The Defense Response of Honeyberry to Root Rot Pathogens: Evidence Based on Pathogen Identification and Host Mechanism
Abstract
1. Introduction
2. Results
2.1. Field Disease Symptoms
2.2. Identification of Pathogenic Fungi
2.3. Genus Fusarium as the Main Pathogenic Fungi
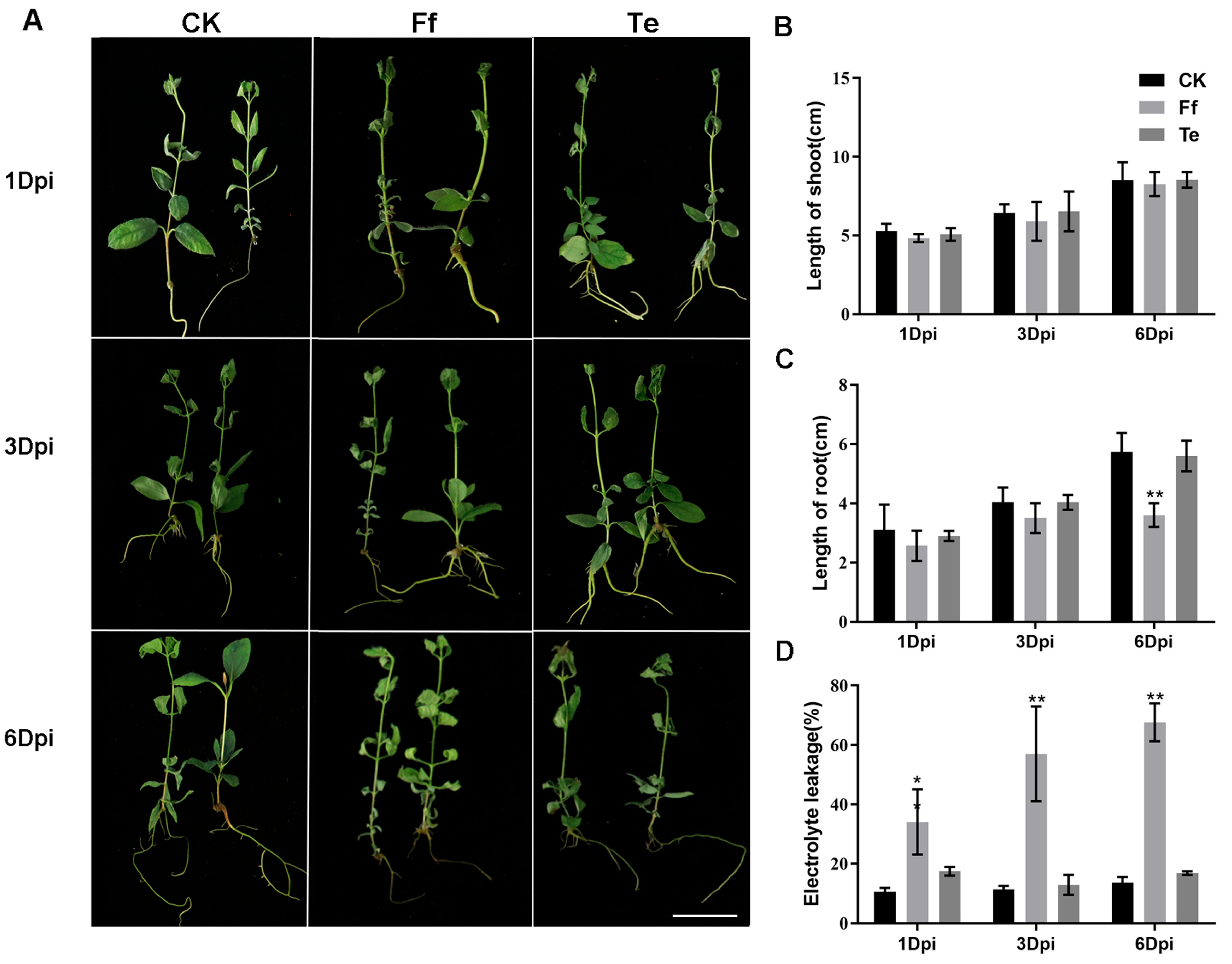
2.4. Phenotype and Electrolyte Leakage Analysis
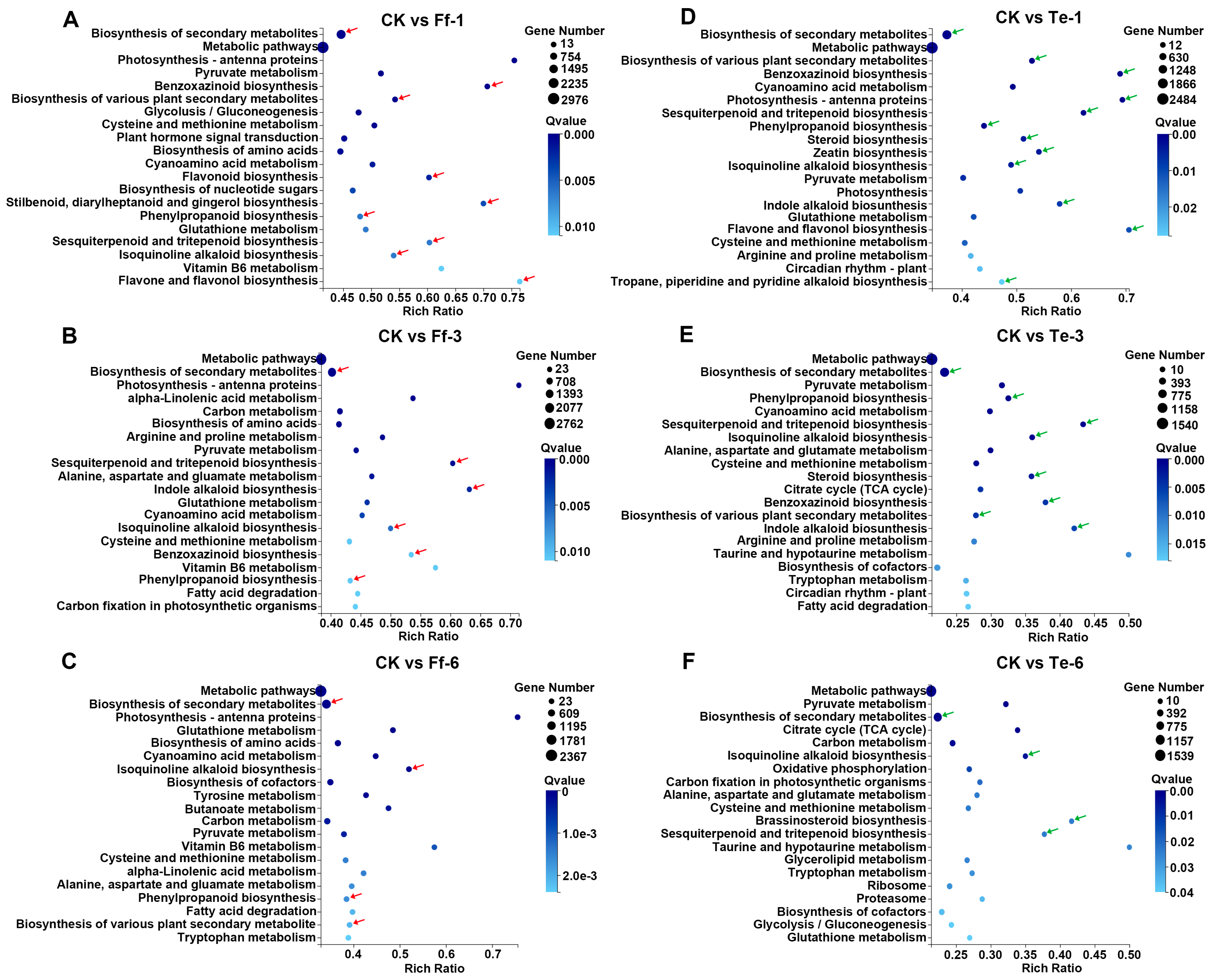
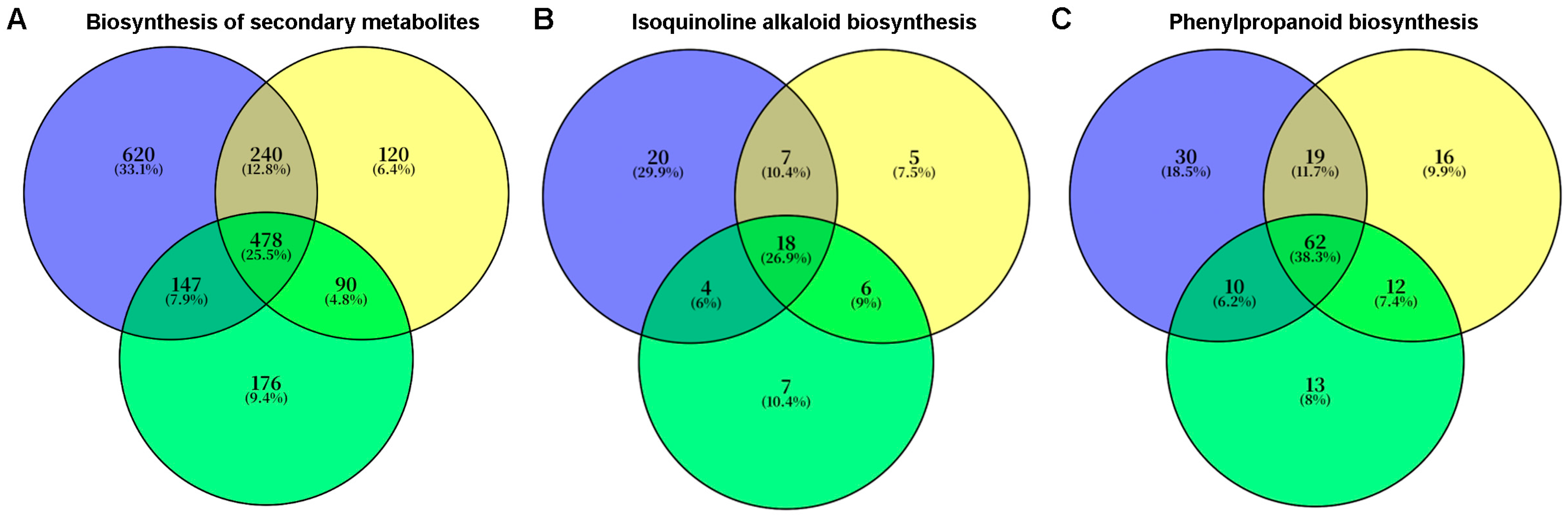
2.5. Transcriptomic Analysis
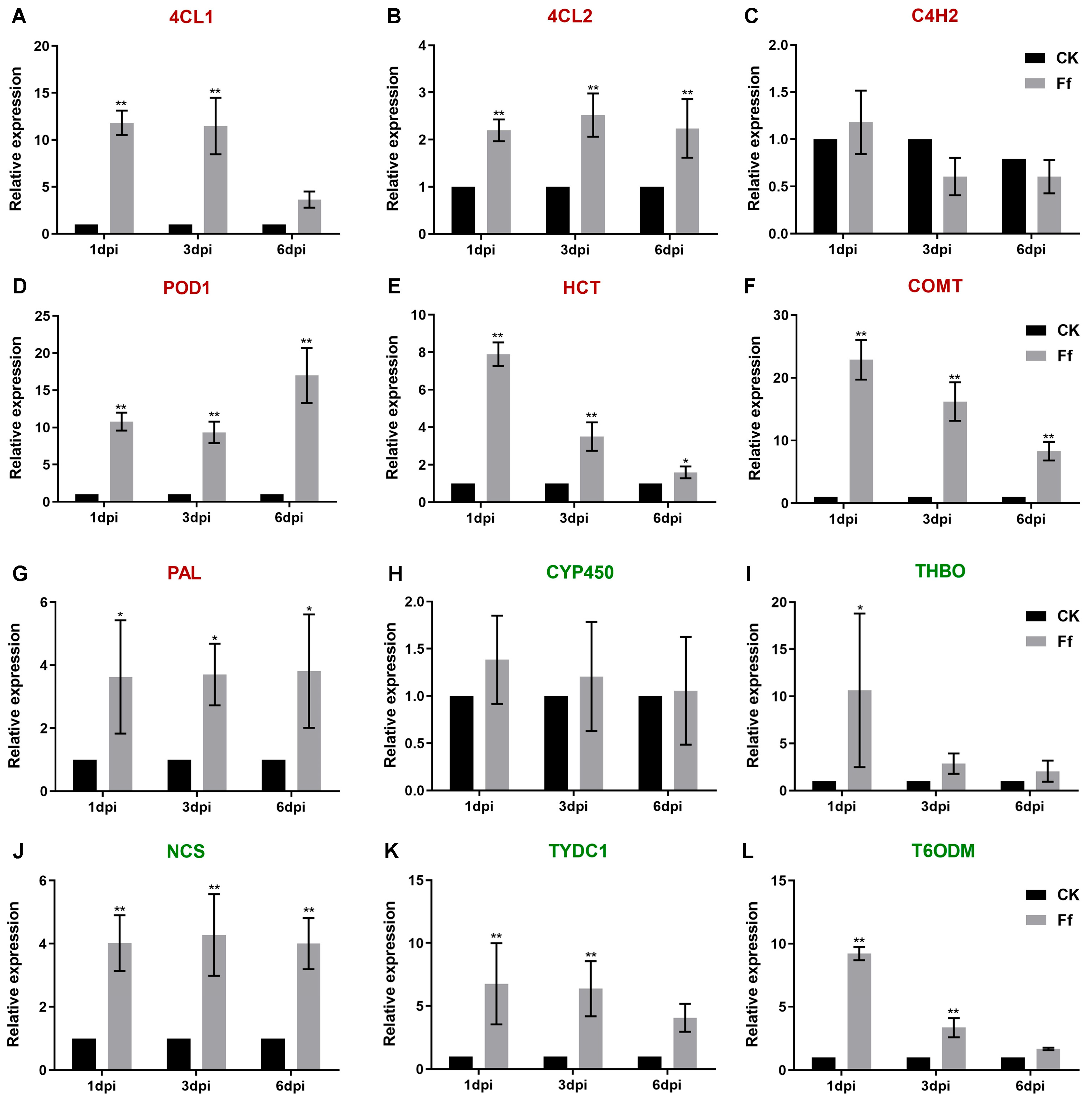
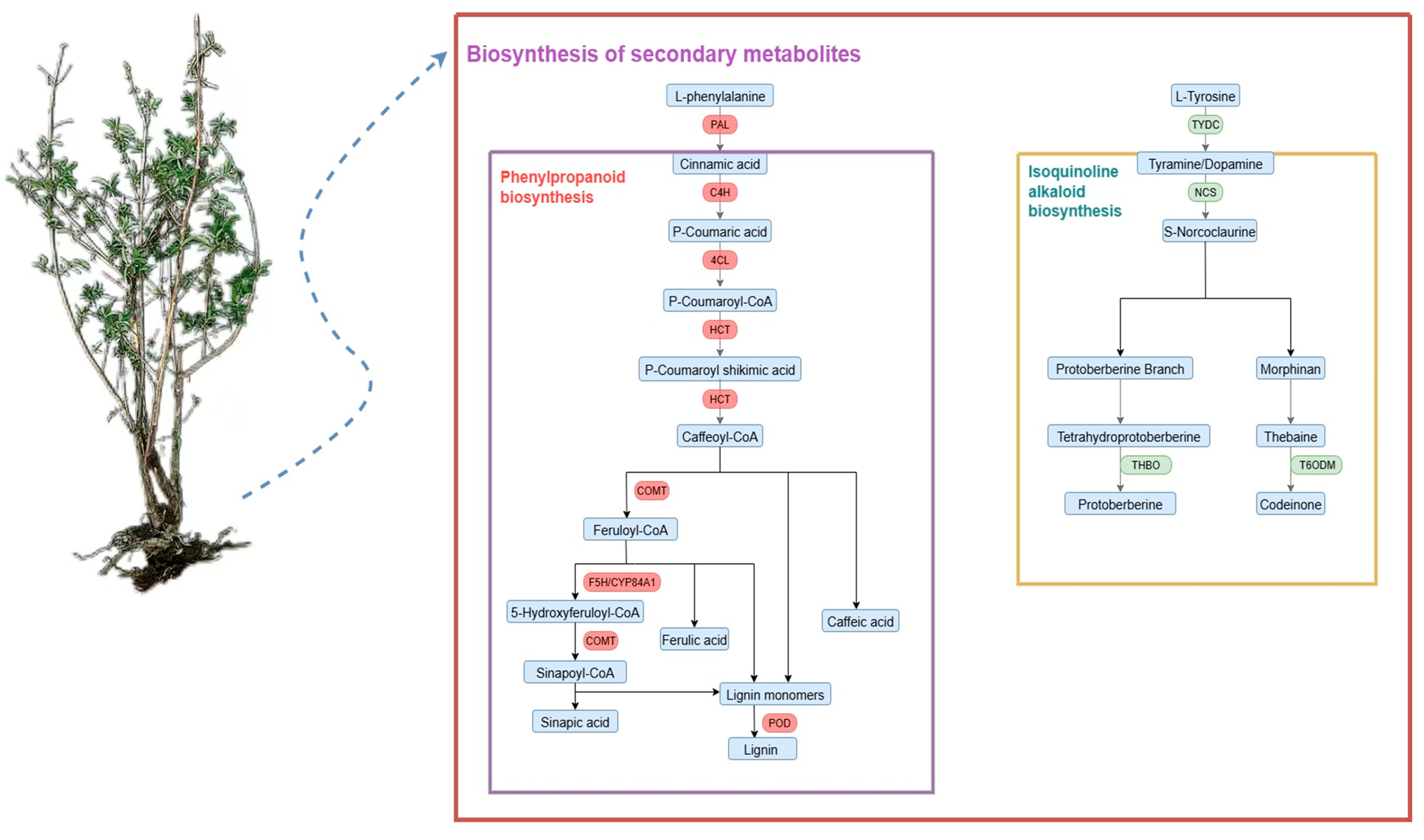
2.6. Differential Expression of Genes in Honeyberry Infected by F. foetens
3. Discussion
4. Materials and Methods
4.1. Collection and Preservation of Diseased Samples
4.2. Isolation and Purification of Pathogenic Fungi
4.3. Morphological and Molecular Identification
4.4. Pathogenicity Assay
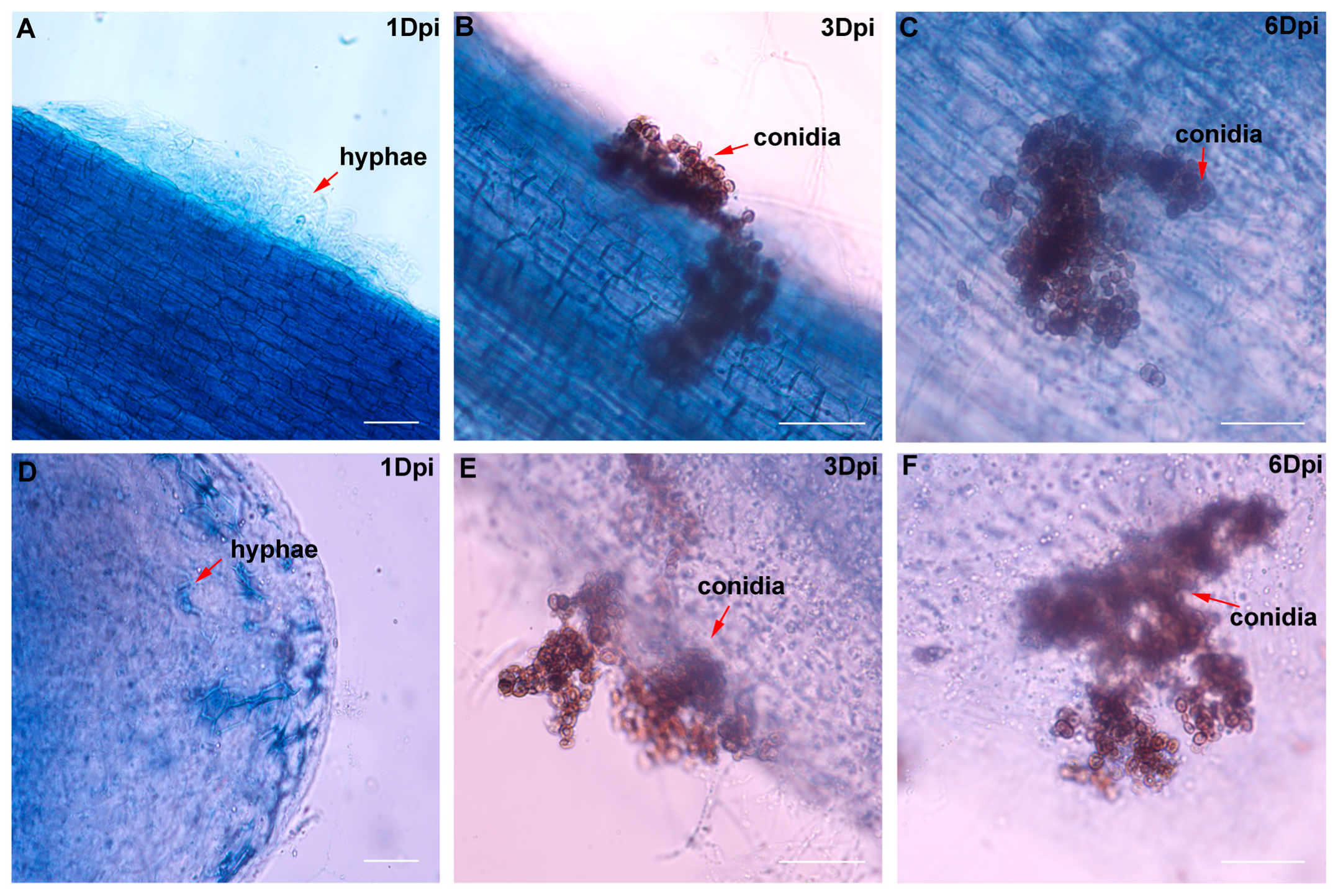
4.5. Infection Kinetics and Histological Observation
4.6. Phenotypic and Physiological Indicators
4.7. RNA Extraction
4.8. Library Preparation and Sequencing
4.9. Quality Control and De Novo Assembly
4.10. Differential Expression Analysis and Functional Enrichment
4.11. Real-Time Quantitative PCR (RT-qPCR)
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jakiene, E.; Kause, E.; Vaitkeviciene, N. Impact of picking time on leaf quality parameters of three varieties of honeysuckle (Lonicera caerulea L.). Agriculture 2025, 15, 257. [Google Scholar] [CrossRef]
- Chen, J.; Ren, B.B.; Bian, C.Y.; Qin, D.; Zhang, L.J.; Li, J.C.; Wei, J.; Wang, A.X.; Huo, J.W.; Gang, H.X. Transcriptomic and metabolomic analyses reveal molecular mechanisms associated with the natural abscission of blue honeysuckle (Lonicera caerulea L.) ripe fruits. Plant Physiol. Biochem. 2023, 199, 107740. [Google Scholar] [CrossRef]
- Guo, L.C.; Qiao, J.L.; Mikhailovich, M.S.; Chen, Y.X.; Ji, X.F.; She, H.H.; Zhang, L.J.; Zhang, Y.; Huo, J.W. Comprehensive structural analysis of anthocyanins in blue honeysuckle (Lonicera caerulea L.), bilberry (Vaccinium uliginosum L.), cranberry (Vaccinium macrocarpon Ait.), and antioxidant capacity comparison. Food Chem. X 2024, 23, 101734. [Google Scholar] [CrossRef]
- Negreanu-Pirjol, B.S.; Oprea, O.C.; Negreanu-Pirjol, T.; Roncea, F.N.; Prelipcean, A.M.; Craciunescu, O.; Iosageanu, A.; Artem, V.; Ranca, A.; Motelica, L.; et al. Health benefits of antioxidant bioactive compounds in the fruits and leaves of Lonicera caerulea L. and Aronia melanocarpa (Michx.) Elliot. Antioxidants 2023, 12, 951. [Google Scholar] [CrossRef]
- Jia, M.J.; Luo, J.Y.; Zhao, Z.H.; Jiang, L.; Bao, Y.H.; Huo, J.W. Co-analysis of network pharmacology and metabolomics: Revealing the mechanism of using mixed strains to produce multi-stage fermentation products of fresh blue honeysuckle (Lonicera caerulea L.) to against hyperlipidemia. Food Chem. X 2025, 29, 102852. [Google Scholar] [CrossRef]
- Li, X.N.; Fan, L.L.; Zhu, Q.; Liu, J.H.; Xie, Z.Y.; Cao, J.L.; Tan, J.Y.; Lin, L.; Li, X.S.; Wei, X.H. Blue honeysuckle (Lonicera caerulea L.)-anthocyanins and cyanidin-3-O-glucoside protect dopaminergic neurons against ferroptosis by activating the Nrf2-GPX7 axis. Free Radic. Biol. Med. 2025, 239, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Zhang, L.; Gao, Y.; Qin, D.; Huo, J. Two novel blue honeysuckle (Lonicera caerulea L.) cultivars: Lanjingling and Wulan. HortScience 2022, 57, 1145–1147. [Google Scholar] [CrossRef]
- Zhou, L.P.; Wang, H.; Yi, J.J.; Yang, B.R.; Li, M.S.; He, D.R.; Yang, W.; Zhang, Y.; Ni, H.W. Anti-tumor properties of anthocyanins from Lonicera caerulea ‘Beilei’ fruit on human hepatocellular carcinoma: In vitro and in vivo study. Biomed. Pharmacother. 2018, 104, 520–529. [Google Scholar] [CrossRef]
- Yan, H.H.; Li, Y.X.; Lv, Y.S.; Zhao, X.; Man, Z.J.; Chen, Z.X.; Guo, L.C.; Huo, J.W.; Sang, M.Y.; Li, C.Y.; et al. Postharvest fruit rot on blue honeysuckle (Lonicera caerulea L.) caused by Penicillium oxalicum newly reported in China. Crop Prot. 2025, 193, 107195. [Google Scholar] [CrossRef]
- Qi, H.X.; Qi, X.X.; Xu, W.H.; An, Z.L.; Fan, T.Y. First report of root rot caused by Fusarium avenaceum and Fusarium flocciferum on Astragalus membranaceus var. mongholicus in China. J. Phytopathol. 2024, 172, e13397. [Google Scholar] [CrossRef]
- Dita, M.; Barquero, M.; Heck, D.; Mizubuti, E.S.G.; Staver, C.P. Fusarium wilt of banana: Current knowledge on epidemiology and research needs toward sustainable disease management. Front. Plant Sci. 2018, 9, 1468. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Gardiner, D.M. Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: Recent progress and future prospects. Mol. Plant Pathol. 2018, 19, 1547–1562. [Google Scholar] [CrossRef]
- Naqvi, S.A.H.; Farhan, M.; Ahmad, M.; Kiran, R.; Shahbaz, M.; Abbas, A.; Hakim, F.; Shabbir, M.; Tan, Y.S.; Seelan, J.S.S. Fungicide resistance in Fusarium species: Exploring environmental impacts and sustainable management strategies. Arch. Microbiol. 2025, 207, 31. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Yang, Z.; Liu, J.G.; Li, X.D.; Wang, X.X.; Dai, C.C.; Zhang, T.L.; Carrión, V.J.; Wei, Z.; Cao, F.L.; et al. Crop rotation and native microbiome inoculation restore soil capacity to suppress a root disease. Nat. Commun. 2023, 14, 8126. [Google Scholar] [CrossRef]
- Li, Z.F.; Bai, X.L.; Jiao, S.; Li, Y.M.; Li, P.R.; Yang, Y.; Zhang, H.; Wei, G.H. A simplified synthetic community rescues Astragalus mongholicus from root rot disease by activating plant-induced systemic resistance. Microbiome 2021, 9, 217. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Yang, W.H.; Li, Y.; Zhao, Q.; Dong, Y. Wheat-faba bean intercropping can control Fusarium wilt in faba bean under F. commune and ferulic acid stress as revealed by histopathological analysis. Physiol. Mol. Plant Pathol. 2023, 124, 101965. [Google Scholar] [CrossRef]
- Romanu, R.; Liga, S.; Tripon, M.R.; Huiban, F.; Iliescu, D.; Dehelean, C.A.; Camelia, T. Chelidonium majus L.: A current perspective on isoquinoline alkaloids, emerging phytochemicals, alkaloid biosynthesis, and biological activities. Plants 2025, 14, 2627. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.Y.; Han, J.Y.; Li, X.J.; Zhou, J.Z.; Zhao, L.M.; Ying, Y.L.; Kai, G.Y. Atractylodes macrocephala root rot affects microbial communities in various root-associated niches. Agronomy 2024, 14, 2662. [Google Scholar] [CrossRef]
- Zhu, T.; Luo, X.Z.; Hao, C.X.; Zhu, Z.M.; Liu, L.; Deng, Z.N.; Cao, Y.L.; Ma, X.F. Isolation, identification and pathogenicity of two root rot pathogens Fusarium solani in Citrus. Hortic. Plant J. 2024, 9, 8. [Google Scholar] [CrossRef]
- Tian, L.X.; Ou, J.R.; Sun, X.; Miao, Y.J.; Pei, J.; Zhao, L.; Huang, L.F. The discovery of pivotal fungus and major determinant factor shaping soil microbial community composition associated with rot root of American ginseng. Plant Signal. Behav. 2021, 16, 1952372. [Google Scholar] [CrossRef]
- Pu, W.L.; Hu, Y.C.; Wu, T.; Luan, F.G.; Zheng, Y.M.; Zhou, L.F.; Zhou, X.D. Fusarium decemcellulare brick causes root rot of Cinnamomum camphora (Linn) Presl. For. Pathol. 2024, 54, e12867. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, Z.Y.; Yang, Y.G.; Ma, Q.H.; Zheng, Y.X.; Xu, C.X.; Gao, X.H.; Gao, W.N.; Huang, Z.Q.; Liu, X.L. Characterization of Fusarium species causing soybean root rot in Heilongjiang, China, and mechanism underlying the differences in sensitivity to DMI fungicides. Pestic. Biochem. Physiol. 2024, 200, 105828. [Google Scholar] [CrossRef]
- Shang, M.; Zhang, X.; Zhang, X.; Yan, J.; Liu, Q.; Zhai, H.; Gao, S.; Zhao, N.; He, S.; Zhang, H. A chromosome-level genome assembly of Fusarium foetens that causes sweet potato root rot facilitates the identification of a key Fusarium-specific pathogenicity factor. Plant Commun. 2025, 6, 101421. [Google Scholar] [CrossRef] [PubMed]
- Yi, B.L.; Ma, J.; Luo, L.; Ghani, M.I.; Siddique, J.A.; Tang, X.L.; Cernava, T.; Chen, X.Y.L. First report of Fusarium foetens causing tobacco root and stem rots in Guizhou, China. J. Phytopathol. 2025, 173, e70104. [Google Scholar] [CrossRef]
- Wei, X.J.; Jia, B.G.; Wang, X.; Li, N.Y.; Wang, S.N.; Li, L.; Zhang, A.; Zhang, H.T.; Wang, L.P. First report of Fusarium foetens causing root rot on lavender (Lavandula angustifolia) in China. J. Plant Pathol. 2023, 105, 1173–1174. [Google Scholar] [CrossRef]
- Imara, D.A.; Ghebrial, E.W.R.; El-Abeid, S.E.; Hussein, E.M.A.; Elsayed, M.I.E.; Yousef, R.S. Reduction of oxidative damage caused by Fusarium falciforme and Fusarium foetens in schefflera plants using chitosan nanoparticles loaded with l-proline or indole butyric acid. Chem. Biol. Technol. Agric. 2024, 11, 167. [Google Scholar] [CrossRef]
- Yang, K.K.; Zhou, G.; Chen, C.; Liu, X.H.; Wei, L.; Zhu, F.Y.; Liang, Z.H.; Chen, H.M. Joint metabolomic and transcriptomic analysis identify unique phenolic acid and flavonoid compounds associated with resistance to Fusarium wilt in cucumber (Cucumis sativus L.). Front. Plant Sci. 2024, 15, 1447860. [Google Scholar] [CrossRef] [PubMed]
- Fung, S.M.; Razali, Z.; Somasundram, C. Involvement of phenolic compounds and their composition in the defense response of Fusarium oxysporum infected berangan banana plants. Sains Malays. 2021, 50, 23–33. [Google Scholar] [CrossRef]
- Song, X.H.; Mei, P.Y.; Dou, T.; Liu, Q.D.; Li, L.Y. Multi-omics analysis reveals the resistance mechanism and the pathogens causing root rot of Coptis chinensis. Microbiol. Spectr. 2023, 11, 04803–04822. [Google Scholar] [CrossRef]
- Liu, X.M.; Tan, J.P.; Cheng, S.Y.; Chen, Z.X.; Ye, J.B.; Zheng, J.R.; Xu, F.; Zhang, W.W.; Liao, Y.L.; Yang, X.Y. Comparative transcriptome analysis provides novel insights into the molecular mechanism of berberine biosynthesis in Coptis chinensis. Sci. Hortic. 2022, 291, 110585. [Google Scholar] [CrossRef]
- Matsushima, Y.; Minami, H.; Hori, K.; Sato, F. Pathway engineering of benzylisoquinoline alkaloid biosynthesis in transgenic California poppy cells with ectopic expression of tetrahydroberberine oxidase from Coptis japonica. Plant Biotechnol. 2012, 29, 473–481. [Google Scholar] [CrossRef]
- Awal, M.A.; Abdullah, N.S.; Prismantoro, D.; Dwisandi, R.F.; Safitri, R.; Mohd-Yusuf, Y.; Suhaimi, N.S.M.; Doni, F. Mechanisms of action and biocontrol potential of Trichoderma against Fusarium in horticultural crops. Cogent Food Agric. 2024, 10, 2394685. [Google Scholar] [CrossRef]
- Niu, J.P.; Yan, X.; Bai, Y.G.; Li, W.D.; Lu, G.L.; Wang, Y.Y.; Liu, H.J.; Shi, Z.Y.; Liang, J.P. Integration of Transcriptomics and WGCNA to Characterize Trichoderma harzianum-Induced Systemic Resistance in Astragalus mongholicus for Defense against Fusarium solani. Genes 2024, 15, 1180. [Google Scholar] [CrossRef]
- Sehim, A.E.; Hewedy, O.A.; Altammar, K.A.; Alhumaidi, M.S.; Abd Elghaffar, R.Y. Trichoderma asperellum empowers tomato plants and suppresses Fusarium oxysporum through priming responses. Front. Microbiol. 2023, 14, 1140378. [Google Scholar] [CrossRef] [PubMed]
- Cuervo-Parra, J.A.; España, V.H.P.; Zavala-González, E.A.; Peralta-Gil, M.; Burgos, J.E.A.; Romero-Cortes, T. Trichoderma Asperellum strains as potential biological control agents against Fusarium verticillioides and Ustilago maydis in maize. Biocontrol Sci. Technol. 2022, 32, 624–647. [Google Scholar] [CrossRef]
- Nofal, A.M.; Abd El-Rahman, M.; Abdelghany, T.M.; Abd El-Mongy, M. Mycoparasitic nature of Egyptian Trichoderma isolates and their impact on suppression Fusarium wilt of tomato. Egypt. J. Biol. Pest Control 2021, 31, 103. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Smith-Unna, R.; Boursnell, C.; Patro, R.; Hibberd, J.M.; Kelly, S. TransRate: Reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 2016, 26, 1134–1144. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]

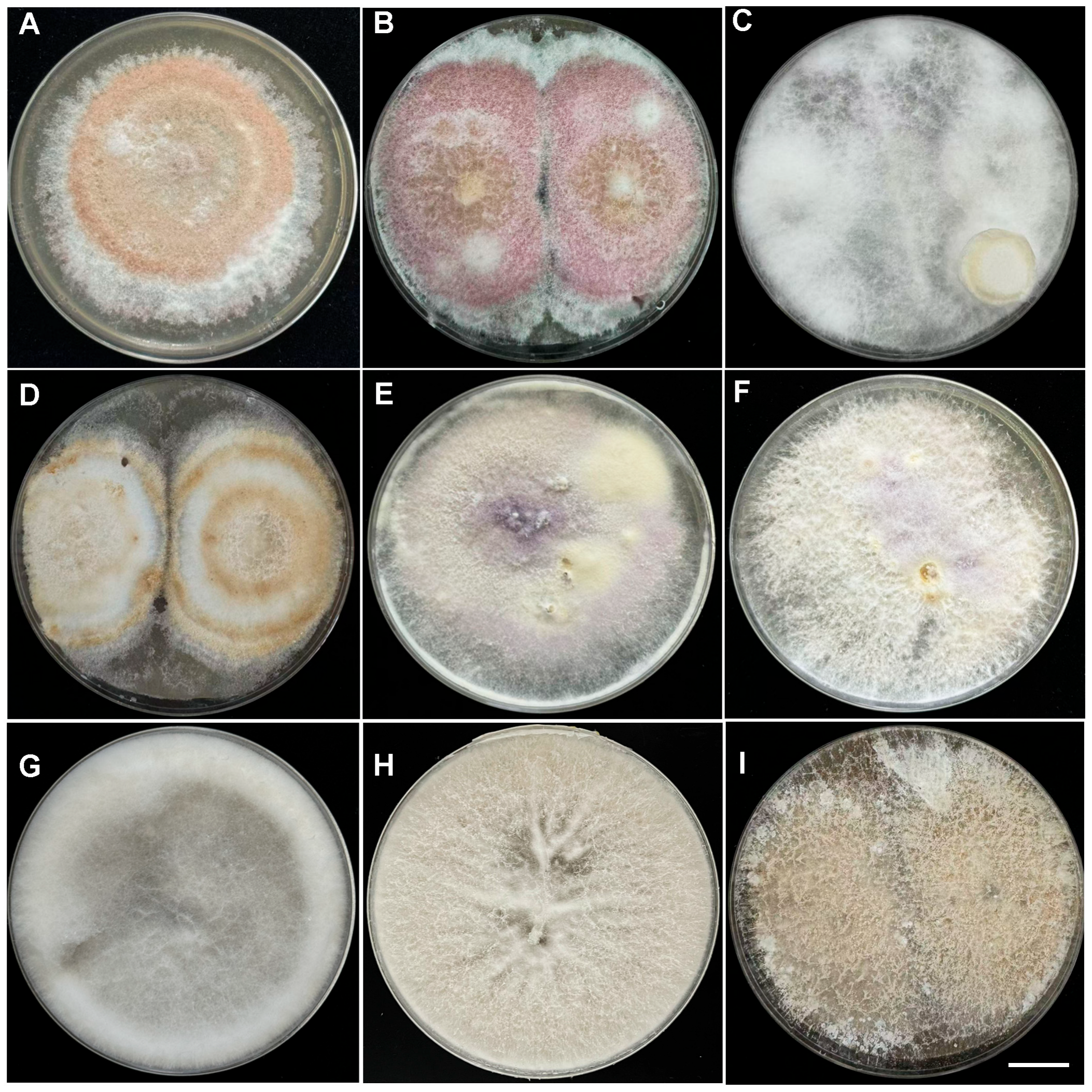


| Species | Colony Morphology | Microscopic Morphology | ||||
|---|---|---|---|---|---|---|
| Form | Elevation | Texture | Pigmentation | Microconidia Shape/Width | Macroconidia Shape/Width | |
| Lc1 | Irregular | Raised | Velvety | Light brown to honey-colored, and the edges are rose-brown to brick red | Ovoid, ellipsoid to subcylindrical/4–8 × 2–3 µm | Curved fusiform/ 31 × 4 µm |
| Lc2 | Irregular | Raised | Cottony | Red and white colors with a fluffy mycelium, and the edges are white | No Microconidia | Falcate to straight /38 × 5 µm |
| Lc3 | Circular | Raised | Velvety | White mycelium | Ellipsoidal to ovoid /5–12 × 2–3.5 µm | Spindle-shaped to lanceolate/25 × 5 µm |
| Lc4 | Irregular | Raised | Velvety | White mycelium which later produced a pale brown to dark brown pigmentation | Aseptate, hyaline, ovoid, fusiform (ellipsoidal), or slightly curved/8.2–13.8 × 2.0–4.5 µm | Slightly curved/18.1–41.6 × 2.5–4.9 µm |
| Lc5 | Irregular | Raised | Velvety | White to purple | Obovoid with flatted base, reniform, fusiform/4.2–13.9× 0.9–3.8 μm | Ring shape and straight/12.5–65.6 × 0.9–4.9 μm |
| Lc6 | Filamentous | Raised | Cottony | White to light purple | Ovoid to kidney shape/4.2–13.9 × 0.9–3.8 μm | Cylindrical/20.0–33.0 μm × 3.2–4.3 μm |
| Lc7 | Circular | Flat | Velvety | White | Globalose, subglobose or broadly/3–4.5 × 2.5–3.5 μm | Ellipsoidal or obovoid/5–7 × 3–4 μm |
| Lc8 | Circular | Flat | Cottony | White | Globalose or subglobose/3–4 × 2.5–3.5 μm | Ellipsoidal to Cylindrical/8–12 × 4–5.5 μm |
| Lc9 | Irregular | Raised | Cottony | Light yellow | No Microconidia | No Microconidia |
| Closest Type Strain | Isolate Number | Similarity | Accession Number † |
|---|---|---|---|
| Fusarium sp. | Lc1 | 91.18% | PX419195 |
| Fusarium kyushuense | Lc2 | 96.96% | PX419178 |
| Fusarium solani | Lc3 | 98.69% | PX421328 |
| Fusarium equiseti | Lc4 | 98.25% | PX421525 |
| Fusarium annulatum | Lc5 | 100.00% | PX421526 |
| Fusarium foetens | Lc6 | 98.92% | PX421528 |
| Trichoderma hamatum | Lc7 | 98.85% | PX421549 |
| Trichoderma evansii | Lc8 | 99.55% | PX421563 |
| Ceratobasidium sp. | Lc9 | 84.42% | PX421570 |
| Gene ID | Gene Symbol | Primers |
|---|---|---|
| Unigene34927-S21 | 4-coumarate--CoA ligase 1 | F: GCGATTGCTAAAAGTCCGATT R: TCATTCCATAACCCTGTCCAAG |
| Unigene20850-S18 | 4-coumarate--CoA ligase 2 | F: CAGAGAAGGAGAGTTGGCGG R: CACCACTTTCATCTCCTCACG |
| Unigene56302-S5 | cinnamate 4-hydroxylase 2 | F: TAGCCAAACAAGTCCTCCACAC R: ATGCTTTGACCATTACCCGTGA |
| Unigene31805-S17 | polyphenol oxidase 1 | F: CACCAAAATCACCAACACGCTC R: GTTGCCGCACAAGATAAAGAGC |
| Unigene12199-S5 | hydroxycinnamoyltransferase | F: TTTCCATACGCCGAGTGTCTAC R: ATAAAAGGGAACTAAGGCTCGG |
| Unigene50287-S21 | caffeic acid 3-O-methyltransferase | F: AGAAACACAAATCACCCCCCTA R: CACCATAGGTAAGACTGAGGCG |
| Unigene2458-S7 | prephenate aminotransferase | F: TCTCTACAGACCTCACCACAACC R: CTGAAGCAAGAATCTGAATGAGG |
| Unigene10300-S19 | cytochrome P450, family 706, subfamily A, polypeptide 2 | F: TACCTCGCCTGCCCTACCTTA R: GTGTAGCCCATCACCTCGCAT |
| Unigene39881-S8 | tetrahydroprotoberberine oxidase | F: TATCTGTGAGTGCGTTGGTGTC R: CAATGCCAAAGTTATGTCCTGC |
| Unigene57218-S10 | S-norcoclaurine synthase | F: TAGAAGTGAAGGTGGGAGCAGG R: GCCAGTTGAAGGGTTCCATACA |
| Unigene46037-S12 | Tyrosine decarboxylase 1 | F: TCTAATAGCAGTGTTGCGGGA R: TTACTCCACCACCTTTCCCTG |
| Unigene32362-S5 | thebaine 6-O-demethylase | F: GAGCAGTGACAAGCGAAACAAG R: TTGTAGGTTGCGGGTTTTTGTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, S.; Wei, D.; Chen, H.; Yu, J.; Gong, S.; Niu, Z.; Zhou, A.; Qiao, K.; Wang, J. The Defense Response of Honeyberry to Root Rot Pathogens: Evidence Based on Pathogen Identification and Host Mechanism. Plants 2025, 14, 3820. https://doi.org/10.3390/plants14243820
Qiao S, Wei D, Chen H, Yu J, Gong S, Niu Z, Zhou A, Qiao K, Wang J. The Defense Response of Honeyberry to Root Rot Pathogens: Evidence Based on Pathogen Identification and Host Mechanism. Plants. 2025; 14(24):3820. https://doi.org/10.3390/plants14243820
Chicago/Turabian StyleQiao, Siyu, Dianwen Wei, Hui Chen, Jinghua Yu, Shufang Gong, Zhiyong Niu, Aimin Zhou, Kun Qiao, and Jingang Wang. 2025. "The Defense Response of Honeyberry to Root Rot Pathogens: Evidence Based on Pathogen Identification and Host Mechanism" Plants 14, no. 24: 3820. https://doi.org/10.3390/plants14243820
APA StyleQiao, S., Wei, D., Chen, H., Yu, J., Gong, S., Niu, Z., Zhou, A., Qiao, K., & Wang, J. (2025). The Defense Response of Honeyberry to Root Rot Pathogens: Evidence Based on Pathogen Identification and Host Mechanism. Plants, 14(24), 3820. https://doi.org/10.3390/plants14243820






