scholar] [CrossRef] [PubMed]
- Yoo, M.J.; Szadkowski, E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 2013, 110, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Rambani, A.; Page, J.T.; Udall, J.A. Polyploidy and the petal transcriptome of Gossypium. BMC Plant Biol. 2014, 14, 3. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef]
- Cheng, S.; Chen, P.; Su, Z.; Ma, L.; Hao, P.; Zhang, J.; Ma, Q.; Liu, G.; Liu, J.; Wang, H.; et al. High-resolution temporal dynamic transcriptome landscape reveals a GhCAL-mediated flowering regulatory pathway in cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 2021, 19, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wang, H.; Wei, H.; Gu, L.; Hao, P.; Sun, H.; Wu, A.; Cheng, S.; Yu, S. The MADS transcription factor GhAP1.7 coordinates the flowering regulatory pathway in upland cotton (Gossypium hirsutum L.). Gene 2021, 769, 145235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ren, Z.; Hu, G.; Zhao, S.; Wei, H.; Fan, S.; Ma, Q. Functional divergence of GhAP1.1 and GhFUL2 associated with flowering regulation in upland cotton (Gossypium hirsutum L.). J. Plant Physiol. 2022, 275, 153757. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wei, J.; Fan, S.; Song, M.; Pang, C.; Wei, H.; Wang, C.; Yu, S. Functional characterization of GhSOC1 and GhMADS42 homologs from upland cotton (Gossypium hirsutum L.). Plant Sci. 2016, 242, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yan, Y. GhSOC1s evolve to respond differently to the environmental cues and promote flowering in partially independent ways. Front. Plant Sci. 2022, 13, 882946. [Google Scholar] [CrossRef]
- de Moura, S.M.; Artico, S.; Lima, C.; Nardeli, S.M.; Berbel, A.; Oliveira-Neto, O.B.; Grossi-de-Sa, M.F.; Ferrandiz, C.; Madueno, F.; Alves-Ferreira, M. Functional characterization of AGAMOUS-subfamily members from cotton during reproductive development and in response to plant hormones. Plant Reprod. 2017, 30, 19–39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Nardeli, S.M.; Arge, L.W.P.; Artico, S.; de Moura, S.M.; Tschoeke, D.A.; de Freitas Guedes, F.A.; Grossi-de-Sa, M.F.; Martinelli, A.P.; Alves-Ferreira, M. Global gene expression profile and functional analysis reveal the conservation of reproduction-associated gene networks in Gossypium hirsutum. Plant Reprod. 2024, 37, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.S.; Irish, V.F. Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc. Natl. Acad. Sci. USA 2003, 100, 6558–6563. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Krizek, B.A.; Meyerowitz, E.M. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 1996, 93, 4793–4798. [Google Scholar] [CrossRef] [PubMed]
- de Martino, G.; Pan, I.; Emmanuel, E.; Levy, A.; Irish, V.F. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 2006, 18, 1833–1845. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Lopez, R.; Wegier, A.; Alavez, V.; Perez-Lopez, J.; Vazquez-Barrios, V.; Arroyo-Lambaer, D.; Ponce-Mendoza, A.; Kunin, W.E. The Mating System of the Wild-to-Domesticated Complex of Gossypium hirsutum L. Is Mixed. Front. Plant Sci. 2018, 9, 574. [Google Scholar] [CrossRef] [PubMed]
- Grover, C.E.; Yoo, M.J.; Lin, M.; Murphy, M.D.; Harker, D.B.; Byers, R.L.; Lipka, A.E.; Hu, G.; Yuan, D.; Conover, J.L.; et al. Genetic Analysis of the Transition from Wild to Domesticated Cotton (Gossypium hirsutum L.). G3 2020, 10, 731–754. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, E.W.; Mohamed, A.; Litt, A. Functional Divergence of APETALA1 and FRUITFULL is due to Changes in both Regulation and Coding Sequence. Front. Plant Sci. 2015, 6, 1076. [Google Scholar] [CrossRef] [PubMed]
- Litt, A. An evaluation of A function: Evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Plant Sci. 2007, 168, 73–91. [Google Scholar] [CrossRef]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Pelaz, S.; Gustafson-Brown, C.; Kohalmi, S.E.; Crosby, W.L.; Yanofsky, M.F. APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 2001, 26, 385–394. [Google Scholar] [CrossRef]
- Chen, L.; Yan, Y.; Ke, H.; Zhang, Z.; Meng, C.; Ma, L.; Sun, Z.; Chen, B.; Liu, Z.; Wang, G.; et al. SEP-like genes of Gossypium hirsutum promote flowering via targeting different loci in a concentration-dependent manner. Front. Plant Sci. 2022, 13, 990221. [Google Scholar] [CrossRef]
- Dreni, L.; Zhang, D. Flower development: The evolutionary history and functions of the AGL6 subfamily MADS-box genes. J. Exp. Bot. 2016, 67, 1625–1638. [Google Scholar] [CrossRef]
- Viaene, T.; Vekemans, D.; Becker, A.; Melzer, S.; Geuten, K. Expression divergence of the AGL6 MADS domain transcription factor lineage after a core eudicot duplication suggests functional diversification. BMC Plant Biol. 2010, 10, 148. [Google Scholar] [CrossRef] [PubMed]
- Rijpkema, A.S.; Zethof, J.; Gerats, T.; Vandenbussche, M. The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J. 2009, 60, 1–9. [Google Scholar] [CrossRef]
- Song, B.; Chen, J.; Lev-Yadun, S.; Niu, Y.; Gao, Y.; Ma, R.; Armbruster, W.S.; Sun, H. Multifunctionality of angiosperm floral bracts: A review. Biol. Rev. Camb. Philos. Soc. 2024, 99, 1100–1120. [Google Scholar] [CrossRef]
- Elmore, C.D. Contributions of the capsule wall and bracts to the developing cotton fruit. Crop Sci. 1973, 13, 751–752. [Google Scholar] [CrossRef]
- Zou, J.; Hu, W.; Loka, D.A.; Snider, J.L.; Zhu, H.; Li, Y.; He, J.; Wang, Y.; Zhou, Z. Carbon assimilation and distribution in cotton photosynthetic organs is a limiting factor affecting boll weight formation under drought. Front. Plant Sci. 2022, 13, 1001940. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.Y.; Zhang, Y.L.; Luo, H.H.; Li, W.; Oguchi, R.; Fan, D.Y.; Chow, W.S.; Zhang, W.F. Important photosynthetic contribution from the non-foliar green organs in cotton at the late growth stage. Planta 2012, 235, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of Phenylpropanoid Pathway and the Role of Polyphenols in Plants under Abiotic Stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [PubMed]
- Shomali, A.; Das, S.; Arif, N.; Sarraf, M.; Zahra, N.; Yadav, V.; Aliniaeifard, S.; Chauhan, D.K.; Hasanuzzaman, M. Diverse Physiological Roles of Flavonoids in Plant Environmental Stress Responses and Tolerance. Plants 2022, 11, 3158. [Google Scholar] [CrossRef]
- Zhuang, W.B.; Li, Y.H.; Shu, X.C.; Pu, Y.T.; Wang, X.J.; Wang, T.; Wang, Z. The Classification, Molecular Structure and Biological Biosynthesis of Flavonoids, and Their Roles in Biotic and Abiotic Stresses. Molecules 2023, 28, 3599. [Google Scholar] [CrossRef]
- Lv, Y.P.; Zhao, G.; Xie, Y.F.; Owusu, A.G.; Wu, Y.; Gao, J.S. Transcriptome and Metabolome Profiling Unveil Pigment Formation Variations in Brown Cotton Lines (Gossypium hirsutum L.). Int. J. Mol. Sci. 2023, 24, 5249. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.H.; Yan, Q.; Ding, H.; Luo, M.; Hou, L.; Zhang, M.; Yao, D.; Liu, H.S.; Li, X.; Zhao, J.; et al. Transcriptome and biochemical analyses revealed a detailed proanthocyanidin biosynthesis pathway in brown cotton fiber. PLoS ONE 2014, 9, e86344. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Li, Y.; Wang, S.; Zhang, L.; Liu, Y.; Xue, F.; Sun, Y.; Wang, Y.; Sun, J. Molecular analysis of proanthocyanidins related to pigmentation in brown cotton fibre (Gossypium hirsutum L.). J. Exp. Bot. 2014, 65, 5759–5769. [Google Scholar] [CrossRef]
- Cascallares, M.; Setzes, N.; Marchetti, F.; Lopez, G.A.; Distefano, A.M.; Cainzos, M.; Zabaleta, E.; Pagnussat, G.C. A Complex Journey: Cell Wall Remodeling, Interactions, and Integrity During Pollen Tube Growth. Front. Plant Sci. 2020, 11, 599247. [Google Scholar] [CrossRef]
- Mollet, J.C.; Leroux, C.; Dardelle, F.; Lehner, A. Cell Wall Composition, Biosynthesis and Remodeling during Pollen Tube Growth. Plants 2013, 2, 107–147. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Li, P.F.; Chen, M.K.; Lee, Y.I.; Yang, C.H. FOREVER YOUNG FLOWER negatively regulates ethylene response DNA-binding factors by activating an ethylene-responsive factor to control Arabidopsis floral organ senescence and abscission. Plant Physiol. 2015, 168, 1666–1683. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, J.; Wang, H.; Fu, Z.; Liu, J.; Yu, Y. Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments. J. Exp. Bot. 2011, 62, 825–840. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.; Ma, N.; Tian, J.; Luo, J.; Chen, J.; Li, J.; Zheng, Y.; Chen, X.; Fei, Z.; Gao, J. An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiol. 2013, 163, 775–791. [Google Scholar] [CrossRef]
- Althiab-Almasaud, R.; Chen, Y.; Maza, E.; Djari, A.; Frasse, P.; Mollet, J.C.; Mazars, C.; Jamet, E.; Chervin, C. Ethylene signaling modulates tomato pollen tube growth through modifications of cell wall remodeling and calcium gradient. Plant J. 2021, 107, 893–908. [Google Scholar] [CrossRef]
- Jia, H.; Yang, J.; Liesche, J.; Liu, X.; Hu, Y.; Si, W.; Guo, J.; Li, J. Ethylene promotes pollen tube growth by affecting actin filament organization via the cGMP-dependent pathway in Arabidopsis thaliana. Protoplasma 2018, 255, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Wang, M.; Tu, L.; Nie, Y.; Lin, Y.; Zhang, X. The flavonoid pathway regulates the petal colors of cotton flower. PLoS ONE 2013, 8, e72364. [Google Scholar] [CrossRef]
- Wilson, Z.A.; Song, J.; Taylor, B.; Yang, C. The final split: The regulation of anther dehiscence. J. Exp. Bot. 2011, 62, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Rieu, I.; Wolters-Arts, M.; Derksen, J.; Mariani, C.; Weterings, K. Ethylene regulates the timing of anther dehiscence in tobacco. Planta 2003, 217, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; He, H.; Li, Y.; Wang, F.; Yu, D. Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 2014, 164, 249–258. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, B.H.; Jung, J.H.; Park, S.K.; Song, J.T.; Kim, J.H. GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR Specify Meristematic Cells of Gynoecia and Anthers. Plant Physiol. 2018, 176, 717–729. [Google Scholar] [CrossRef]
- Monfared, M.M.; Simon, M.K.; Meister, R.J.; Roig-Villanova, I.; Kooiker, M.; Colombo, L.; Fletcher, J.C.; Gasser, C.S. Overlapping and antagonistic activities of BASIC PENTACYSTEINE genes affect a range of developmental processes in Arabidopsis. Plant J. 2011, 66, 1020–1031. [Google Scholar] [CrossRef] [PubMed]
- Simonini, S.; Roig-Villanova, I.; Gregis, V.; Colombo, B.; Colombo, L.; Kater, M.M. Basic pentacysteine proteins mediate MADS domain complex binding to the DNA for tissue-specific expression of target genes in Arabidopsis. Plant Cell 2012, 24, 4163–4172. [Google Scholar] [CrossRef]
- Ezquer, I.; Mizzotti, C.; Nguema-Ona, E.; Gotte, M.; Beauzamy, L.; Viana, V.E.; Dubrulle, N.; Costa de Oliveira, A.; Caporali, E.; Koroney, A.S.; et al. The Developmental Regulator SEEDSTICK Controls Structural and Mechanical Properties of the Arabidopsis Seed Coat. Plant Cell 2016, 28, 2478–2492. [Google Scholar] [CrossRef]
- Zhang, T.; Li, C.; Li, D.; Liu, Y.; Yang, X. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants. J. Plant Res. 2020, 133, 751–763. [Google Scholar] [CrossRef]
- Finet, C.; Floyd, S.K.; Conway, S.J.; Zhong, B.; Scutt, C.P.; Bowman, J.L. Evolution of the YABBY gene family in seed plants. Evol. Dev. 2016, 18, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Ye, W.; Song, Q.; Han, F.; Zhang, T.; Chen, Z.J. Histone Modifications Define Expression Bias of Homoeologous Genomes in Allotetraploid Cotton. Plant Physiol. 2016, 172, 1760–1771. [Google Scholar] [CrossRef]
- Han, J.; Yu, G.; Zhang, X.; Dai, Y.; Zhang, H.; Zhang, B.; Wang, K. Histone Maps in Gossypium darwinii Reveal Epigenetic Regulation Drives Subgenome Divergence and Cotton Domestication. Int. J. Mol. Sci. 2023, 24, 10607. [Google Scholar] [CrossRef]
- Guan, X.; Song, Q.; Chen, Z.J. Polyploidy and small RNA regulation of cotton fiber development. Trends Plant Sci. 2014, 19, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Masonbrink, R.E.; Gallagher, J.P.; Jareczek, J.J.; Renny-Byfield, S.; Grover, C.E.; Gong, L.; Wendel, J.F. CenH3 evolution in diploids and polyploids of three angiosperm genera. BMC Plant Biol. 2014, 14, 383. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, C.L.; Wendel, J.F. Reevaluating the origin of domesticated cotton (Gossypium hirsutum Malvaceae) using nuclear restriction fragment length polymorphisms (RFLPs). Am. J. Bot. 1994, 81, 1309–1326. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Jung, S.; Cheng, C.H.; Lee, T.; Zheng, P.; Buble, K.; Crabb, J.; Humann, J.; Hough, H.; Jones, D.; et al. CottonGen: The Community Database for Cotton Genomics, Genetics, and Breeding Research. Plants 2021, 10, 2805. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Fisher, R.A. On the Interpretation of χ2 from Contingency Tables, and the Calculation of P. J. R. Stat. Soc. B. Stat. Methodol. 1922, 85, 87. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
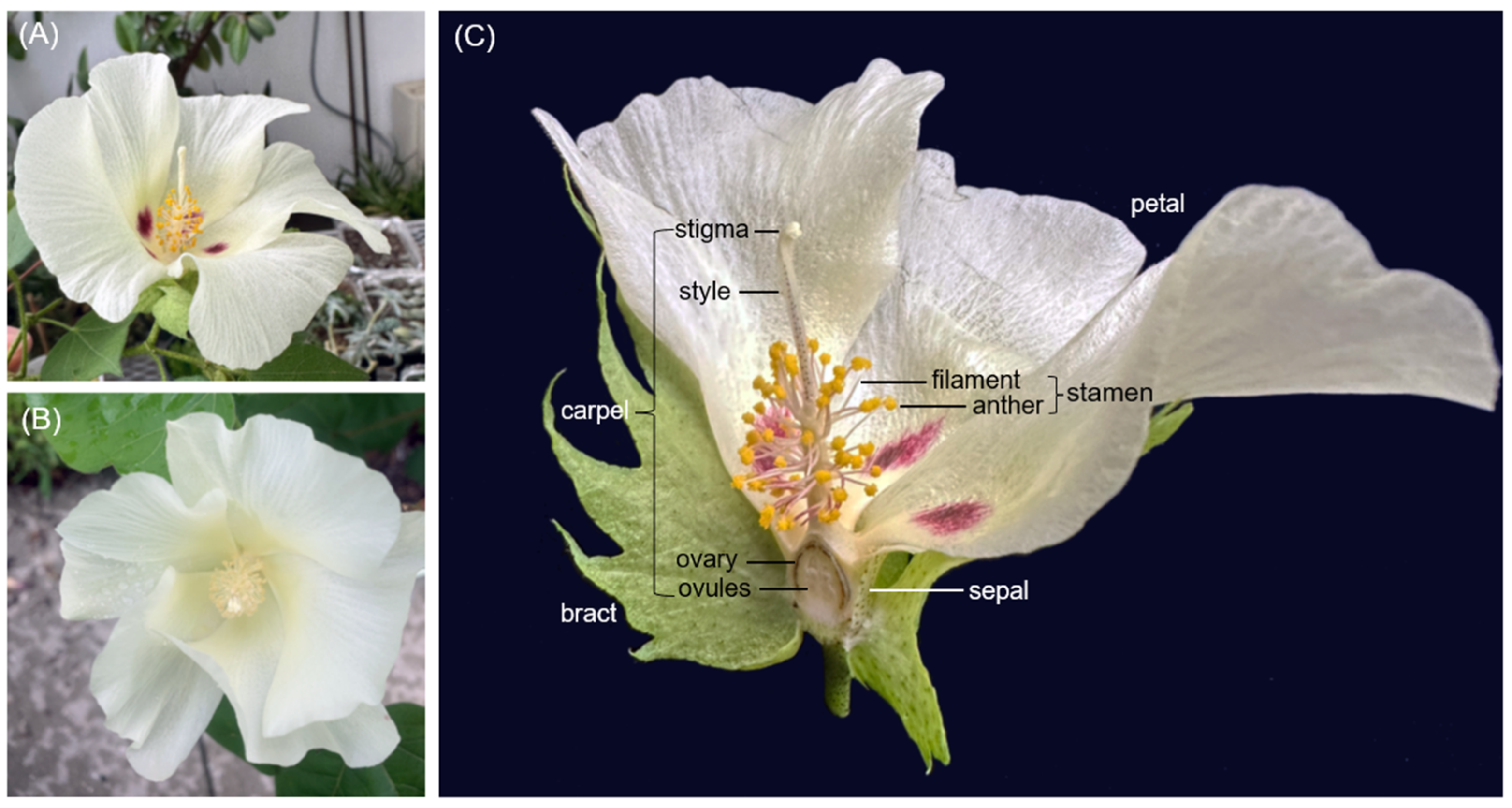
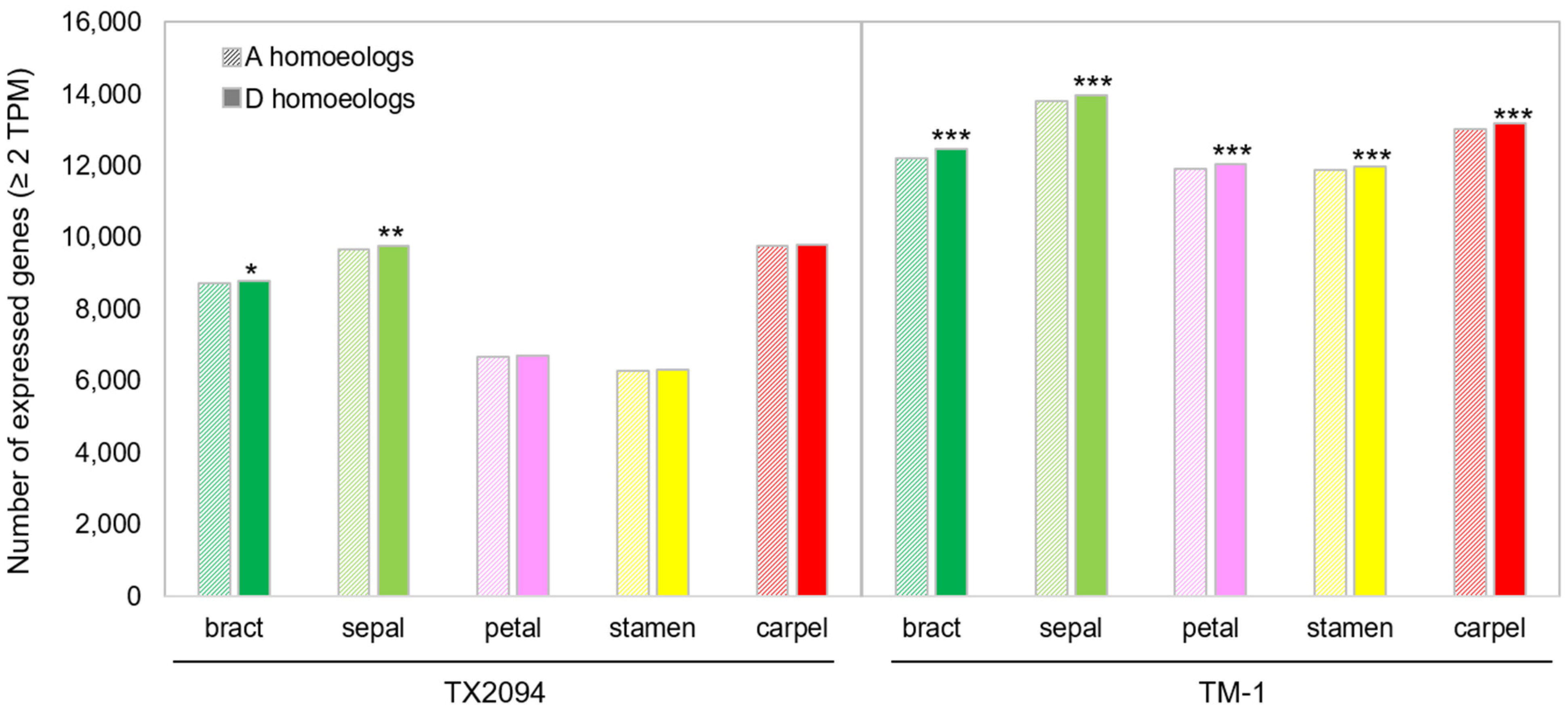
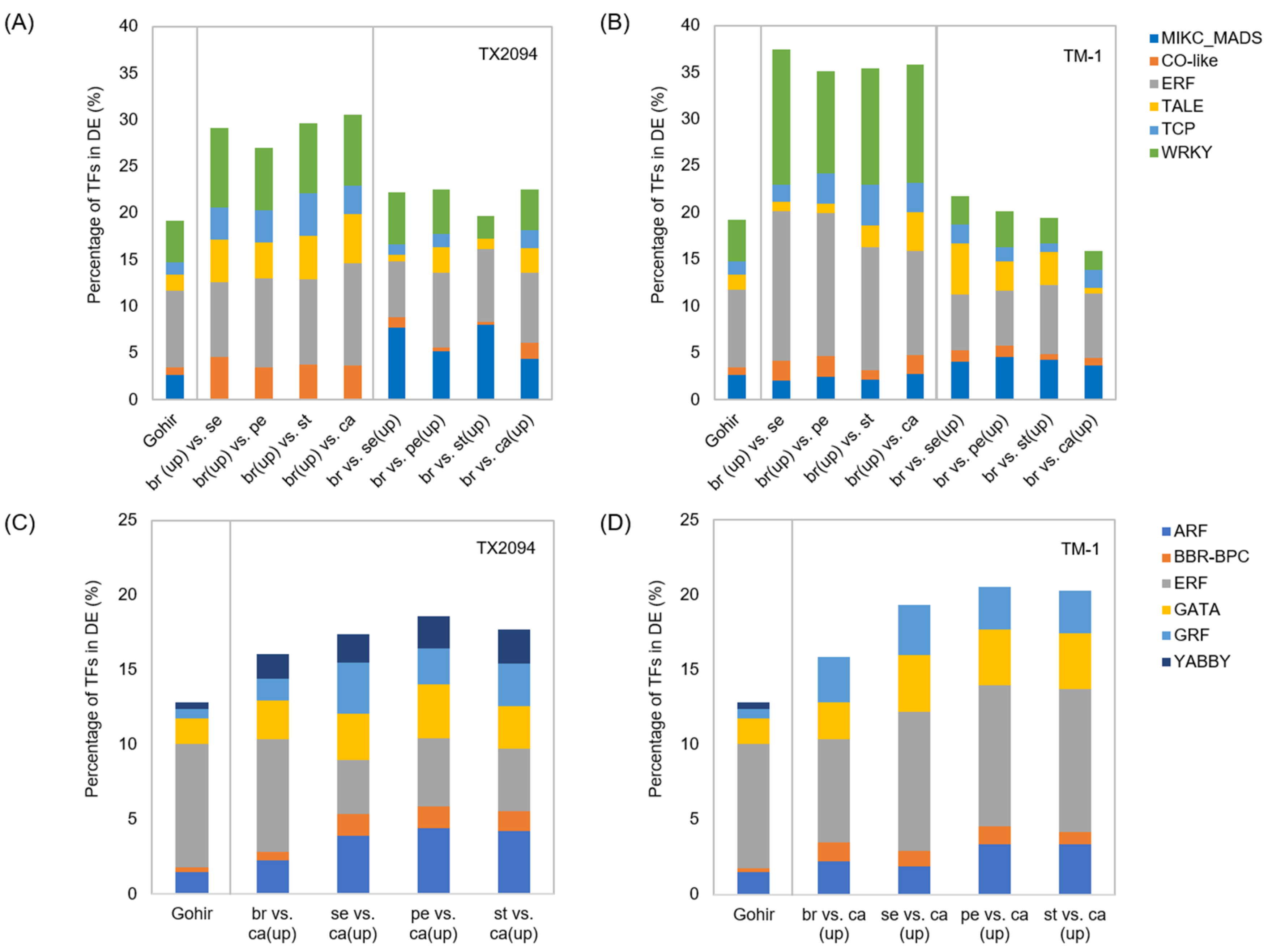
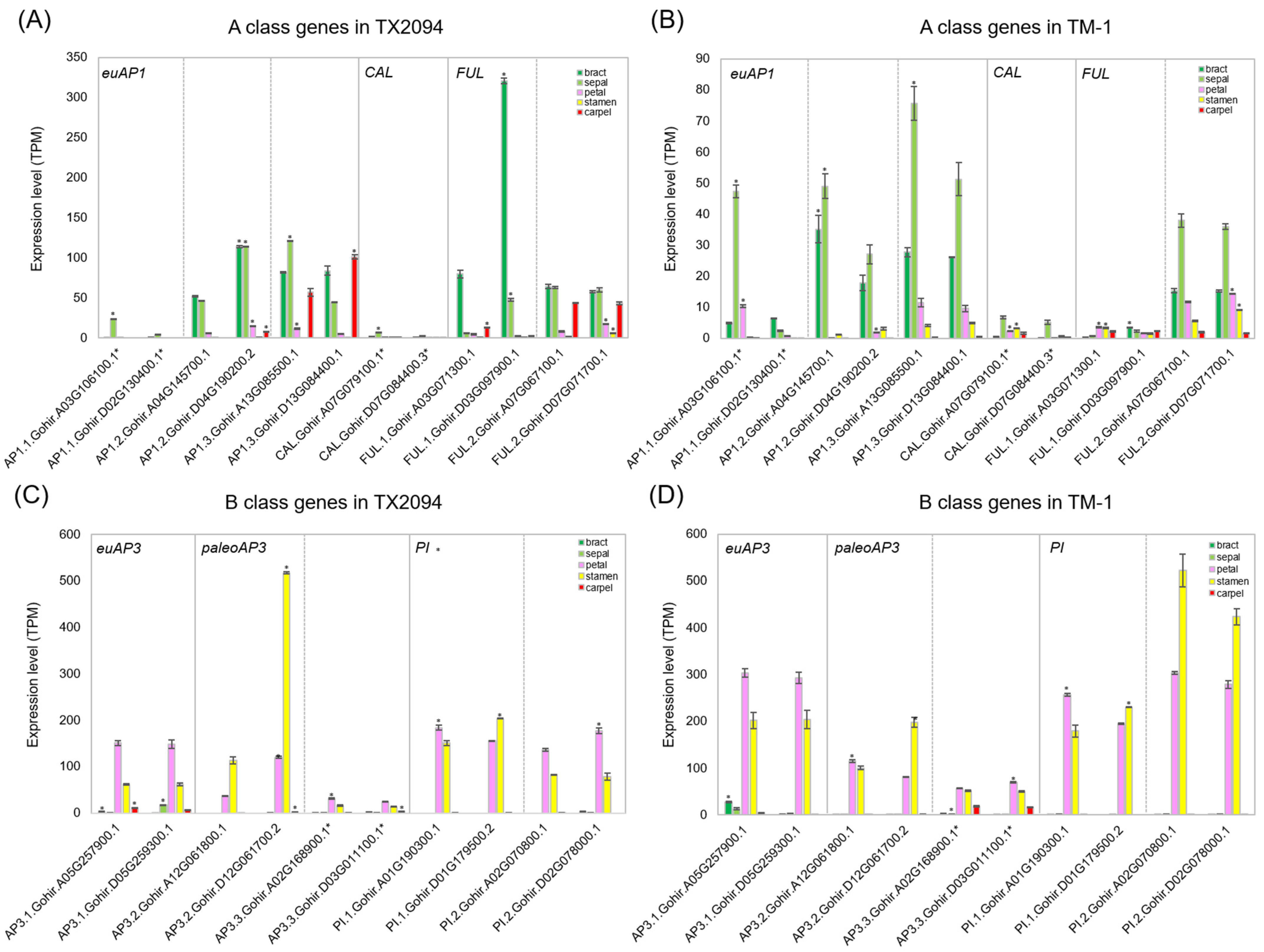
| Category | Bias | Bracts | Sepals | Petals | Stamens | Carpels |
|---|---|---|---|---|---|---|
| TX2094_only | A | 510 | 622 | 395 | 440 | 604 |
| D | 508 | 586 | 390 | 503 | 623 | |
| TM1_only | A | 1704 | 2711 | 2737 | 2727 | 2448 |
| D | 1742 | 2691 | 2702 | 2726 | 2523 | |
| Shared | A | 440 (7.9%) | 1151 (12.4%) | 1072 (12.5%) | 716 (9.1%) | 542 (7.2%) |
| D | 537 (9.7%) | 1311 (14.1%) | 1132 (13.2%) | 675 (8.6%) | 595 (7.9%) | |
| Changes from TX2094 to TM-1 | A→D | 62 | 106 | 89 | 49 | 78 |
| D→A | 44 | 104 | 86 | 55 | 82 |
| A > D | A < D | A = D | Low Expression (<2 TPM) | Total * | |
|---|---|---|---|---|---|
| TX2094 | 50 | 46 | 29 | 80 | 205 |
| TM-1 | 49 | 34 | 34 | 88 | 205 |
| MIKC-MADS Box Genes | HEB in TX2094 Only | HEB in TM-1 Only | Shared HEB | |||
|---|---|---|---|---|---|---|
| A Bias | D Bias | A Bias | D Bias | A Bias | D Bias | |
| A class | 3 | 5 | 7 | 0 | 2 | 4 |
| B class | 2 | 5 | 2 | 1 | 2 | 2 |
| C/D/Bsister class | 1 | 3 | 5 | 4 | 0 | 0 |
| E class | 10 | 2 | 0 | 8 | 4 | 5 |
| AGL6 | 0 | 1 | 3 | 0 | 9 | 4 |
| Total | 16 | 16 | 17 | 13 | 18 | 14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nobles, A.; Wendel, J.F.; Yoo, M.-J. Comparative Analysis of Floral Transcriptomes in Gossypium hirsutum (Malvaceae). Plants 2025, 14, 502. https://doi.org/10.3390/plants14040502
Nobles A, Wendel JF, Yoo M-J. Comparative Analysis of Floral Transcriptomes in Gossypium hirsutum (Malvaceae). Plants. 2025; 14(4):502. https://doi.org/10.3390/plants14040502
Chicago/Turabian StyleNobles, Alexander, Jonathan F. Wendel, and Mi-Jeong Yoo. 2025. "Comparative Analysis of Floral Transcriptomes in Gossypium hirsutum (Malvaceae)" Plants 14, no. 4: 502. https://doi.org/10.3390/plants14040502
APA StyleNobles, A., Wendel, J. F., & Yoo, M.-J. (2025). Comparative Analysis of Floral Transcriptomes in Gossypium hirsutum (Malvaceae). Plants, 14(4), 502. https://doi.org/10.3390/plants14040502






