Genome-Wide Discovery of SSR Markers Based on Whole-Genome Resequencing Data of Dendrobium officinale
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification and Characterization of SSRs in D. officinale
2.2. Identification of Unique SSRs with Polymorphisms
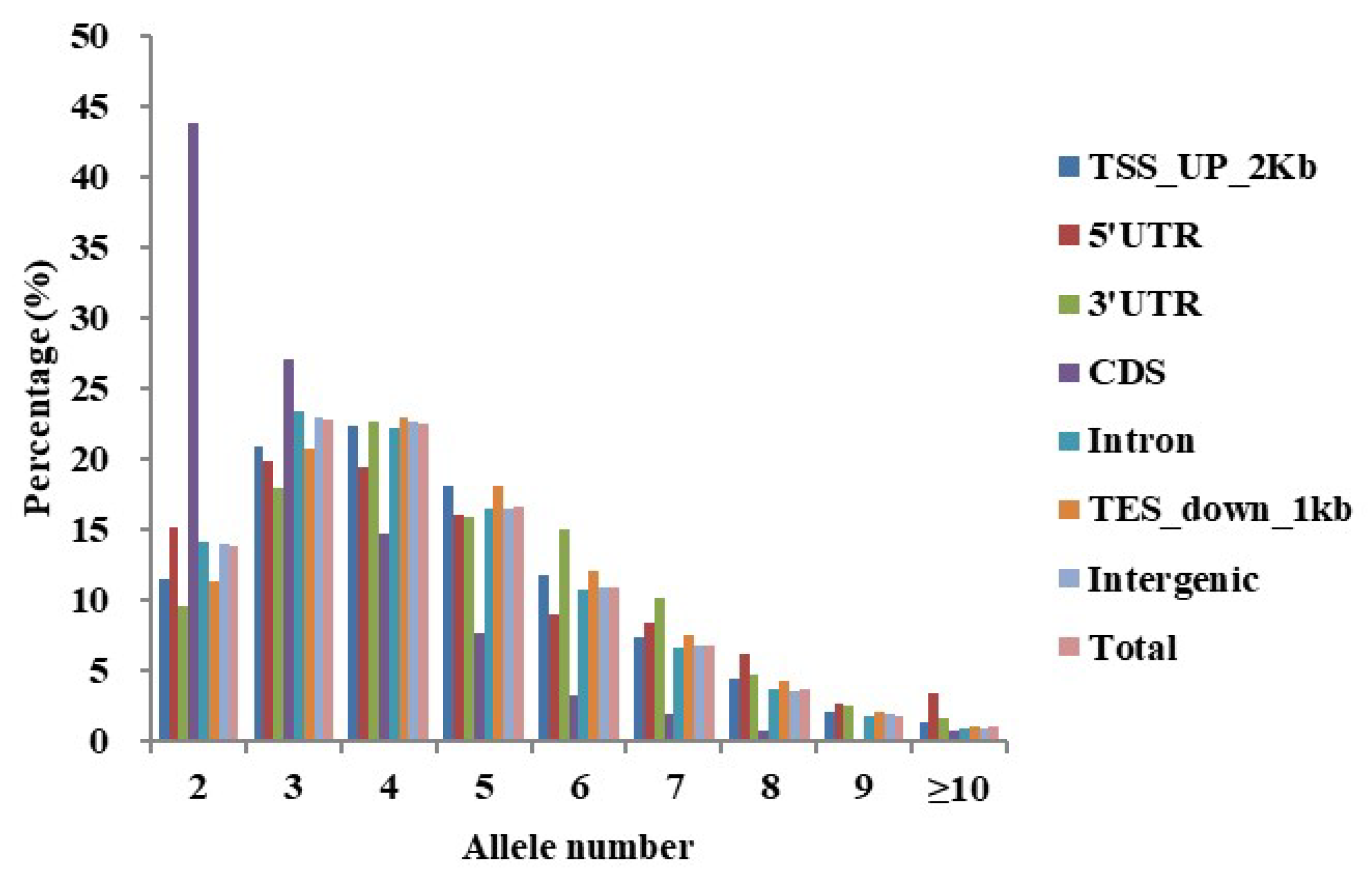
2.3. Analysis of the Frequency and Distribution of Polymorphic SSRs
2.4. Development of SSR Markers
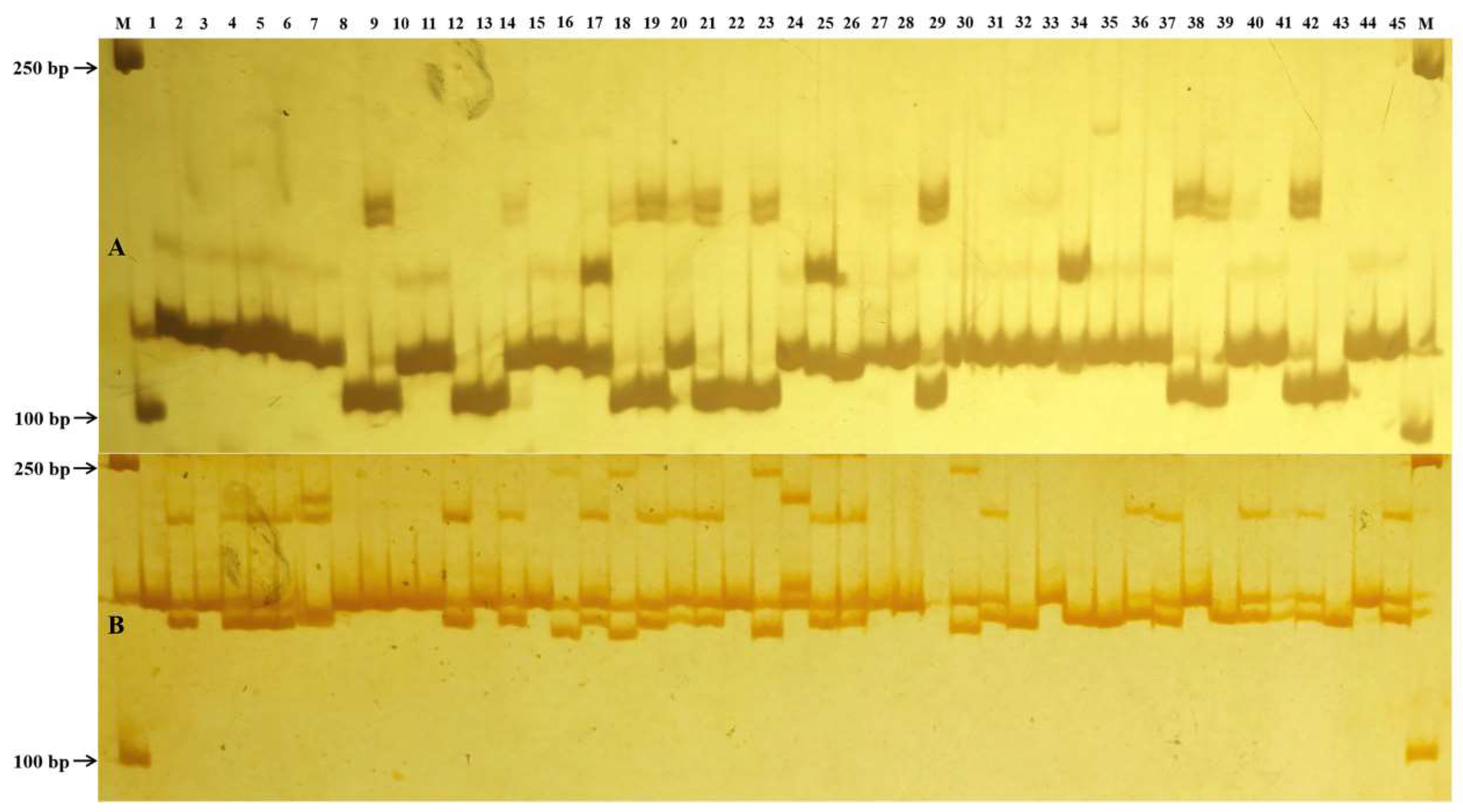
2.5. Validation of SSR Markers for Amplification Efficiency and Polymorphism
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Genome Sequencing
4.2. SSR Screening and Identification of Unique Loci
4.3. SSR Variation in D. officinale Genomes
4.4. Primer Design and Experimental Validation of Polymorphic SSRs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- He, Y.; Li, L.; Chang, H.; Cai, B.; Gao, H.; Chen, G.; Hou, W.; Jappar, Z.; Yan, Y. Research progress on extraction, purification, structure and biological activity of Dendrobium officinale polysaccharides. Front. Nutr. 2022, 9, 965073. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, M.; Cui, H.; Li, J.; Wang, M. Transcriptomic landscape of medicinal Dendrobium reveals genes associated with the biosynthesis of bioactive components. Front. Plant Sci. 2020, 11, 391. [Google Scholar] [CrossRef]
- Xing, X.; Cui, S.W.; Nie, S.; Phillips, G.O.; Goff, H.D.; Wang, Q. A review of isolation process, structural characteristics, and bioactivities of water-soluble polysaccharides from Dendrobium plants. Bioact. Carbohydr. Diet. Fibre 2013, 1, 131–147. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, L.; Zhang, Y.; Liu, X.; Wu, Z.; Gilbert, R.G.; Deng, B.; Wang, K. Dendrobium officinale polysaccharide ameliorates diabetic hepatic glucose metabolism via glucagon-mediated signaling pathways and modifying liver-glycogen structure. J. Ethnopharmacol. 2020, 248, 112308. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Wang, X.; Liu, H.; Tian, Y.; Lian, J.; Yang, R.; Hao, S.; Wang, X.; Yang, S.; Li, Q.; et al. The genome of Dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol. Plant. 2015, 8, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hu, B.; Zhang, F.; Luo, X.; Xie, J. Cytological observation and transcriptome comparative analysis of self-pollination and pross-pollination in Dendrobium officinale. Genes 2021, 12, 432. [Google Scholar] [CrossRef]
- Niu, Z.; Zhu, F.; Fan, Y.; Li, C.; Zhang, B.; Zhu, S.; Hou, Z.; Wang, M.; Yang, J.; Xue, Q.; et al. The chromosome-level reference genome assembly for Dendrobium officinale and its utility of functional genomics research and molecular breeding study. Acta Pharm. Sin. B 2021, 11, 2080–2092. [Google Scholar] [CrossRef]
- Huang, K.; Li, Y.; Tao, S.; Wei, G.; Huang, Y.; Chen, D.; Wu, C. Purification, characterization and biological activity of polysaccharides from Dendrobium officinale. Molecules 2016, 21, 701. [Google Scholar] [CrossRef]
- Chen, X.M.; Wang, F.F.; Wang, Y.Q.; Li, X.L.; Wang, A.R.; Wang, C.L.; Guo, S.X. Discrimination of the rare medicinal plant Dendrobium officinale based on naringenin, bibenzyl, and polysaccharides. Sci. China Life Sci. 2012, 55, 1092–1099. [Google Scholar] [CrossRef]
- Zhou, C.; Xie, Z.; Lei, Z.; Huang, Y.; Wei, G. Simultaneous identification and determination of flavonoids in Dendrobium officinale. Chem. Cent. J. 2018, 12, 40. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, A.; He, B.; Zhang, X.; Yu, Q.; Si, J. Quantitative variation of total alkaloids contents in Dendrobium. China J. Chin. Mater. Med. 2010, 35, 2388–2391. [Google Scholar]
- Zhang, A.L.; Wei, T.; Si, J.P.; Jin, L.Y.; Mo, Y.N. Study on basic amino acid contents in Dendrobium officinale. China J. Chin. Mater. Med. 2011, 36, 2632–2635. [Google Scholar]
- Liang, J.; Li, H.; Chen, J.; He, L.; Du, X.; Zhou, L.; Xiong, Q.; Lai, X.; Yang, Y.; Huang, S.; et al. Dendrobium officinale polysaccharides alleviate colon tumorigenesis via restoring intestinal barrier function and enhancing anti-tumor immune response. Pharmacol. Res. 2019, 148, 104417. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, H.; Liu, Y.; Shui, W.; Wang, J.; Cao, P.; Wang, H.; You, R.; Zhang, Y. Dendrobium officinale polysaccharide attenuates type 2 diabetes mellitus via the regulation of PI3K/Akt-mediated glycogen synthesis and glucose metabolism. J. Funct. Foods. 2018, 40, 261–271. [Google Scholar] [CrossRef]
- Zheng, Q.; Qiu, D.; Liu, X.; Zhang, L.; Cai, S.; Zhang, X. Antiproliferative effect of Dendrobium catenatum Lindley polypeptides against human liver, gastric and breast cancer cell lines. Food Funct. 2015, 6, 1489–1495. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Chen, S.; Hu, Y.; Yang, Y.; Yuan, J.; Wu, Y.; Li, S.; Lin, J.; He, L.; Hou, S.; et al. Protective roles and mechanisms of Dendrobium officinale polysaccharides on secondary liver injury in acute colitis. Int. J. Biol. Macromol. 2018, 107, 2201–2210. [Google Scholar] [CrossRef]
- Cakova, V.; Bonte, F.; Lobstein, A. Dendrobium: Sources of active ingredients to treat age-related pathologies. Aging Dis. 2017, 8, 827–849. [Google Scholar] [CrossRef]
- Li, X.L.; Hong, M. Aqueous extract of Dendrobium officinale confers neuroprotection against hypoxic-ischemic brain damage in neonatal rats. Kaohsiung J. Med. Sci. 2020, 36, 43–53. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, X.; Zhu, X.; Hua, Y. Chemical constituents, bioactivities, and pharmacological mechanisms of Dendrobium officinale: A review of the past decade. J. Agric. Food Chem. 2023, 71, 14870–14889. [Google Scholar] [CrossRef]
- Zheng, T.; Li, P.; Li, L.; Zhang, Q. Research advances in and prospects of ornamental plant genomics. Hortic. Res. 2021, 8, 65. [Google Scholar] [CrossRef]
- Gu, S.; Ding, X.Y.; Wang, Y.; Zhou, Q.; Ding, G.; Li, X.X.; Qian, L. Isolation and characterization of microsatellite markers in Dendrobium officinale, an endangered herb endemic to China. Mol. Ecol. Notes 2007, 7, 1166–1168. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, F.X.; Zheng, F.S.; Zhang, F.X.; Wang, J.L. Research progress on cultivation of Dendrobium candidum in greenhouse. Seed Sci. Technol. 2020, 38, 9–10. [Google Scholar]
- Adams, P.B. Systematics of Dendrobiinae (Orchidaceae), with special reference to Australian taxa. Bot. J. Linn. Soc. 2011, 166, 105–126. [Google Scholar] [CrossRef]
- Morris, M.W.; Stern, W.L.; Judd, W.S. Vegetative anatomy and systematics of subtribe Dendrobiinae (Orchidaceae). Bot. J. Linn. Soc. 1996, 120, 89–144. [Google Scholar] [CrossRef]
- Yukawa, T.; Uehara, K. Vegetative diversification and radiation in subtribe Dendrobiinae (Orchidaceae): Evidence from chloroplast DNA phylogeny and anatomical characters. Plant Syst. Evol. 1996, 201, 1–14. [Google Scholar] [CrossRef]
- Xiang, X.G.; Schuiteman, A.; Li, D.Z.; Huang, W.C.; Chung, S.W.; Li, J.W.; Zhou, H.; Jin, W.; Lai, Y.J.; Li, Z.Y.; et al. Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Mol. Phylogenet. Evol. 2013, 69, 950–960. [Google Scholar] [CrossRef]
- Ding, G.; Ding, X.Y.; Shen, J.; Tang, F.; Liu, D.Y.; He, J.; Li, X.X.; Chu, B.H. Genetic diversity and molecular authentication of wild populations of Dendrobium officinale by RAPD. Acta Pharm. Sin. 2005, 40, 1028–1032. [Google Scholar]
- Ding, G.; Li, X.; Ding, X.; Qian, L. Genetic diversity across natural populations of Dendrobium officinale, the endangered medicinal herb endemic to China, revealed by ISSR and RAPD markers. Russ. J. Genet. 2009, 45, 327–334. [Google Scholar] [CrossRef]
- Yuan, H.; Lin, E.P.; Zhu, B.; Yu, Q.X.; Si, J.P. Genetic diversity in cultivated populations of Dendrobium officinale. Chin. Tradit. Herb. Drugs. 2011, 42, 566–569. [Google Scholar]
- Li, X.X.; Ding, X.Y.; Chu, B.H.; Zhou, Q.; Ding, G.; Gu, S. Genetic diversity analysis and conservation of the endangered Chinese endemic herb Dendrobium officinale Kimura et Migo (Orchidaceae) based on AFLP. Genetica 2008, 133, 159–166. [Google Scholar] [CrossRef]
- Song, S.; Zhou, Y.; Liu, Z.; Zhao, M.; Yang, J.; Xu, S.; Wen, G. Genetic diversity among Dendrobium with known origins based on the ISSR and AFLP markers. J. Yunnan Agric. Univ. (Nat. Sci.) 2016, 31, 688–695. [Google Scholar]
- Xu, L.; Liu, L.; Peng, S.D.; Li, W.Y.; Zhang, T.; Li, Z.M.; Wang, H.M.; Li, S.J.; Lin, L.B. Genetic diversity of Dendrobium officinale revealed by SSR markers. Mol. Plant Breed. 2015, 13, 1616–1622. [Google Scholar]
- Zhang, Z.Y.; Zhou, M.L.; Liang, J.P.; Huang, P.P.; Zhang, C. Genetic diversity analysis of Dendrobium officinale germplasm resources in Guanzhaishan. Mol. Plant Breed. 2019, 17, 6179–6185. [Google Scholar]
- Hu, Z.Y.; Fu, T.; He, Y.Q.; Li, W.; Lin, L. Evaluation, selection and application of Dendrobium officinale EST-SSR primers. Bull. Bot. Res. 2017, 37, 78–87. [Google Scholar]
- Xie, J.K.; Zuo, J.H.; Huang, Y.H.; Li, C.S.; Chen, Y.L. The origin and germplasm collection for cultivated Dendrobium officinale K. Kimura & Migo individuals revealed by EST-SSR markers. Genet. Resour. Crop Evol. 2020, 67, 1209–1219. [Google Scholar]
- Li, Y.Q.; Ye, W.; Jiang, J.L.; Lei, F.G. Analysis of genetic diversity of germplasm resources of Dendrobium officinale by ISSR. Southwest China J. Agric. Sci. 2015, 28, 1530–1534. [Google Scholar]
- Yang, T.W.; Gao, M.R.; Huang, S.Y.; Zhang, S.W.; Zhang, X.J.; Li, T.; Yu, W.H.; Meng, P.; Shi, Q. Genetic diversity and DNA fingerprinting of Dendrobium officinale based on ISSR and SCoT markers. Appl. Ecol. Environ. Res. 2023, 21, 421–438. [Google Scholar] [CrossRef]
- Wang, C.; Huang, S.; Zhou, Y.; Wang, X.; Liu, A.; Hou, B.; Ye, M. Analysis of the genetic relationships among 3 Dendrobium species based on genotyping by sequencing. Chin. Wild Plant Resour. 2021, 40, 19–24. [Google Scholar]
- Tautz, D.; Renz, M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984, 12, 4127–4138. [Google Scholar] [CrossRef]
- Powell, W.; Machray, G.C.; Provan, J. Polymorphism revealed by simple sequence repeat. Trends Plant Sci. 1996, 1, 215–222. [Google Scholar] [CrossRef]
- Kalia, R.K.; Rai, M.K.; Kalia, S.; Singh, R.; Dhawan, A.K. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Zalapa, J.E.; Cuevas, H.; Zhu, H.; Steffan, S.; Senalik, D.; Zeldin, E.; McCown, B.; Harbut, R.; Simon, P. Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am. J. Bot. 2012, 99, 193–208. [Google Scholar] [CrossRef]
- Xie, M.L.; Hou, B.W.; Han, L.; Ma, Y.H.; Ding, X.Y. Development of microsatellites of Dendrobium officinale and its application in purity identification of germplasm. Acta Pharm. Sin. 2010, 45, 667–672. [Google Scholar]
- Qiu, D.S.; Zheng, X.L.; Cai, S.K.; Zheng, J.R.; Luo, H.M.; Zhang, L.; Deng, R.Y.; Li, W.; Liu, X.J. Development and transfer analysis of SSR in Dendrobium. Plant Sci. J. 2013, 31, 500–509. [Google Scholar] [CrossRef]
- Lu, J.J.; Suo, N.N.; Hu, X.; Wang, S.; Liu, J.J.; Wang, H.Z. Development and characterization of 110 novel EST-SSR markers for Dendrobium officinale (Orchidaceae). Am. J. Bot. 2012, 99, e415–e420. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Hu, Z.Y.; He, Y.Q.; Li, W.; Lin, L. Development of EST-SSR molecular markers in Dendrobium officinale. J. Nucl. Agric. Sci. 2017, 31, 663–670. [Google Scholar]
- Xu, M.; Liu, X.; Wang, J.W.; Teng, S.Y.; Shi, J.Q.; Li, Y.Y.; Huang, M.R. Transcriptome sequencing and development of novel genic SSR markers for Dendrobium officinale. Mol. Breed. 2017, 37, 18. [Google Scholar] [CrossRef]
- Xiao, D.; Zhang, Z.; Guan, Y. Microsatellite markers primer designing and screening from Dendrobium officinale. Biotechnol. Bull. 2012, 7, 88–92. [Google Scholar]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef]
- Zhang, Z.; Deng, Y.; Tan, J.; Hu, S.; Yu, J.; Xue, Q. A genome-wide microsatellite polymorphism database for the indica and japonica rice. DNA Res. 2007, 14, 37–45. [Google Scholar] [CrossRef]
- Xu, J.; Liu, L.; Xu, Y.; Chen, C.; Rong, T.; Ali, F.; Zhou, S.; Wu, F.; Liu, Y.; Wang, J.; et al. Development and characterization of simple sequence repeat markers providing genome-wide coverage and high resolution in maize. DNA Res. 2013, 20, 497–509. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Zane, L.; Bargelloni, L.; Patarnello, T. Strategies for microsatellite isolation: A review. Mol. Ecol. 2002, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qu, J.; Hu, K.; Zhang, L.; Li, J.; Wu, B.; Luo, C.; Wei, A.; Han, Y.; Cui, X. Development of genomewide simple sequence repeat fingerprints and highly polymorphic markers in cucumbers based on next-generation sequence data. Plant Breed. 2015, 134, 605–611. [Google Scholar] [CrossRef]
- Zhou, X.; Dong, Y.; Zhao, J.; Huang, L.; Ren, X.; Chen, Y.; Huang, S.; Liao, B.; Lei, Y.; Yan, L.; et al. Genomic survey sequencing for development and validation of single-locus SSR markers in peanut (Arachis hypogaea L.). BMC Genom. 2016, 17, 420. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef]
- Anderson, J.A.; Churchill, G.A.; Autrique, J.E.; Tanksley, S.D.; Sorrells, M.E. Optimizing parental selection for genetic linkage maps. Genome 1993, 36, 181–186. [Google Scholar] [CrossRef]
| Types | Repeat Units | Overall SSRs a | Unique SSRs b | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Count | Length (bp) | GC (%) | Rate c (%) | Count | Length (bp) | GC (%) | Rate d (%) | ||
| Simple SSRs | MNRs | 329,390 | 12.64 | 11.62 | 66.57 | 259,178 | 12.70 | 11.74 | 66.70 |
| DNRs | 91,524 | 34.14 | 17.17 | 18.50 | 71,921 | 35.96 | 15.65 | 18.51 | |
| TNRs | 37,451 | 19.93 | 22.28 | 7.57 | 28,766 | 20.18 | 20.57 | 7.40 | |
| TTRs | 2235 | 21.51 | 11.42 | 0.45 | 1861 | 21.43 | 9.42 | 0.48 | |
| PNRs | 448 | 26.30 | 18.60 | 0.09 | 326 | 26.34 | 18.52 | 0.08 | |
| HNRs | 222 | 32.48 | 44.05 | 0.04 | 165 | 32.50 | 45.37 | 0.04 | |
| Total | 461,270 | 17.57 | 14.78 | 93.23 | 362,217 | 17.98 | 14.10 | 93.22 | |
| Compound SSRs e | -- | 31,384 | 96.40 | 23.50 | 6.34 | 24,590 | 63.03 | 22.02 | 6.33 |
| Compound* SSRs f | -- | 2129 | 63.26 | 22.56 | 0.43 | 1746 | 60.16 | 23.66 | 0.45 |
| Total | -- | 494,783 | 22.76 | 17.22 | 100.00 | 388,553 | 21.02 | 15.73 | 100.00 |
| Genome Regions | Overall SSRs | Unique SSRs | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Count | Interval a (Kbp) | Length (bp) | GC (%) | Count | Interval a (Kbp) | Length (bp) | GC (%) | Rate b (%) | |
| TSS_up_2Kb | 32,278 | 0.63 | 20.80 | 12.94 | 27,848 | 0.54 | 20.82 | 12.25 | 86.28 |
| 5’UTR | 1320 | 0.69 | 18.79 | 36.09 | 1132 | 0.59 | 17.07 | 19.17 | 85.76 |
| 3’UTR | 998 | 0.53 | 15.14 | 11.54 | 944 | 0.50 | 15.23 | 11.16 | 94.59 |
| CDS | 1299 | 0.05 | 17.42 | 54.39 | 1205 | 0.04 | 17.26 | 54.53 | 92.76 |
| Intron | 177,177 | 0.43 | 22.27 | 17.94 | 140,309 | 0.34 | 14.12 | 14.44 | 79.19 |
| TES_down_1kb | 17,603 | 0.69 | 19.11 | 10.49 | 15,583 | 0.61 | 18.74 | 9.85 | 88.52 |
| Intergenic | 264,108 | 0.42 | 23.60 | 18.76 | 201,532 | 0.32 | 14.68 | 14.77 | 76.31 |
| Total | 494,783 | 0.43 | 22.76 | 17.99 | 388,553 | 0.34 | 20.86 | 16.34 | 78.53 |
| Primer Name | SSR Type | e-PCR | PCR Validation | PCR-Based Primer | ||||
|---|---|---|---|---|---|---|---|---|
| Allele Number | PIC | Allele Number | PIC | Forward | Reverse | Product Size (bp) | ||
| Chr1-459196 | (T)10 | 3 | 0.60 | 4 | 0.54 | CATTCACTAACGACCACATTTGA | CCGATAGGCCTAATCCTGTTC | 103 |
| Chr1-463581 | (A)16 | 4 | 0.65 | 3 | 0.59 | AATTGCTAACCAGGCCATCA | TCTTCGATCCCTTGACTGCT | 136 |
| Chr1-59422185 | (TGT)8 | 2 | 0.50 | 6 | 0.51 | AAGGATTCATGTAGCCGACC | GGAACGAAAATATTATGTGCCAA | 112 |
| Chr1-508186 | (TTAT)5 | 4 | 0.56 | 4 | 0.31 | TTTCTAGTGTCGCTCTGAAATG | CCTGATTCACGCTCACATTG | 105 |
| Chr2-4294007 | (T)10 | 4 | 0.68 | 4 | 0.69 | CTAACCACACCCATCCCAAC | CGGCGGTAAGCCTAACTTTT | 135 |
| Chr2-47634991 | (ATTT)5 | 2 | 0.50 | 1 | 0.00 | CAAAGCTCATGAAAGCTTCAAC | TTCAAAGAAAAATACAATCCCCA | 165 |
| Chr2-5701487 | (TCT)5 | 3 | 0.62 | 2 | 0.43 | TTGCTCATAAACACACACTCTCTC | CAACAGGAAGGACTAGAGGATCA | 104 |
| Chr2-78041173 | (CAAAA)5 | 2 | 0.50 | 2 | 0.39 | AAAGAGAACCAATCTACTGAACGG | GCATCTGCCTTTTGAATCTTCT | 129 |
| Chr3-7889288 | (TA)6 | 2 | 0.50 | / | / | AAGATGGTACAGAAAGCAGAGACA | TTTTTCATCTTTCTTTGTTTGAGC | 90 |
| Chr3-3538709 | (TGT)5 | 3 | 0.50 | 3 | 0.54 | CCACGTAGTGGGATAAAGGC | GTATCAGGGGGCATTGATGT | 116 |
| Chr3-5676585 | (ATT)6 | 3 | 0.58 | 3 | 0.58 | CCAGTTTAATTTTTGGGATTCA | TGACCATAAACGTTGCCTGA | 110 |
| Chr3-16813768 | (AAT)10 | 4 | 0.66 | 4 | 0.69 | GGGCTAATGCAAAATGTAGTTG | TGATTCGTTTTATGTTTATTTGGG | 130 |
| Chr4-23399662 | (TTTA)6 | 2 | 0.50 | 4 | 0.52 | CCCAATAATGAATGCAGCTCT | CAAAAATGATTGAAAACAACTCCA | 160 |
| Chr4-41229100 | (CCCT)5 | 3 | 0.60 | 4 | 0.69 | GGGTACCAACCGGCCTAT | AAGAGAGGGAGAGGGAGAGG | 88 |
| Chr4-55668381 | (GGGTG)6 | 4 | 0.71 | 5 | 0.66 | TCACGGCTAGCTTCTTGAGAG | GCCAAATGTCCTCCCTCTTT | 100 |
| Chr4-7402326 | (TGT)8 | 5 | 0.59 | 5 | 0.53 | TTCATGTAGCCGACCCTATG | CAGGTCACCAAAAGTTATTTGC | 118 |
| Chr5-16940219 | (TGT)5 | 2 | 0.50 | 3 | 0.49 | TGGATTCATATAGTCGACCCAA | AAGCTGGCCATAAAGAGGGT | 83 |
| Chr5-2512207 | (AAGC)5 | 3 | 0.57 | 3 | 0.56 | CTAAATTAGTTTGGCAGCACAGT | TCATCCTTCTGCAGTGAGAAAA | 109 |
| Chr5-2550036 | (GAT)8 | 4 | 0.63 | 4 | 0.67 | TTCAGAGTATTCATGGGATTTCA | AATATCACCATTCCAAAGAGTTTC | 100 |
| Chr5-4384422 | (AT)9 | 5 | 0.68 | 4 | 0.74 | TTCAACATTTGACTCGACTGTACT | AAACAAAGTGAAAGGAAGGCT | 121 |
| Sample | Accession Number | Geographic Origin |
|---|---|---|
| 1 | 3 | Huangshan, Anhui |
| 2 | A82 | Pucheng, Fujian |
| 3 | A52 | Taining, Fujian |
| 4 | A44 | Wuyishan, Fujian |
| 5 | GHY | Heyuan, Guangdong |
| 6 | A17 | Renhua, Guangdong |
| 7 | A34 | Rong, Guangxi |
| 8 | A39 | Pingjiang, Hunan |
| 9 | A31 | Guangfeng, Jiangxi |
| 10 | A32 | Longnan, Jiangxi |
| 11 | HY | Hanyuan, Sichuan |
| 12 | LS | Liandu, Zhejiang |
| 13 | XJ1 | Xianju, Zhejiang |
| 14 | A23 | Xingguo, Jiangxi |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, M.; Mi, H.; Zhou, P.; Li, T.; Wang, Y.; Liu, J.; Jiang, W. Genome-Wide Discovery of SSR Markers Based on Whole-Genome Resequencing Data of Dendrobium officinale. Plants 2025, 14, 3589. https://doi.org/10.3390/plants14233589
Zheng M, Mi H, Zhou P, Li T, Wang Y, Liu J, Jiang W. Genome-Wide Discovery of SSR Markers Based on Whole-Genome Resequencing Data of Dendrobium officinale. Plants. 2025; 14(23):3589. https://doi.org/10.3390/plants14233589
Chicago/Turabian StyleZheng, Mingmin, Hang Mi, Pingrong Zhou, Ting Li, Yelin Wang, Jian Liu, and Wei Jiang. 2025. "Genome-Wide Discovery of SSR Markers Based on Whole-Genome Resequencing Data of Dendrobium officinale" Plants 14, no. 23: 3589. https://doi.org/10.3390/plants14233589
APA StyleZheng, M., Mi, H., Zhou, P., Li, T., Wang, Y., Liu, J., & Jiang, W. (2025). Genome-Wide Discovery of SSR Markers Based on Whole-Genome Resequencing Data of Dendrobium officinale. Plants, 14(23), 3589. https://doi.org/10.3390/plants14233589





