Functional Characterization of Double-Bond Reductases in Dihydro-β-Ionone Biosynthesis in Cymbidium sinense
Abstract
1. Introduction
2. Results
2.1. Characterization of Volatile Compounds
2.2. Transcriptome Assembly and Annotation
2.3. Identification of Transcription Factors
2.4. Analysis of Carotenoid Biometabolic Pathway
2.5. Identification, Phylogenetic and Bioinformatic Characterization of CsDBRs
2.6. Spatiotemporal Expression Analysis of CsDBRs
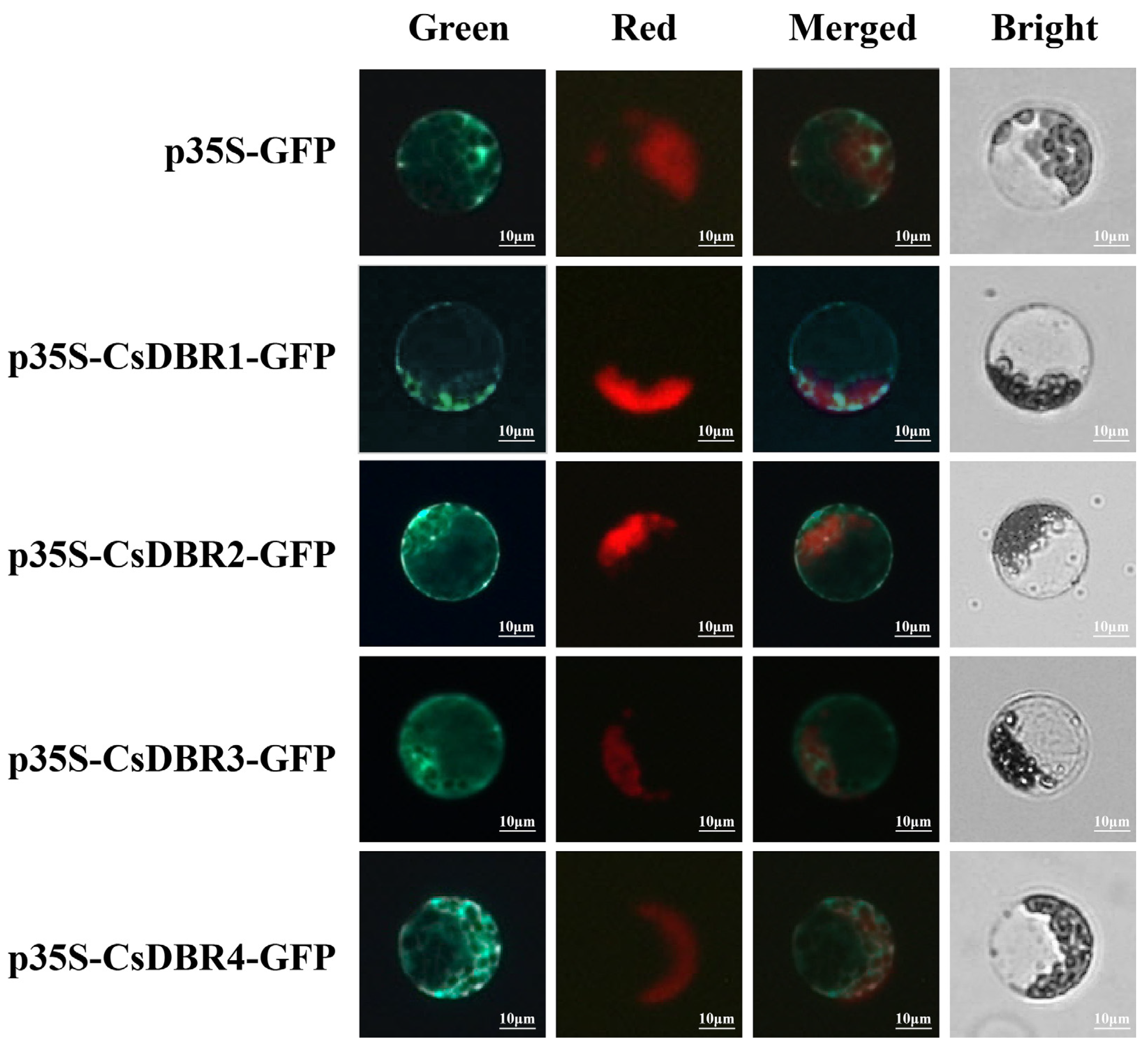
2.7. Cytoplasmic Localization of CsDBR Proteins
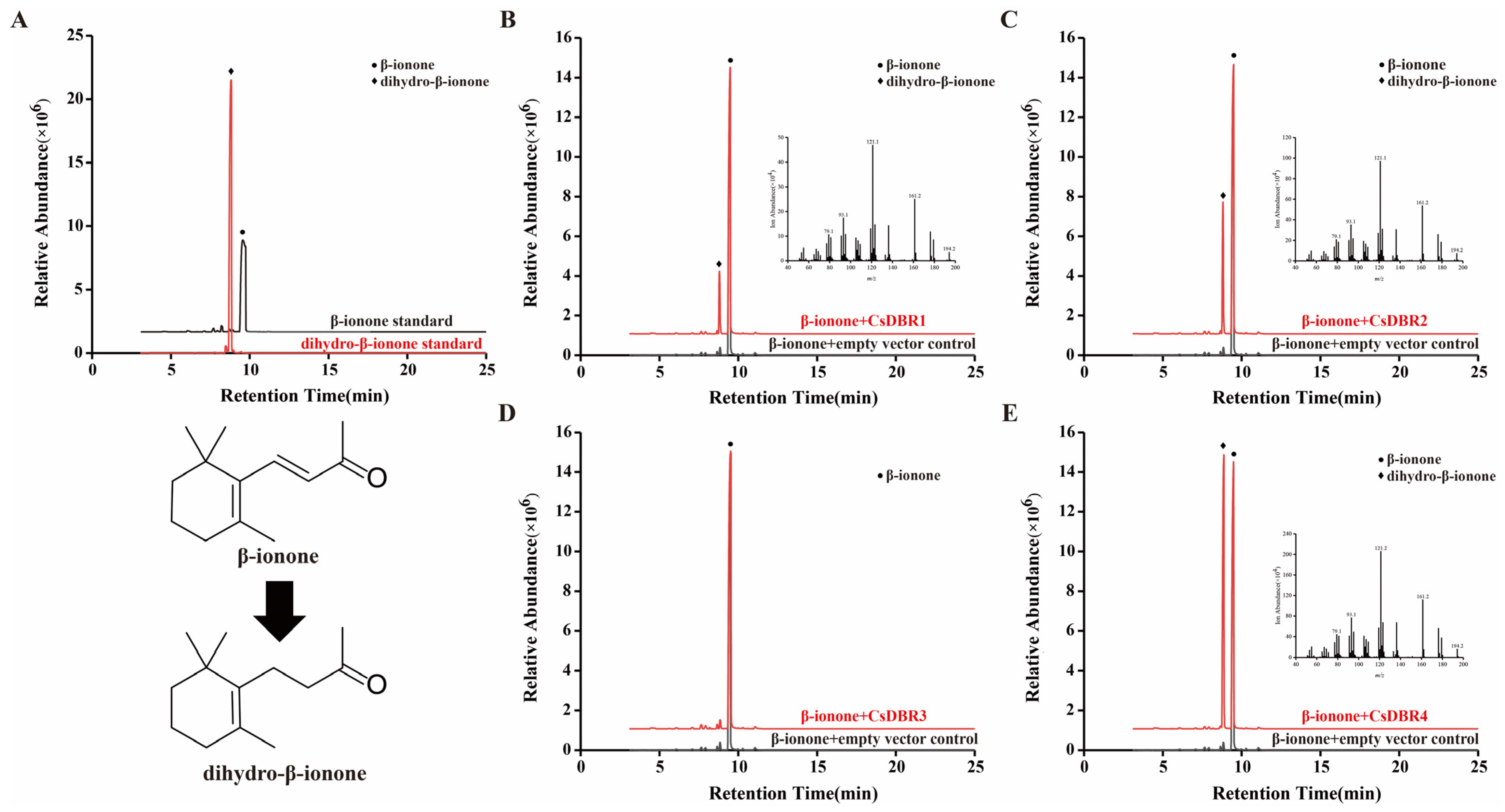
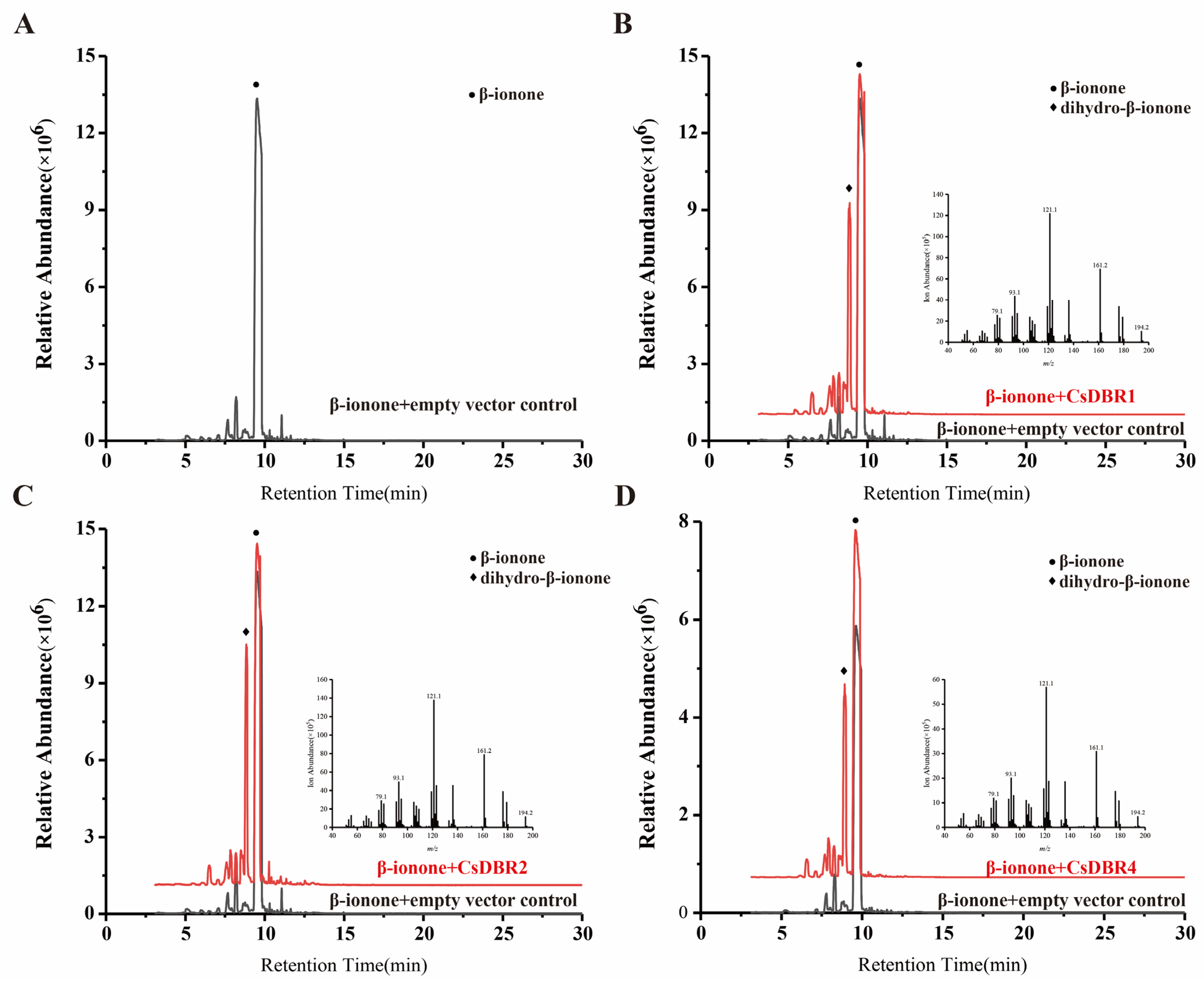
2.8. Heterologous Expression and In Vitro Enzymatic Assays of CsDBRs
3. Discussion
3.1. Analysis of Floral Fragrance Compounds and Release Patterns
3.2. Functional Diversification of CsDBR Enzymes in C. sinense
3.3. Catalytic Efficiency and Metabolic Channeling of CsDBRs in the Cytosol
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. GC-MS Profiling of Volatile Compounds
4.3. RNA Extraction and Transcriptome Sequencing
4.4. Validation of Transcriptomic Data Using qPCR
4.5. Identification and Structural Characterization of DBRs
4.6. Phylogenetic and Structural Comparative Analysis of DBRs
4.7. Subcellular Localization Analysis
4.8. Cloning and Heterologous Expression of CsDBRs in E. coli
4.9. Enzyme Activity Assay and Product Analysis
4.10. Transient Expression of CsDBRs in N. benthamiana
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CDS | Coding sequence |
| DEGs | Differentially expressed genes |
| GC-MS | Gas chromatography-mass spectrometry |
| GO | Gene Ontology |
| HS-SPME | Headspace solid-phase microextraction |
| HS-SPME-GC-MS | Headspace solid-phase microextraction coupled with gas chromatography-mass spectrometry |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KOG | EuKaryotic Orthologous Groups |
| MeJA | Methyl jasmonate |
| NR | Non-redundant protein |
| NT | Non-redundant nucleotide |
| ORF | Open reading frame |
| Pfam | Protein families database |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| TFs | Transcription factors |
| VOCs | Volatile organic compounds |
| MVA | Mevalonic acid |
| MVAP | Mevalonic acid 5-phosphate |
| MVAPP | 5-Diphosphomevalonic acid |
| IPP | Isopentenyl diphosphate |
| FPP | Farnesyl diphosphate |
| DXP | 1-deoxy-D-xylulose 5-phosphate |
| MEPP | 2-C-Methyl-D-erythritol 4-phosphate |
| MCP-ME | 4-(Cytidine 5′-diphospho)-2-C-methyl-D-erythritol |
| CDP-MEP | 2-Phospho-4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol |
| MECP | 2-C-Methyl-D-erythritol 2,4-cyclodiphosphate |
| HMBPP | 1-Hydroxy-2-methyl-2-butenyl 4-diphosphate |
| DMAPP | Dimethylallyl diphosphate |
| GPP | Geranyl pyrophosphate |
| GGPP | Geranylgeranyl diphosphate |
| AACT | Acetyl-CoA acetyltransferase |
| HMGS | Hydroxymethylglutaryl-CoA |
| HMGR | Hydroxymethylglutaryl-CoA reductase synthase |
| MVK | Mevalonate kinase |
| PMK | Phosphomevalonate kinase |
| MVD | Mevalonate diphosphate decarboxylase |
| FPPS | Farnesyl diphosphate synthase |
| DXS | 1-deoxy-D-xylulose-5-phosphate synthase |
| DXR | 1-deoxy-D-xylulose-5-phosphate reductase |
| MCT | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
| CMK | 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase |
| MECS | 2-C-methylerythritol 2,4-cyclodiphosphate synthase |
| HDS | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| HDR | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
| IDI | Isopentenyl pyrophosphate isomerase |
| GPPS | Geranyl pyrophosphate synthase |
| GGPPS | Geranylgeranyl diphosphate synthase |
| PSY | Phytoene synthase |
| PDS | Phytoene desaturase |
| Z-ISO | 15-cis-ζ-carotene isomerase |
| ZDS | ζ-carotene desaturase |
| CRTISO | Carotenoid isomerase |
| LCY-E | Lycopene ε-cyclase |
| LCY-B | Lycopene β-cyclase |
| CHY-E | ɛ-hydroxylase |
| CHY-B | β-hydroxylase |
| VDE | Violaxanthin de-epoxidase |
| ZEP | Zeaxanthin epoxidase |
| NXS | Neoxanthin synthase |
| NCED | Nine-cis-epoxycarotenoid dioxygenase |
| CCD | Carotenoid cleavage dioxygenase |
References
- Yang, F.; Gao, J.; Wei, Y.; Ren, R.; Zhang, G.; Lu, C.; Jin, J.; Ai, Y.; Wang, Y.; Chen, L.; et al. The genome of Cymbidium sinense reveals the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, K.; Zeng, S.; Teixeira da Silva, J.A.; Zhao, X.; Tian, C.-E.; Xia, H.; Duan, J. Transcriptome analysis of Cymbidium sinense and its application to the identification of genes associated with floral development. BMC Genom. 2013, 14, 279. [Google Scholar] [CrossRef]
- Su, S.; Shao, X.; Zhu, C.; Xu, J.; Lu, H.; Tang, Y.; Jiao, K.; Guo, W.; Xiao, W.; Liu, Z.; et al. Transcriptome-wide analysis reveals the origin of peloria in Chinese cymbidium (Cymbidium sinense). Plant Cell Physiol. 2018, 59, 2064–2074. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Que, C.; Teixeira da Silva, J.A.; Xu, S.; Li, D. Comparative transcriptomic and metabolic analyses reveal the molecular mechanism of ovule development in the orchid, Cymbidium sinense. Front. Plant Sci. 2022, 12, 814275. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.-Y.; Zhu, G.-F.; Lu, C.-Q.; Gao, J.; Li, J.; Xie, Q.; Wei, Y.-L.; Jin, J.-P.; Wang, F.-L.; Yang, F.-X. Functional conservation and divergence of SEPALLATA-like genes in floral development in Cymbidium sinense. Front. Plant Sci. 2023, 14, 1209834. [Google Scholar] [CrossRef]
- Zhu, G.; Yang, F.; Shi, S.; Li, D.; Wang, Z.; Liu, H.; Huang, D.; Wang, C. Transcriptome characterization of Cymbidium sinense ‘Dharma’ using 454 pyrosequencing and its application in the identification of genes associated with leaf color variation. PLoS ONE 2015, 10, e0128592. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Ren, R.; Wei, Y.; Jin, J.; Ahmad, S.; Lu, C.; Wu, J.; Zheng, C.; Yang, F.; Zhu, G. Comparative metabolomic analysis reveals distinct flavonoid biosynthesis regulation for leaf color development of Cymbidium sinense ‘Red Sun’. Int. J. Mol. Sci. 2020, 21, 1869. [Google Scholar] [CrossRef]
- Huang, J.H.; Shi, Z.R.; Zhang, Y.X.; Xiang, M.M. First report of anthracnose caused by Colletotrichum gloeosporioides on Cymbidium sinense in China. Plant Dis. 2012, 96, 915. [Google Scholar] [CrossRef]
- Liang, P.; Xu, M.; Jiang, J.; Tan, L.; Li, Q.; Zhou, Y.; Sun, Z. First report of stem rot in Cymbidium sinense caused by Fusarium oxysporum in China. Plant Dis. 2023, 107, 557. [Google Scholar] [CrossRef]
- Li, J.-W.; Chen, X.-D.; Hu, X.-Y.; Ma, L.; Zhang, S.-B. Comparative physiological and proteomic analyses reveal different adaptive strategies by Cymbidium sinense and C. tracyanum to drought. Planta 2018, 247, 69–97. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, Q.; Tu, S.; Ke, S.; Bi, Y.; Ahmad, S.; Zhang, D.; Liu, D.; Lan, S. Genome-wide identification analysis of the R2R3-MYB transcription factor family in Cymbidium sinense for insights into drought stress responses. Int. J. Mol. Sci. 2023, 24, 3235. [Google Scholar] [CrossRef]
- Li, J.; Zhu, G.; Wang, Z. Chemical variation in essential oil of Cymbidium sinense flowers from six cultivars. J. Essent. Oil Bear. Plants 2017, 20, 385–394. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, J.; Wang, Y.; Chingin, K.; Hua, R.; Zhu, L.; Rahman, M.M.; Frankevich, V.; Chen, H. Floral volatiles identification and molecular differentiation of Osmanthus fragrans by neutral desorption extractive atmospheric pressure chemical ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2019, 33, 1861–1869. [Google Scholar] [CrossRef]
- Drioiche, A.; Ailli, A.; Handaq, N.; Remok, F.; Elouardi, M.; Elouadni, H.; Al Kamaly, O.; Saleh, A.; Bouhrim, M.; Elazzouzi, H.; et al. Identification of compounds of Crocus sativus by GC-MS and HPLC/UV-ESI-MS and evaluation of their antioxidant, antimicrobial, anticoagulant, and antidiabetic properties. Pharmaceuticals 2023, 16, 545. [Google Scholar] [CrossRef]
- Wang, Y.; He, Y.; Liu, Y.; Wang, D. Analyzing volatile compounds of young and mature Docynia delavayi fruit by HS-SPME-GC-MS and rOAV. Foods 2022, 12, 59. [Google Scholar] [CrossRef]
- Maia, A.; Gibernau, M.; Dötterl, S.; Navarro, D.; Seifert, K.; Müller, T.; Schlindwein, C. The floral scent of Taccarum ulei (Araceae): Attraction of scarab beetle pollinators to an unusual aliphatic acyloin. Phytochemistry 2013, 93, 71–78. [Google Scholar] [CrossRef]
- Lalko, J.; Lapczynski, A.; McGinty, D.; Bhatia, S.; Letizia, C.S.; Api, A.M. Fragrance material review on dihydro-β-ionone. Food Chem. Toxicol. 2007, 45, S225–S228. [Google Scholar] [CrossRef]
- Lun, T.L.; Iwasaki, A.; Suenaga, K.; Kato-Noguchi, H. Isolation and identification of plant-growth inhibitory constituents from Polygonum chinense Linn and evaluation of their bioherbicidal potential. Plants 2023, 12, 1577. [Google Scholar] [CrossRef]
- Cáceres, L.A.; Lakshminarayan, S.; Yeung, K.K.-C.; McGarvey, B.D.; Hannoufa, A.; Sumarah, M.W.; Benitez, X.; Scott, I.M. Repellent and attractive effects of α-, β-, and dihydro-β-ionone to generalist and specialist herbivores. J. Chem. Ecol. 2016, 42, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, X.; Lindley, N.D.; Too, H. A “plug-n-play” modular metabolic system for the production of apocarotenoids. Biotechnol. Bioeng. 2018, 115, 174–183. [Google Scholar] [CrossRef]
- Qi, Z.; Tong, X.; Zhang, X.; Lin, H.; Bu, S.; Zhao, L. One-pot synthesis of dihydro-β-ionone from carotenoids using carotenoid cleavage dioxygenase and enoate reductase. Bioprocess Biosyst. Eng. 2022, 45, 891–900. [Google Scholar] [CrossRef]
- Kasahara, H.; Jiao, Y.; Bedgar, D.L.; Kim, S.-J.; Patten, A.M.; Xia, Z.-Q.; Davin, L.B.; Lewis, N.G. Pinus taeda phenylpropenal double-bond reductase: Purification, cDNA cloning, heterologous expression in Escherichia coli, and subcellular localization in P. taeda. Phytochemistry 2006, 67, 1765–1780. [Google Scholar] [CrossRef]
- Koeduka, T.; Watanabe, B.; Suzuki, S.; Hiratake, J.; Mano, J.; Yazaki, K. Characterization of raspberry ketone/zingerone synthase, catalyzing the alpha, beta-hydrogenation of phenylbutenones in raspberry fruits. Biochem. Biophys. Res. Commun. 2011, 412, 104–108. [Google Scholar] [CrossRef]
- Rao, S.T.; Rossmann, M.G. Comparison of super-secondary structures in proteins. J. Mol. Biol. 1973, 76, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Taneja, B.; Mande, S.C. Conserved structural features and sequence patterns in the GroES fold family. Protein Eng. 1999, 12, 815–818. [Google Scholar] [CrossRef]
- Zhou, P.; Shao, Y.; Jiang, Z.; Dang, J.; Qu, C.; Wu, Q. The revealing of a novel double bond reductase related to perilla ketone biosynthesis in Perilla frutescens. BMC Plant Biol. 2023, 23, 345. [Google Scholar] [CrossRef]
- Ibdah, M.; Berim, A.; Martens, S.; Valderrama, A.L.H.; Palmieri, L.; Lewinsohn, E.; Gang, D.R. Identification and cloning of an NADPH-dependent hydroxycinnamoyl-CoA double bond reductase involved in dihydrochalcone formation in Malus × domestica Borkh. Phytochemistry 2014, 107, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ng, D.W.-K.; Abeysinghe, J.K.; Kamali, M. Regulating the regulators: The control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 2018, 19, 3737. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [PubMed]
- Meraj, T.A.; Fu, J.; Raza, M.A.; Zhu, C.; Shen, Q.; Xu, D.; Wang, Q. Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes 2020, 11, 346. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Johansson, H.; Lee, K.P.; Bou-Torrent, J.; Stewart, K.; Steel, G.; Rodríguez-Concepción, M.; Halliday, K.J. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 2014, 10, e1004416. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, J.M.; Stanley, L.E.; LaFountain, A.M.; Frank, H.A.; Liu, C.; Yuan, Y.-W. An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 2016, 209, 1049–1057. [Google Scholar] [CrossRef]
- Zhu, F.; Luo, T.; Liu, C.; Wang, Y.; Yang, H.; Yang, W.; Zheng, L.; Xiao, X.; Zhang, M.; Xu, R.; et al. An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulata. New Phytol. 2017, 216, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Sun, Q.; Chen, H.; Mei, X.; Lu, S.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. Ethylene activation of carotenoid biosynthesis by a novel transcription factor CsERF061. J. Exp. Bot. 2021, 72, 3137–3154. [Google Scholar] [CrossRef]
- Yan, X.; Ding, W.; Wu, X.; Wang, L.; Yang, X.; Yue, Y. Insights into the MYB-related transcription factors involved in regulating floral aroma synthesis in sweet osmanthus. Front. Plant Sci. 2022, 13, 765213. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.J.; Tosi, T.; Bell, D.; Hleba, Y.B.; Polizzi, K.M.; Freemont, P.S. A cell-free synthetic biochemistry platform for raspberry ketone production. J. Biol. Eng. 2017, 11, 14. [Google Scholar]
- Caliandro, R.; Polsinelli, I.; Demitri, N.; Musiani, F.; Martens, S.; Benini, S. The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates. Int. J. Biol. Macromol. 2021, 171, 89–99. [Google Scholar] [CrossRef]
- Mansell, D.J.; Toogood, H.S.; Waller, J.; Hughes, J.M.X.; Levy, C.W.; Gardiner, J.M.; Scrutton, N.S. Biocatalytic asymmetric alkene reduction: Crystal structure and characterization of a double bond reductase from Nicotiana tabacum. ACS Catal. 2013, 3, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Youn, B.; Kim, S.-J.; Moinuddin, S.G.A.; Lee, C.; Bedgar, D.L.; Harper, A.R.; Davin, L.B.; Lewis, N.G.; Kang, C. Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970. J. Biol. Chem. 2006, 281, 40076–40088. [Google Scholar] [CrossRef]
- Yu, C.-S.; Cheng, C.-W.; Su, W.-C.; Chang, K.-C.; Huang, S.-W.; Hwang, J.-K.; Lu, C.-H. CELLO2GO: A web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS ONE 2014, 9, e99368. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.W.; Cameron, K.M.; Freudenstein, J.V.; Pridgeon, A.M.; Salazar, G.; van den Berg, C.; Schuiteman, A. An updated classification of Orchidaceae: Updated classification of Orchidaceae. Bot. J. Linn. Soc. 2015, 177, 151–174. [Google Scholar] [CrossRef]
- Ramya, M.; Jang, S.; An, H.-R.; Lee, S.-Y.; Park, P.-M.; Park, P.H. Volatile organic compounds from orchids: From synthesis and function to gene regulation. Int. J. Mol. Sci. 2020, 21, 1160. [Google Scholar] [CrossRef]
- Ramya, M.; Park, P.H.; Chuang, Y.-C.; Kwon, O.K.; An, H.R.; Park, P.M.; Baek, Y.S.; Kang, B.-C.; Tsai, W.-C.; Chen, H.-H. RNA sequencing analysis of Cymbidium goeringii identifies floral scent biosynthesis related genes. BMC Plant Biol. 2019, 19, 337. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Sharma, S.; Kaundal, M.; Sood, S.; Agnihotri, V.K. Variation in essential oil content and composition of damask rose (Rosa damascena Mill) flowers by salt application under mid hills of the Western Himalayas. J. Essent. Oil Bear. Plants 2016, 19, 297–306. [Google Scholar] [CrossRef]
- Schade, F.; Legge, R.; Thompson, J. Fragrance volatiles of developing and senescing carnation flowers. Phytochemistry 2001, 56, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Baek, Y.-S.; Ramya, M.; An, H.-R.; Park, P.-M.; Lee, S.-Y.; Baek, N.-I.; Park, P.-H. Volatiles profile of the floral organs of a new hybrid cymbidium, ‘Sunny Bell’ using headspace solid-phase microextraction gas chromatography-mass spectrometry analysis. Plants 2019, 8, 251. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xu, Y.; Wu, H.; Zhao, R.; Wang, X.; Wang, F.; Fu, Q.; Tang, T.; Shi, X.; Wang, B. Flavor characterization of native Xinjiang flat peaches based on constructing aroma fingerprinting and stoichiometry analysis. Foods 2023, 12, 2554. [Google Scholar] [CrossRef] [PubMed]
- Cha, D.H.; Powell, T.H.Q.; Feder, J.L.; Linn, C.E. Identification of a new blend of host fruit volatiles from red downy hawthorn, Crataegus mollis, attractive to Rhagoletis pomonella flies from the northeastern United States. J. Chem. Ecol. 2018, 44, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Aihaiti, K.; Li, J.; Xu, N.-N.; Tang, D.; Aisa, H.A. Monoterpenoid derivatives from Hyssopus cuspidatus Boriss. and their bioactivities. Fitoterapia 2023, 165, 105432. [Google Scholar] [CrossRef] [PubMed]
- Ômura, H.; Honda, K.; Hayashi, N. Floral scent of Osmanthus fragrans discourages foraging behavior of cabbage butterfly, Pieris rapae. J. Chem. Ecol. 2000, 26, 655–666. [Google Scholar] [CrossRef]
- Gonçalves-Souza, P.; Schlindwein, C.; Dötterl, S.; Paiva, E.A.S. Unveiling the osmophores of Philodendron adamantinum (Araceae) as a means to understanding interactions with pollinators. Ann. Bot. 2017, 119, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Hu, H.; Ma, L.; Zhou, Q.; Yu, L.; Zeng, S. Carbon-carbon double-bond reductases in nature. Drug Metab. Rev. 2014, 46, 362–378. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Wang, L.; Sun, J.; Jiang, X.; Cong, H.; Sun, H.; Qiao, F. Functional characterization of a Colchicum autumnale L. double-bond reductase (CaDBR1) in colchicine biosynthesis. Planta 2022, 256, 95. [Google Scholar] [CrossRef]
- Ringer, K.L.; McConkey, M.E.; Davis, E.M.; Rushing, G.W.; Croteau, R. Monoterpene double-bond reductases of the (−)-menthol biosynthetic pathway: Isolation and characterization of cDNAs encoding (−)-isopiperitenone reductase and (+)-pulegone reductase of peppermint. Arch. Biochem. Biophys. 2003, 418, 80–92. [Google Scholar] [CrossRef]
- Fisher, K.E.; Tillett, R.L.; Footohi, M.; Caldwell, C.; Petereit, J.; Schlauch, K.; Tittiger, C.; Blomquist, G.J.; MacLean, M. RNA-seq used to identify ipsdienone reductase (IDONER): A novel monoterpene carbon-carbon double bond reductase central to Ips confusus pheromone production. Insect Biochem. Mol. Biol. 2021, 129, 103513. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Wuzhang, K.; Peng, B.; Chen, T.; Zhang, Y.; Liu, H.; Li, L.; Fu, X.; Tang, K. Engineering the expression of plant secondary metabolites-genistein and scutellarin through an efficient transient production platform in Nicotiana benthamiana L. Front. Plant Sci. 2022, 13, 994792. [Google Scholar] [CrossRef]
- Yue, Y.; Wang, L.; Yu, R.; Chen, F.; He, J.; Li, X.; Yu, Y.; Fan, Y. Coordinated and high-level expression of biosynthetic pathway genes is responsible for the production of a major floral scent compound methyl benzoate in Hedychium coronarium. Front. Plant Sci. 2021, 12, 650582. [Google Scholar] [CrossRef]
- Huang, M.; Ma, C.; Yu, R.; Mu, L.; Hou, J.; Yu, Y.; Fan, Y. Concurrent changes in methyl jasmonate emission and the expression of its biosynthesis-related genes in Cymbidium ensifolium flowers. Physiol. Plant. 2015, 153, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Abbas, F.; Zhou, Y.; He, J.; Ke, Y.; Qin, W.; Yu, R.; Fan, Y. Metabolite and transcriptome profiling analysis revealed that melatonin positively regulates floral scent production in Hedychium coronarium. Front. Plant Sci. 2021, 12, 808899. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Chu, Z.; Wang, H.; Wang, G.; Wu, S.; Yang, Y. Selection and validation of reference genes for quantitative real-time PCR in Cymbidium sinense. BioTechniques 2022, 72, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree visualization by one table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Yu, R.; Fan, Y. Characterization of two monoterpene synthases involved in floral scent formation in Hedychium coronarium. Planta 2014, 240, 745–762. [Google Scholar] [CrossRef]
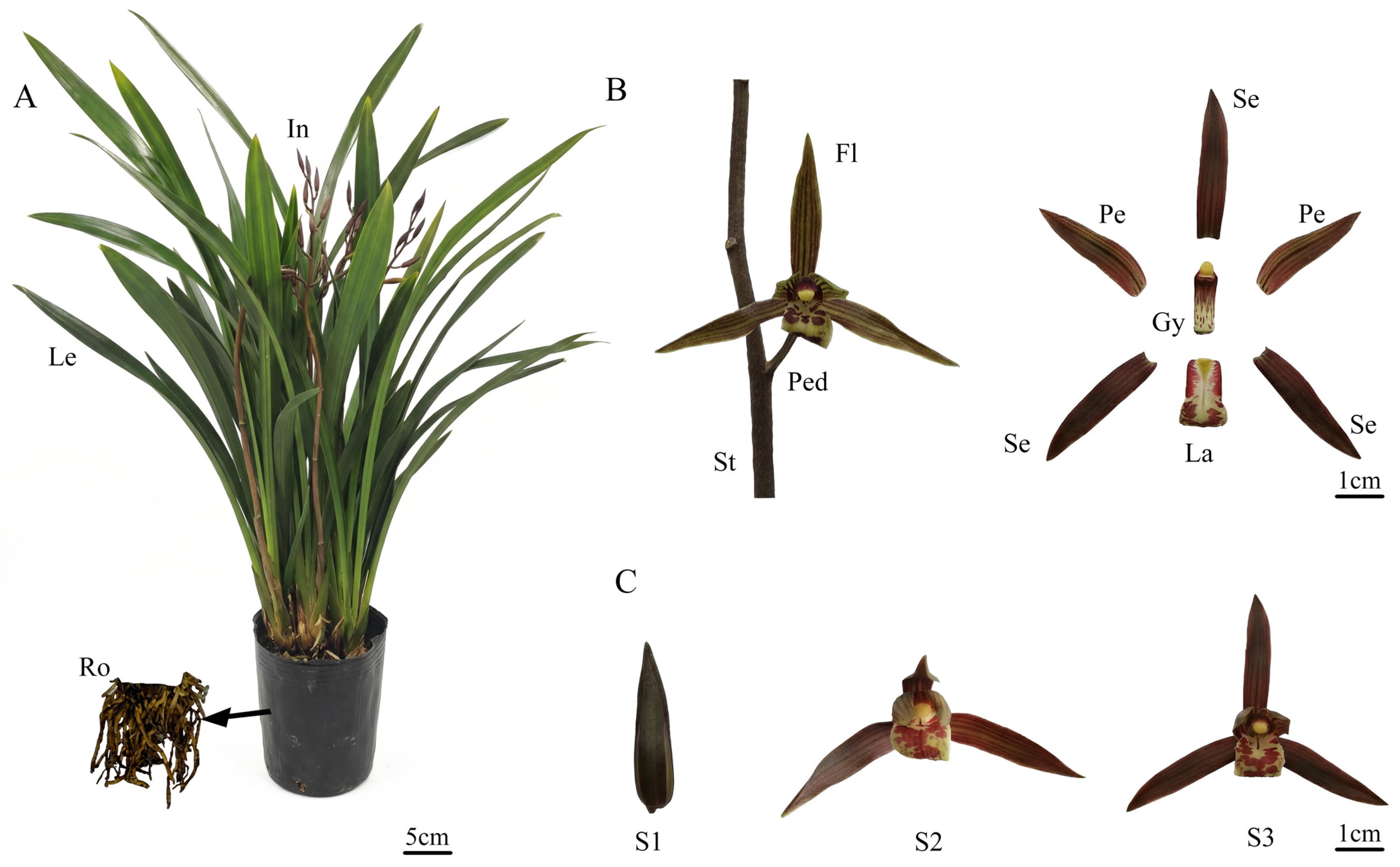
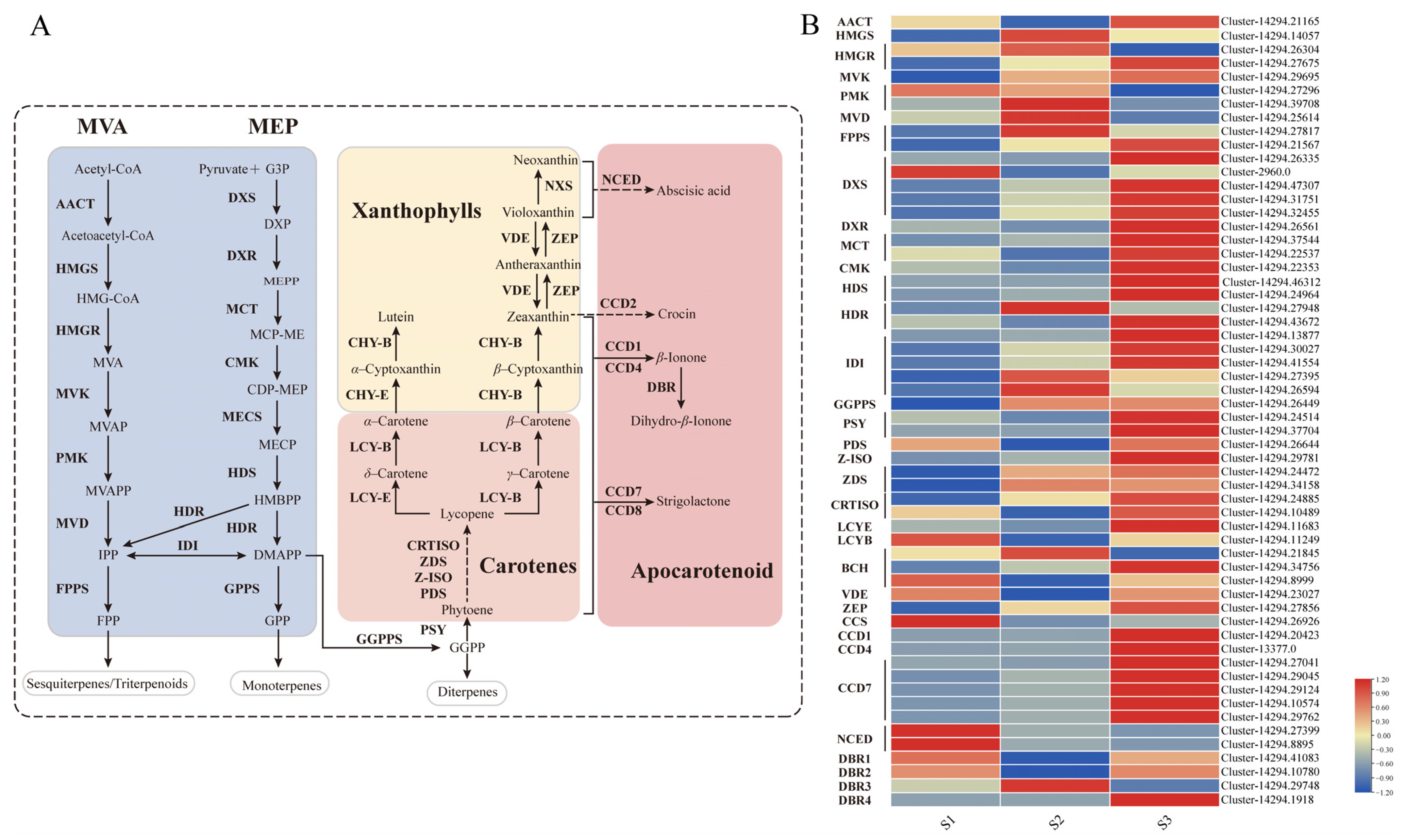
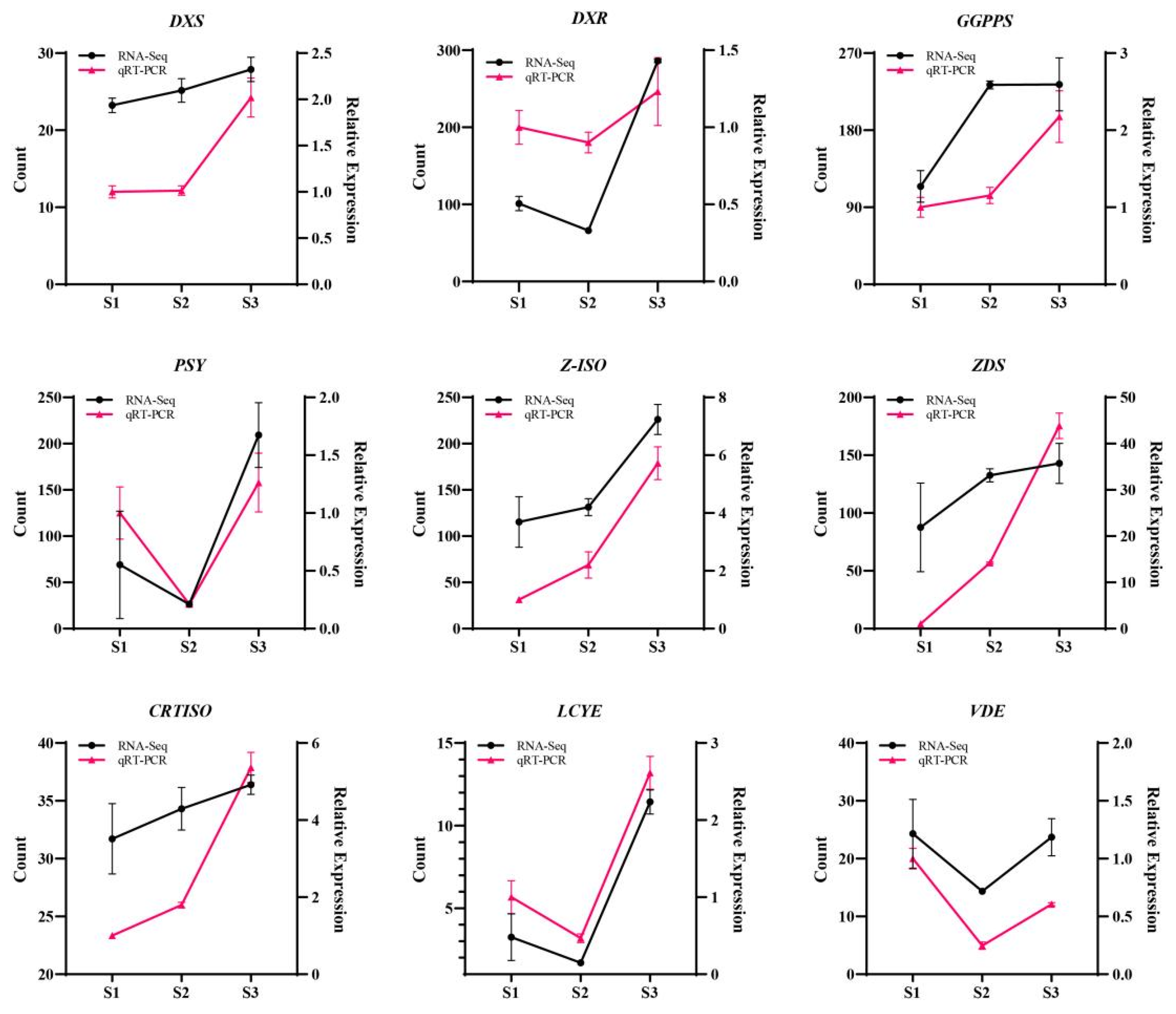
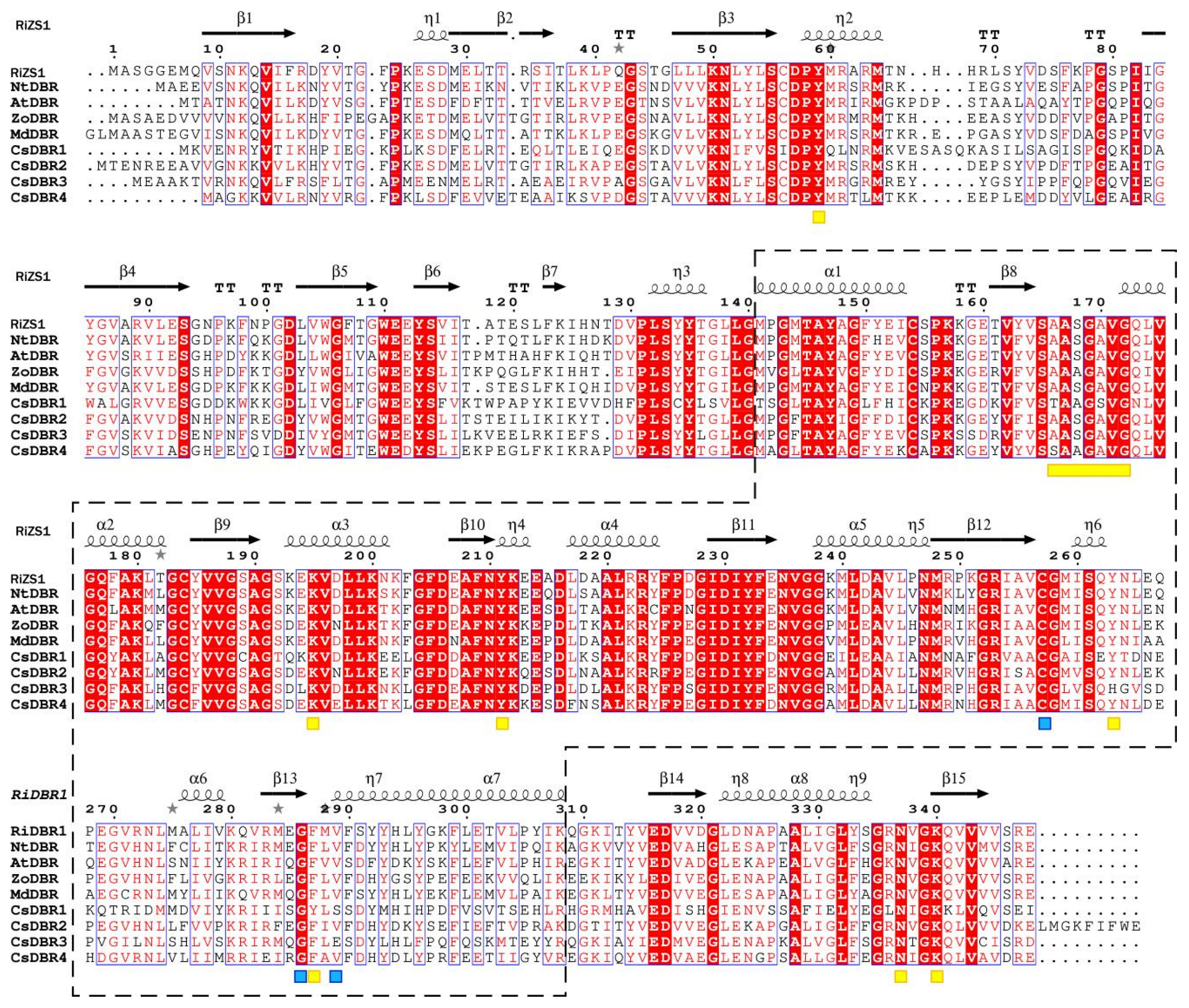
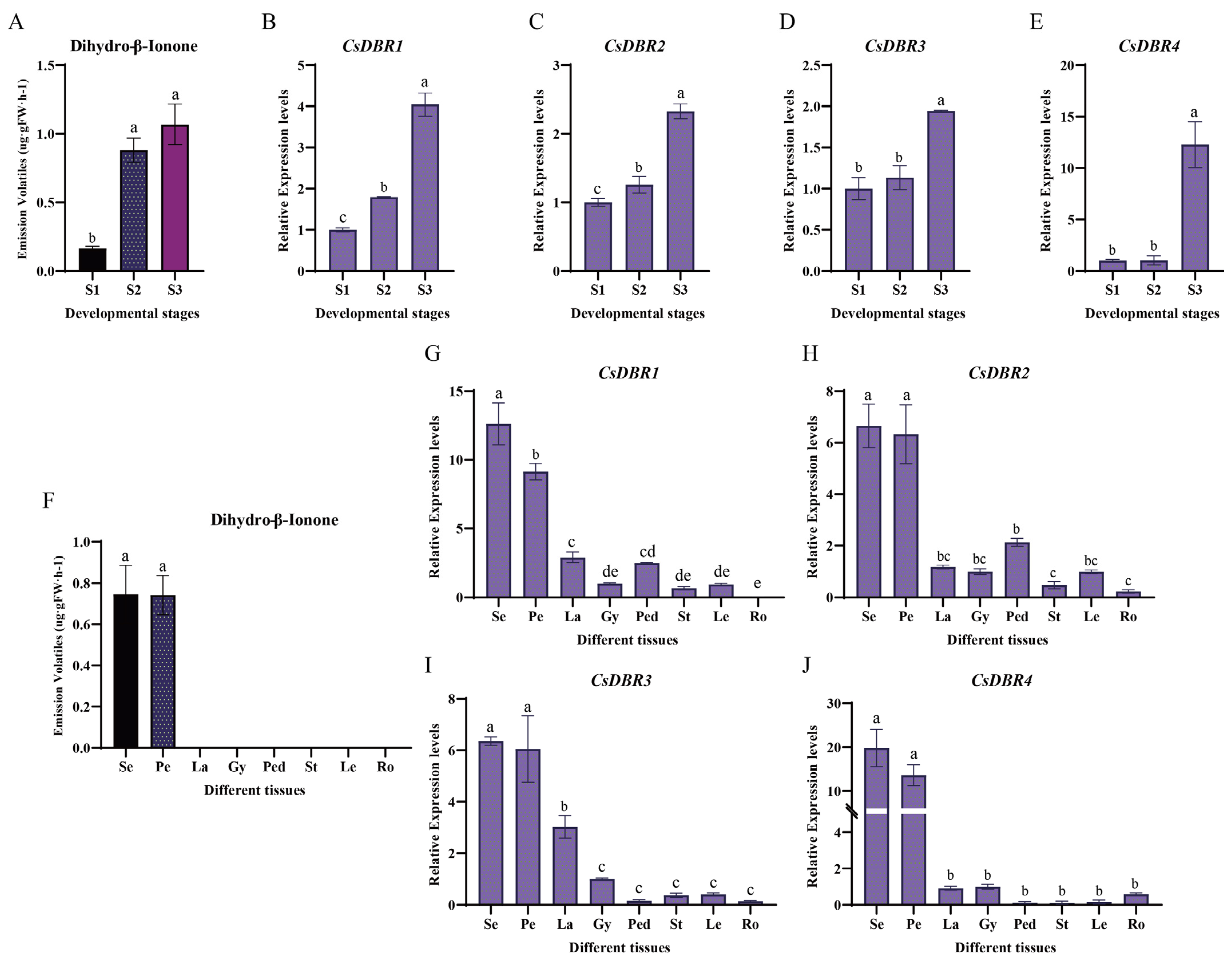
| ID | Name | RT | CAS | MS | Foluma | Type | Relative Content (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | Sepal | Petal | Labellum | Gynandrium | |||||||
| 1 | (E)-β-Ionone | 7.38 | 79-77-6 | 96 | C13H20O | Apocarotenoid | 0.02 ± 0.01c | 0.11 ± 0.04bc | 0.33 ± 0.07a | 0.23 ± 0.079ab | 0.24 ± 0.026ab | — | — |
| 2 | Megastigma-4,6(E),8(E)-triene | 7.61 | 51468-85-0 | 96 | C13H20 | Apocarotenoid | — | — | 0.03 ± 0.001 | — | — | — | — |
| 3 | α-Ionone | 8.43 | 79-76-5 | 95 | C13H20O | Apocarotenoid | — | 0.04 ± 0.003 | — | — | — | — | — |
| 4 | Dihydro-β-ionone | 8.55 | 17283-81-7 | 98 | C13H22O | Apocarotenoid | 0.16 ± 0.02c | 0.88 ± 0.09ab | 1.07 ± 0.15a | 0.746 ± 0.14b | 0.742 ± 0.096b | — | — |
| 5 | (E)-β-Farnesene | 8.77 | 18794-84-8 | 98 | C15H24 | Sesquiterpenoid | — | — | 0.1 ± 0.01 | — | — | — | — |
| 6 | β-Ionone | 9.16 | 14901-07-6 | 98 | C13H20O | Apocarotenoid | 0.71 ± 0.15d | 5.58 ± 0.15a | 6.48 ± 0.03a | 4.27 ± 0.71b | 4.00 ± 0.25b | 2.16 ± 0.56c | — |
| 7 | α-Farnesene | 9.42 | 502-61-4 | 94 | C15H24 | Sesquiterpenoid | — | — | 0.04 ± 0.003 | — | — | — | — |
| 8 | β-Bisabolene | 9.53 | 495-61-4 | 91 | C15H24 | Sesquiterpenoid | — | — | 0.02 ± 0.001 | — | — | — | — |
| 9 | Farnesol | 13.3 | 4602-84-0 | 91 | C15H26O | Sesquiterpenoid | — | — | 0.2 ± 0.06 | — | — | — | — |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Li, X.; Jia, Y.; Huang, M.; Yue, Y.; Wang, L.; Fan, Y.; Yu, Y. Functional Characterization of Double-Bond Reductases in Dihydro-β-Ionone Biosynthesis in Cymbidium sinense. Plants 2025, 14, 3804. https://doi.org/10.3390/plants14243804
Gao X, Li X, Jia Y, Huang M, Yue Y, Wang L, Fan Y, Yu Y. Functional Characterization of Double-Bond Reductases in Dihydro-β-Ionone Biosynthesis in Cymbidium sinense. Plants. 2025; 14(24):3804. https://doi.org/10.3390/plants14243804
Chicago/Turabian StyleGao, Xueqian, Xinyue Li, Yunpeng Jia, Meimei Huang, Yuechong Yue, Lan Wang, Yanping Fan, and Yunyi Yu. 2025. "Functional Characterization of Double-Bond Reductases in Dihydro-β-Ionone Biosynthesis in Cymbidium sinense" Plants 14, no. 24: 3804. https://doi.org/10.3390/plants14243804
APA StyleGao, X., Li, X., Jia, Y., Huang, M., Yue, Y., Wang, L., Fan, Y., & Yu, Y. (2025). Functional Characterization of Double-Bond Reductases in Dihydro-β-Ionone Biosynthesis in Cymbidium sinense. Plants, 14(24), 3804. https://doi.org/10.3390/plants14243804






