Molecular Network Analysis in Model and Non-Model Legumes: Challenges in Omics Data Interpretation Across Species, with a Focus on Glycine max, Lupinus albus and Medicago truncatula
Abstract
1. Introduction
2. Interpretation Challenges in Molecular Data
2.1. Non-Model Legumes (Soybean, Lupin)
2.2. Model Plants (Arabidopsis)
2.3. Cross-Species and Computational Barriers
2.4. Computational Network Analysis Tools (WGCNA and ARACNE)
3. Bridging Discovery and Application
4. Omics Integration Across Species
4.1. At the Genome Level
4.2. At the Transcriptome Level
4.3. At the Proteome Level
4.4. At the Metabolome Level
4.5. Phenomics and Computation
4.6. Emerging Technologies
5. Cell Signaling Networks in Legume Responses
5.1. Signaling Pathways in Soybean
5.2. Aluminum/Phosphorus Signaling in Lupin
5.3. Nodulation and Stress Signals in M. truncatula
5.4. Applications of Signaling Networks for Crop Improvement
6. Toward Sustainable Agriculture: Genetic Diversification
6.1. Climate Resilience and Low-Input Agriculture
6.2. Limitations of AI and Multi-Omics Approaches
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| DEGs | Differentially Expressed Genes |
| GO | Gene Ontology |
| GWAS | Genome-Wide Association Study |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PCA | Principal Component Analysis |
| PPI | Protein–Protein Interaction |
| QTL | Quantitative Trait Loci |
| QTN | Quantitative trait nucleotide |
| R | R Statistical Software |
| RNA-seq | RNA sequencing |
| SNP | Single-Nucleotide Polymorphism |
| TF | Transcription Factor |
| WGCNA | Weighted Gene Co-expression Network Analysis |
References
- Brady, S.; Auge, G.; Ayalew, M.; Balasubramanian, S.; Hamann, T.; Inze, D.; Saito, K.; Brychkova, G.; Berardini, T.Z.; Friesner, J. Arabidopsis research in 2030: Translating the computable plant. Plant J. 2025, 121, e70047. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Tosoroni, A.; Di Vittori, V.; Nanni, L.; Musari, E.; Papalini, S.; Bitocchi, E.; Bellucci, E.; Pieri, A.; Ghitarrini, S.; Susek, K. Recent Advances in Molecular Tools and Pre-Breeding Activities in White Lupin (Lupinus albus). Plants 2025, 14, 914. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2011, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Jin, J.; He, K.; Tang, X.; Li, Z.; Lv, L.; Zhao, Y.; Luo, J.; Gao, G. An arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Mol. Biol. Evol. 2015, 32, 1767–1773. [Google Scholar] [CrossRef]
- Li, P.; Chen, B.; Zhang, G.; Chen, L.; Dong, Q.; Wen, J.; Mysore, K.S.; Zhao, J. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 2016, 210, 905–921. [Google Scholar] [CrossRef]
- Do Amaral, M.N.; Souza, G.M. The challenge to translate OMICS data to whole plant physiology: The context matters. Front. Plant Sci. 2017, 8, 2146. [Google Scholar] [CrossRef]
- Inzé, D.; Nelissen, H. The translatability of genetic networks from model to crop species: Lessons from the past and perspectives for the future. New Phytol. 2022, 236, 43–48. [Google Scholar] [CrossRef]
- Mudge, J.; Cannon, S.B.; Kalo, P.; Oldroyd, G.E.; Roe, B.A.; Town, C.D.; Young, N.D. Highly syntenic regions in the genomes of soybean, Medicago truncatula, and Arabidopsis thaliana. BMC Plant Biol. 2005, 5, 15. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Xing, L.; Guo, M.; Liu, X.; Wang, C.; Wang, L.; Zhang, Y. An improved Bayesian network method for reconstructing gene regulatory network based on candidate auto selection. BMC Genom. 2017, 18, 844. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, Z.; Ge, H.; Liu, Y.; Chen, H. Weighted gene co-expression network analysis identifies genes related to anthocyanin biosynthesis and functional verification of hub gene SmWRKY44. Plant Sci. 2021, 309, 110935. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, M.; Yang, S.; Chen, S.; Chen, X.; Liu, C.; Wang, S.; Wang, H.; Zhang, B.; Liu, H. A global coexpression network of soybean genes gives insights into the evolution of nodulation in nonlegumes and legumes. New Phytol. 2019, 223, 2104–2119. [Google Scholar] [CrossRef] [PubMed]
- Valliyodan, B.; Cannon, S.B.; Bayer, P.E.; Shu, S.; Brown, A.V.; Ren, L.; Jenkins, J.; Chung, C.Y.L.; Chan, T.-F.; Daum, C.G.; et al. Construction and comparison of three reference-quality genome assemblies for soybean. Plant J. 2019, 100, 1066–1082. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, W.; Wei, X.; Sun, Y.; Dong, S. Molecular mechanism of drought resistance in soybean roots revealed using physiological and multi-omics analyses. Plant Physiol. Biochem. 2024, 208, 108451. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Zhang, X.; Xing, Y.; Sun, K.; Guo, Y. OmiEmbed: A unified multi-task deep learning framework for multi-omics data. Cancers 2021, 13, 3047. [Google Scholar] [CrossRef]
- Hufnagel, B.; Marques, A.; Soriano, A.; Marquès, L.; Divol, F.; Doumas, P.; Sallet, E.; Mancinotti, D.; Carrere, S.; Marande, W. High-quality genome sequence of white lupin provides insight into soil exploration and seed quality. Nat. Commun. 2020, 11, 492. [Google Scholar] [CrossRef]
- Hufnagel, B.; Soriano, A.; Taylor, J.; Divol, F.; Kroc, M.; Sanders, H.; Yeheyis, L.; Nelson, M.; Péret, B. Pangenome of white lupin provides insights into the diversity of the species. Plant Biotechnol. J. 2021, 19, 2532–2543. [Google Scholar] [CrossRef]
- Aslam, M.M.; Karanja, J.K.; Zhang, Q.; Lin, H.; Xia, T.; Akhtar, K.; Liu, J.; Miao, R.; Xu, F.; Xu, W. In vitro regeneration potential of white lupin (Lupinus albus) from cotyledonary nodes. Plants 2020, 9, 318. [Google Scholar] [CrossRef]
- Pathi, K.M.; Sprink, T. Lupins in the Genome Editing Era: Advances in Plant Cell Culture, Double Haploid Technology and Genetic Transformation for Crop Improvement. Front. Plant Sci. 2025, 16, 1601216. [Google Scholar] [CrossRef]
- Zhu, X.; Xu, W.; Liu, B.; Zhan, Y.; Xia, T. Adaptation of high-efficiency CRISPR/Cas9-based multiplex genome editing system in white lupin by using endogenous promoters. Physiol. Plant. 2023, 175, e13976. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.J. Medicago truncatula as a model for understanding plant interactions with other organisms, plant development and stress biology: Past, present and future. Funct. Plant Biol. 2008, 35, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sun, Y.; Qiu, X.; Lu, H.; Hwang, I.; Wang, T. Tolerant mechanism of model legume plant Medicago truncatula to drought, salt, and cold stresses. Front. Plant Sci. 2022, 13, 847166. [Google Scholar] [CrossRef] [PubMed]
- Irshad, A.; Rehman, R.N.U.; Kareem, H.A.; Yang, P.; Hu, T. Addressing the challenge of cold stress resilience with the synergistic effect of Rhizobium inoculation and exogenous melatonin application in Medicago truncatula. Ecotoxicol. Environ. Saf. 2021, 226, 112816. [Google Scholar] [CrossRef]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Mundodi, S.; Reiser, L.; Huala, E.; Garcia-Hernandez, M.; Zhang, P.; Mueller, L.A.; Yoon, J.; Doyle, A.; Lander, G. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 2004, 135, 745–755. [Google Scholar] [CrossRef]
- Yilmaz, A.; Mejia-Guerra, M.K.; Kurz, K.; Liang, X.; Welch, L.; Grotewold, E. AGRIS: The Arabidopsis gene regulatory information server, an update. Nucleic Acids Res. 2010, 39, D1118–D1122. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 2018, 162, 2–12. [Google Scholar] [CrossRef]
- Luo, L.; Yu, L.; Yang, J.; Wang, E. Peptide Signals Regulate Nitrogen Deficiency Adaptation of Dicotyledonous Model Plants. Plant Cell Environ. 2024, 47, 1–15. [Google Scholar] [CrossRef]
- Hyung, D.; Lee, C.; Kim, J.-H.; Yoo, D.; Seo, Y.-S.; Jeong, S.-C.; Lee, J.-H.; Chung, Y.; Jung, K.-H.; Cook, D.R. Cross-family translational genomics of abiotic stress-responsive genes between Arabidopsis and Medicago truncatula. PLoS ONE 2014, 9, e91721. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Pei, X.; Kong, F.; Zhao, L.; Lin, X. Divergence of functions and expression patterns of soybean bZIP transcription factors. Front. Plant Sci. 2023, 14, 1150363. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, J.; Long, Y.; Zhang, C.; Wang, D.; Zhang, X.; Dong, W.; Zhao, L.; Liu, C.; Zhai, J. Single-nucleus transcriptomes reveal spatiotemporal symbiotic perception and early response in Medicago. Nat. Plants 2023, 9, 1734–1748. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Song, M.; Shen, H.; Hong, H.; Gong, P.; Deng, H.-W.; Zhang, C. Deep learning methods for omics data imputation. Biology 2023, 12, 1313. [Google Scholar] [CrossRef]
- Van Dijk, A.D.J.; Kootstra, G.; Kruijer, W.; de Ridder, D. Machine learning in plant science and plant breeding. Iscience 2021, 24, 101890. [Google Scholar] [CrossRef]
- Zeng, N.; Yang, Z.; Zhang, Z.; Hu, L.; Chen, L. Comparative transcriptome combined with proteome analyses revealed key factors involved in alfalfa (Medicago sativa) response to waterlogging stress. Int. J. Mol. Sci. 2019, 20, 1359. [Google Scholar] [CrossRef]
- Moore, B.M.; Wang, P.; Fan, P.; Lee, A.; Leong, B.; Lou, Y.-R.; Schenck, C.A.; Sugimoto, K.; Last, R.; Lehti-Shiu, M.D. Within-and cross-species predictions of plant specialized metabolism genes using transfer learning. silico Plants 2020, 2, diaa005. [Google Scholar] [CrossRef]
- MacNish, T.R.; Danilevicz, M.F.; Bayer, P.E.; Bestry, M.S.; Edwards, D. Application of machine learning and genomics for orphan crop improvement. Nat. Commun. 2025, 16, 982. [Google Scholar] [CrossRef]
- Dick, K.; Samanfar, B.; Barnes, B.; Cober, E.R.; Mimee, B.; Tan, L.H.; Molnar, S.J.; Biggar, K.K.; Golshani, A.; Dehne, F. PIPE4: Fast PPI predictor for comprehensive inter-and cross-species interactomes. Sci. Rep. 2020, 10, 1390. [Google Scholar] [CrossRef]
- Casal, U.; González-Domínguez, J.; Martín, M.J. Parallelization of aracne, an algorithm for the reconstruction of gene regulatory networks. Proceedings 2019, 21, 25. [Google Scholar] [CrossRef]
- Wang, K.; Abid, M.A.; Rasheed, A.; Crossa, J.; Hearne, S.; Li, H. DNNGP, a deep neural network-based method for genomic prediction using multi-omics data in plants. Mol. Plant 2023, 16, 279–293. [Google Scholar] [CrossRef]
- Fu, M.; Chen, L.; Cai, Y.; Su, Q.; Chen, Y.; Hou, W. CRISPR/Cas9-mediated mutagenesis of GmFAD2-1A and/or GmFAD2-1B to create high-oleic-acid soybean. Agronomy 2022, 12, 3218. [Google Scholar] [CrossRef]
- Meng, Y.; Hou, Y.; Wang, H.; Ji, R.; Liu, B.; Wen, J.; Niu, L.; Lin, H. Targeted mutagenesis by CRISPR/Cas9 system in the model legume Medicago truncatula. Plant Cell Rep. 2017, 36, 371–374. [Google Scholar] [CrossRef]
- Che, P.; Chang, S.; Simon, M.K.; Zhang, Z.; Shaharyar, A.; Ourada, J.; O’Neill, D.; Torres-Mendoza, M.; Guo, Y.; Marasigan, K.M. Developing a rapid and highly efficient cowpea regeneration, transformation and genome editing system using embryonic axis explants. Plant J. 2021, 106, 817–830. [Google Scholar] [CrossRef]
- Venezia, M.; Creasey Krainer, K.M. Current advancements and limitations of gene editing in orphan crops. Front. Plant Sci. 2021, 12, 742932. [Google Scholar] [CrossRef]
- Beyene, Y.; Semagn, K.; Mugo, S.; Tarekegne, A.; Babu, R.; Meisel, B.; Sehabiague, P.; Makumbi, D.; Magorokosho, C.; Oikeh, S. Genetic gains in grain yield through genomic selection in eight bi-parental maize populations under drought stress. Crop. Sci. 2015, 55, 154–163. [Google Scholar] [CrossRef]
- Pecetti, L.; Annicchiarico, P.; Crosta, M.; Notario, T.; Ferrari, B.; Nazzicari, N. White lupin drought tolerance: Genetic variation, trait genetic architecture, and genome-enabled prediction. Int. J. Mol. Sci. 2023, 24, 2351. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, X.; Li, H.; Zheng, H.; Zhang, J.; Olsen, M.S.; Varshney, R.K.; Prasanna, B.M.; Qian, Q. Smart breeding driven by big data, artificial intelligence, and integrated genomic-enviromic prediction. Mol. Plant 2022, 15, 1664–1695. [Google Scholar] [CrossRef] [PubMed]
- Bayer, P.E.; Valliyodan, B.; Hu, H.; Marsh, J.I.; Yuan, Y.; Vuong, T.D.; Patil, G.; Song, Q.; Batley, J.; Varshney, R.K.; et al. Sequencing the USDA core soybean collection reveals gene loss during domestication and breeding. Plant Genome 2021, 15, e20109. [Google Scholar] [CrossRef]
- Dash, S.; Campbell, J.D.; Cannon, E.K.; Cleary, A.M.; Huang, W.; Kalberer, S.R.; Karingula, V.; Rice, A.G.; Singh, J.; Umale, P.E. Legume information system (LegumeInfo. org): A key component of a set of federated data resources for the legume family. Nucleic Acids Res. 2016, 44, D1181–D1188. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.V.; Conners, S.I.; Huang, W.; Wilkey, A.P.; Grant, D.; Weeks, N.T.; Cannon, S.B.; Graham, M.A.; Nelson, R.T. A new decade and new data at SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2021, 49, D1496–D1501. [Google Scholar] [CrossRef] [PubMed]
- Dyer, S.C.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Barrera-Enriquez, V.P.; Becker, A.; Bennett, R.; Beracochea, M.; Berry, A. Ensembl 2025. Nucleic Acids Res. 2025, 53, D948–D957. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021, 50, D20–D26. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.-A.; Zhang, H.; Liu, Z.; Shi, M. Pan-genome of wild and cultivated soybeans. Cell 2020, 182, 162–176.e113. [Google Scholar] [CrossRef]
- Torkamaneh, D.; Lemay, M.A.; Belzile, F. The pan-genome of the cultivated soybean (PanSoy) reveals an extraordinarily conserved gene content. Plant Biotechnol. J. 2021, 19, 1852–1862. [Google Scholar] [CrossRef]
- Xu, W.; Wang, Q.; Zhang, W.; Zhang, H.; Liu, X.; Song, Q.; Zhu, Y.; Cui, X.; Chen, X.; Chen, H. Using transcriptomic and metabolomic data to investigate the molecular mechanisms that determine protein and oil contents during seed development in soybean. Front. Plant Sci. 2022, 13, 1012394. [Google Scholar] [CrossRef]
- Zafeiriou, I.; Polidoros, A.N.; Baira, E.; Kasiotis, K.M.; Machera, K.; Mylona, P.V. Mediterranean white lupin landraces as a valuable genetic reserve for breeding. Plants 2021, 10, 2403. [Google Scholar] [CrossRef]
- Rychel-Bielska, S.; Bielski, W.; Surma, A.; Annicchiarico, P.; Belter, J.; Kozak, B.; Galek, R.; Harzic, N.; Książkiewicz, M. A GWAS study highlights significant associations between a series of indels in a FLOWERING LOCUS T gene promoter and flowering time in white lupin (Lupinus albus L.). BMC Plant Biol. 2024, 24, 722. [Google Scholar] [CrossRef]
- Schiessl, K.; Jhu, M.Y. From roots to nodules: Regulation of organogenesis in nitrogen-fixing symbiosis. Curr. Opin. Plant Biol. 2025, 86, 102755. [Google Scholar] [CrossRef]
- Kang, Y.; Li, M.; Sinharoy, S.; Verdier, J. A snapshot of functional genetic studies in Medicago truncatula. In The Model Legume Medicago truncatula; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2019; pp. 7–30. [Google Scholar] [CrossRef]
- Curtin, S.J.; Tiffin, P.; Guhlin, J.; Trujillo, D.I.; Burghardt, L.T.; Atkins, P.; Baltes, N.J.; Denny, R.; Voytas, D.F.; Stupar, R.M. Validating genome-wide association candidates controlling quantitative variation in nodulation. Plant Physiol. 2017, 173, 921–931. [Google Scholar] [CrossRef]
- Michno, J.M.; Liu, J.; Jeffers, J.R.; Stupar, R.M.; Myers, C.L. Identification of nodulation-related genes in Medicago truncatula using genome-wide association studies and co-expression networks. Plant Direct 2020, 4, e00220. [Google Scholar] [CrossRef]
- Epstein, B.; Burghardt, L.T.; Heath, K.D.; Grillo, M.A.; Kostanecki, A.; Hämälä, T.; Young, N.D.; Tiffin, P. Combining GWAS and population genomic analyses to characterize coevolution in a legume-rhizobia symbiosis. Mol. Ecol. 2023, 32, 3798–3811. [Google Scholar] [CrossRef]
- Chen, Z.; Lancon-Verdier, V.; Le Signor, C.; She, Y.-M.; Kang, Y.; Verdier, J. Genome-wide association study identified candidate genes for seed size and seed composition improvement in M. truncatula. Sci. Rep. 2021, 11, 4224. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.I.; Vodkin, L.O. Using RNA-Seq to profile soybean seed development from fertilization to maturity. PLoS ONE 2013, 8, e59270. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, J.; Yuan, X.; Chen, X.; Cui, X. Comparative transcriptome analysis reveals key pathways and regulatory networks in early resistance of Glycine max to soybean mosaic virus. Front. Microbiol. 2023, 14, 1241076. [Google Scholar] [CrossRef] [PubMed]
- Severin, A.J.; Woody, J.L.; Bolon, Y.-T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef]
- Nakayama, T.J.; A Rodrigues, F.; Neumaier, N.; Marcolino-Gomes, J.; Molinari, H.B.C.; Santiago, T.R.; Formighieri, E.F.; Basso, M.F.; Farias, J.R.B.; Emygdio, B.M.; et al. Insights into soybean transcriptome reconfiguration under hypoxic stress: Functional, regulatory, structural, and compositional characterization. PLoS ONE 2017, 12, e0187920. [Google Scholar] [CrossRef]
- Li, M.; Li, H.; Sun, A.; Wang, L.; Ren, C.; Liu, J.; Gao, X. Transcriptome analysis reveals key drought-stress-responsive genes in soybean. Front. Genet. 2022, 13, 1060529. [Google Scholar] [CrossRef]
- Le Thanh, T.; Hufnagel, B.; Soriano, A.; Divol, F.; Brottier, L.; Casset, C.; Péret, B.; Doumas, P.; Marquès, L. Dynamic development of white lupin rootlets along a cluster root. Front. Plant Sci. 2021, 12, 738172. [Google Scholar] [CrossRef]
- Pueyo, J.J.; Quiñones, M.A.; Coba de la Peña, T.; Fedorova, E.E.; Lucas, M.M. Nitrogen and phosphorus interplay in lupin root nodules and cluster roots. Front. Plant Sci. 2021, 12, 644218. [Google Scholar] [CrossRef]
- Osorio, C.E.; Till, B.J. A bitter-sweet story: Unraveling the genes involved in quinolizidine alkaloid synthesis in Lupinus albus. Front. Plant Sci. 2022, 12, 795091. [Google Scholar] [CrossRef]
- Burchardt, S.; Czernicka, M.; Kućko, A.; Pokora, W.; Kapusta, M.; Domagalski, K.; Jasieniecka-Gazarkiewicz, K.; Karwaszewski, J.; Wilmowicz, E. Exploring the response of yellow lupine (Lupinus luteus L.) root to drought mediated by pathways related to phytohormones, lipid, and redox homeostasis. BMC Plant Biol. 2024, 24, 1049. [Google Scholar] [CrossRef] [PubMed]
- Carrere, S.; Verdier, J.; Gamas, P. MtExpress, a comprehensive and curated RNAseq-based gene expression atlas for the model legume Medicago truncatula. Plant Cell Physiol. 2021, 62, 1494–1500. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Benedito, V.A.; Wang, M.; Murray, J.D.; Zhao, P.X.; Tang, Y.; Udvardi, M.K. The Medicago truncatula gene expression atlas web server. BMC Bioinform. 2009, 10, 441. [Google Scholar] [CrossRef] [PubMed]
- Boschiero, C.; Dai, X.; Lundquist, P.K.; Roy, S.; Christian de Bang, T.; Zhang, S.; Zhuang, Z.; Torres-Jerez, I.; Udvardi, M.K.; Scheible, W.-R. MtSSPdb: The Medicago truncatula small secreted peptide database. Plant Physiol. 2020, 183, 399–413. [Google Scholar] [CrossRef]
- Chakraborty, S.; Driscoll, H.E.; Abrahante, J.E.; Zhang, F.; Fisher, R.F.; Harris, J.M. Salt stress enhances early symbiotic gene expression in Medicago truncatula and induces a stress-specific set of rhizobium-responsive genes. Mol. Plant-Microbe Interact. 2021, 34, 904–921. [Google Scholar] [CrossRef]
- Hajduch, M.; Ganapathy, A.; Stein, J.W.; Thelen, J.J. A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol. 2005, 137, 1397–1419. [Google Scholar] [CrossRef]
- Herman, E.M. Soybean seed proteome rebalancing. Front. Plant Sci. 2014, 5, 437. [Google Scholar] [CrossRef]
- Huang, Y.; Meng, B.; Qin, Y.; Liu, J.; Lu, A.; Dai, X.; Zhao, Y.; Ge, L. Comparative Proteomic Atlas of Two Soybean Varieties with Contrasting Seed Oil and Protein Content. J. Agric. Food Chem. 2025, 73, 2279–2288. [Google Scholar] [CrossRef]
- Islam, N.; Krishnan, H.B.; Natarajan, S. Quantitative proteomic analyses reveal the dynamics of protein and amino acid accumulation during soybean seed development. Proteomics 2022, 22, 2100143. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Huang, Y.; Lu, A.; Liao, H.; Zhai, R.; Gong, X.; Dong, L.; Jiang, Y.; Dai, X.; Fang, X. Enhanced Analysis of Low-Abundance Proteins in Soybean Seeds Using Advanced Mass Spectrometry. Int. J. Mol. Sci. 2025, 26, 949. [Google Scholar] [CrossRef] [PubMed]
- Mulalapele, L.T.; Xi, J. Detection and inactivation of allergens in soybeans: A brief review of recent research advances. Grain Oil Sci. Technol. 2021, 4, 191–200. [Google Scholar] [CrossRef]
- Islam, N.; Krishnan, H.B.; Slovin, J.; Li, Z.; Fakir, T.; Luthria, D.; Natarajan, S. High-Resolution Mass Spectrometry Approach for Proteomic and Metabolomic Analyses of High-Protein Soybean Seeds. J. Agric. Food Chem. 2025, 73, 6993–7002. [Google Scholar] [CrossRef]
- Dong, S.; Zhou, Q.; Yan, C.; Song, S.; Wang, X.; Wu, Z.; Wang, X.; Ma, C. Comparative protein profiling of two soybean genotypes with different stress tolerance reveals major components in drought tolerance. Front. Sustain. Food Syst. 2023, 7, 1200608. [Google Scholar] [CrossRef]
- Wai, P.P.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Komatsu, S. Morphological and Proteomic Analyses to Reveal Salt-Tolerant Mechanisms in Soybean Seedlings Treated with Titanium-Oxide Nanoparticles. Oxygen 2025, 5, 4. [Google Scholar] [CrossRef]
- Angermann, C.; Heinemann, B.; Hansen, J.; Töpfer, N.; Braun, H.-P.; Hildebrandt, T.M. Proteome reorganization and amino acid metabolism during germination and seedling establishment in Lupinus albus. J. Exp. Bot. 2024, 75, 4891–4903. [Google Scholar] [CrossRef]
- Spina, A.; Saletti, R.; Fabroni, S.; Natalello, A.; Cunsolo, V.; Scarangella, M.; Rapisarda, P.; Canale, M.; Muccilli, V. Multielemental, nutritional, and proteomic characterization of different Lupinus spp. genotypes: A source of nutrients for dietary use. Molecules 2022, 27, 8771. [Google Scholar] [CrossRef]
- De Bang, T.C.; Lundquist, P.K.; Dai, X.; Boschiero, C.; Zhuang, Z.; Pant, P.; Torres-Jerez, I.; Roy, S.; Nogales, J.; Veerappan, V. Genome-wide identification of Medicago peptides involved in macronutrient responses and nodulation. Plant Physiol. 2017, 175, 1669–1689. [Google Scholar] [CrossRef]
- Pagano, A.; Kunz, L.; Dittmann, A.; Araújo, S.D.S.; Macovei, A.; Shridhar Gaonkar, S.; Sincinelli, F.; Wazeer, H.; Balestrazzi, A. Changes in Medicago truncatula seed proteome along the rehydration–dehydration cycle highlight new players in the genotoxic stress response. Front. Plant Sci. 2023, 14, 1188546. [Google Scholar] [CrossRef] [PubMed]
- Tareq, F.S.; Kotha, R.R.; Natarajan, S.; Sun, J.; Luthria, D.L. An untargeted metabolomics approach to study the variation between wild and cultivated soybeans. Molecules 2023, 28, 5507. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, J.; Xia, N.; Liu, Y.; Qu, Y.; Ming, M.; Zhan, Y.; Han, Y.; Zhao, X.; Li, Y. Combined analysis of the metabolome and transcriptome provides insight into seed oil accumulation in soybean. Biotechnol. Biofuels Bioprod. 2023, 16, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhao, Y.; Yang, J.; He, Y.; Zhang, X.; Li, L.; Wang, H.; Hong, G. Spatial metabolome mass spectrometry imaging reveals the flavonoid distribution change in soybean nodules under drought and alkaline stresses. Plant Stress 2025, 16, 100880. [Google Scholar] [CrossRef]
- Gai, Y.; Liu, S.; Zhang, Z.; Wei, J.; Wang, H.; Liu, L.; Bai, Q.; Qin, Q.; Zhao, C.; Zhang, S. Integrative Approaches to Soybean Resilience, Productivity, and Utility: A Review of Genomics, Computational Modeling, and Economic Viability. Plants 2025, 14, 671. [Google Scholar] [CrossRef]
- Namdar, D.; Mulder, P.P.; Ben-Simchon, E.; Hacham, Y.; Basheer, L.; Cohen, O.; Sternberg, M.; Shelef, O. New analytical approach to quinolizidine alkaloids and their assumed biosynthesis pathways in lupin seeds. Toxins 2024, 16, 163. [Google Scholar] [CrossRef]
- Khedr, T.; Gao, L.L.; Kamphuis, L.G.; Bose, U.; Juhász, A.; Colgrave, M.L. Evaluation of alkaloid levels in commercial and wild genotypes of narrow-leafed lupin. J. Food Compos. Anal. 2024, 135, 106600. Available online: https://www.sciencedirect.com/science/article/pii/S0889157524006343 (accessed on 1 November 2025). [CrossRef]
- Keller, C.; Maeda, J.; Jayaraman, D.; Chakraborty, S.; Sussman, M.R.; Harris, J.M.; Ané, J.-M.; Li, L. Comparison of vacuum MALDI and AP-MALDI platforms for the mass spectrometry imaging of metabolites involved in salt stress in Medicago truncatula. Front. Plant Sci. 2018, 9, 1238. [Google Scholar] [CrossRef]
- Kranawetter, C.; Zeng, S.; Joshi, T.; Sumner, L.W. A Medicago truncatula metabolite atlas enables the visualization of differential accumulation of metabolites in root tissues. Metabolites 2021, 11, 238. [Google Scholar] [CrossRef]
- Liu, L.; Si, L.; Zhang, L.; Guo, R.; Wang, R.; Dong, H.; Guo, C. Metabolomics and transcriptomics analysis revealed the response mechanism of alfalfa to combined cold and saline-alkali stress. Plant J. 2024, 119, 1900–1919. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Cruz De Carvalho, M.H.; Torres-Jerez, I.; Kang, Y.; Allen, S.N.; Huhman, D.V.; Tang, Y.; Murray, J.; Sumner, L.W.; Udvardi, M.K. Global reprogramming of transcription and metabolism in M edicago truncatula during progressive drought and after rewatering. Plant Cell Environ. 2014, 37, 2553–2576. [Google Scholar] [CrossRef]
- Kumar, J.; Pratap, A.; Kumar, S. Phenomics in Crop Plants: Trends, Options and Limitations; Springer: New Delhi, India, 2015. [Google Scholar]
- Morrison, M.J.; Gahagan, A.C.; Lefebvre, M.B. Measuring canopy height in soybean and wheat using a low-cost depth camera. Plant Phenome J. 2021, 4, e20019. [Google Scholar] [CrossRef]
- Sarkar, S.; Sagan, V.; Bhadra, S.; Rhodes, K.; Pokharel, M.; Fritschi, F.B. Soybean seed composition prediction from standing crops using PlanetScope satellite imagery and machine learning. ISPRS J. Photogramm. Remote Sens. 2023, 204, 257–274. [Google Scholar] [CrossRef]
- Kemeshi, J.; Chang, Y.; Yadav, P.K.; Maimaitijiang, M.; Reicks, G. Development and Evaluation of a Multiaxial Modular Ground Robot for Estimating Soybean Phenotypic Traits Using an RGB-Depth Sensor. AgriEngineering 2025, 7, 76. [Google Scholar] [CrossRef]
- Chen, Y.L.; Dunbabin, V.M.; Postma, J.A.; Diggle, A.J.; Palta, J.A.; Lynch, J.P.; Siddique, K.H.; Rengel, Z. Phenotypic variability and modelling of root structure of wild Lupinus angustifolius genotypes. Plant Soil 2011, 348, 345–364. [Google Scholar] [CrossRef]
- Alkemade, J.A.; Messmer, M.M.; Arncken, C.; Leska, A.; Annicchiarico, P.; Nazzicari, N.; Książkiewicz, M.; Voegele, R.T.; Finckh, M.R.; Hohmann, P. A high-throughput phenotyping tool to identify field-relevant anthracnose resistance in white lupin. Plant Dis. 2021, 105, 1719–1727. [Google Scholar] [CrossRef]
- Sinde-González, I.; Murgueitio-Herrera, E.; Falconí, C.E.; Gil-Docampo, M.; Toulkeridis, T. Spectral Variability Analysis of Lupinus mutabilis Sweet Under Nanofertilizer and Chelate Application Through Spectroscopy and Unmanned Aerial Vehicle (UAV) Multispectral Images. Agronomy 2025, 15, 469. [Google Scholar] [CrossRef]
- Tran, B.T.; Cavagnaro, T.R.; Jewell, N.; Brien, C.; Berger, B.; Watts-Williams, S.J. High-throughput phenotyping reveals growth of Medicago truncatula is positively affected by arbuscular mycorrhizal fungi even at high soil phosphorus availability. Plants People Planet 2021, 3, 600–613. [Google Scholar] [CrossRef]
- Jeudy, C.; Adrian, M.; Baussard, C.; Bernard, C.; Bernaud, E.; Bourion, V.; Busset, H.; Cabrera-Bosquet, L.; Cointault, F.; Han, S. RhizoTubes as a new tool for high throughput imaging of plant root development and architecture: Test, comparison with pot grown plants and validation. Plant Methods 2016, 12, 31. [Google Scholar] [CrossRef]
- Jubery, T.Z.; Carley, C.N.; Singh, A.; Sarkar, S.; Ganapathysubramanian, B.; Singh, A.K. Using machine learning to develop a fully automated soybean nodule acquisition pipeline (snap). Plant Phenomics 2021, 2021, 9834746. [Google Scholar] [CrossRef]
- Bersanelli, M.; Mosca, E.; Remondini, D.; Giampieri, E.; Sala, C.; Castellani, G.; Milanesi, L. Methods for the integration of multi-omics data: Mathematical aspects. BMC Bioinform. 2016, 17, S15. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B.; Langefeld, C.; Olivier, M.; Cox, L.A. Integrated omics: Tools, advances and future approaches. J. Mol. Endocrinol. 2019, 62, R21–R45. [Google Scholar] [CrossRef]
- Naik, H.S.; Zhang, J.; Lofquist, A.; Assefa, T.; Sarkar, S.; Ackerman, D.; Singh, A.; Singh, A.K.; Ganapathysubramanian, B. A real-time phenotyping framework using machine learning for plant stress severity rating in soybean. Plant Methods 2017, 13, 23. [Google Scholar] [CrossRef]
- Nobori, T. Exploring the untapped potential of single-cell and spatial omics in plant biology. New Phytol. 2025, 247, 1098–1116. [Google Scholar] [CrossRef]
- Xia, K.; Sun, H.-X.; Li, J.; Li, J.; Zhao, Y.; Chen, L.; Qin, C.; Chen, R.; Chen, Z.; Liu, G. The single-cell stereo-seq reveals region-specific cell subtypes and transcriptome profiling in Arabidopsis leaves. Dev. Cell 2022, 57, 1299–1310.e1294. [Google Scholar] [CrossRef]
- Dorrity, M.W.; Alexandre, C.M.; Hamm, M.O.; Vigil, A.-L.; Fields, S.; Queitsch, C.; Cuperus, J.T. The regulatory landscape of Arabidopsis thaliana roots at single-cell resolution. Nat. Commun. 2021, 12, 3334. [Google Scholar] [CrossRef]
- Rhaman, M.S.; Ali, M.; Ye, W.; Li, B. Opportunities and Challenges in Advancing Plant Research with Single-cell Omics. Genom. Proteom. Bioinform. 2024, 22, qzae026. [Google Scholar] [CrossRef]
- Yang, M.; Du, C.; Li, M.; Wang, Y.; Bao, G.; Huang, J.; Zhang, Q.; Zhang, S.; Xu, P.; Teng, W. The transcription factors GmVOZ1A and GmWRI1a synergistically regulate oil biosynthesis in soybean. Plant Physiol. 2025, 197, kiae485. [Google Scholar] [CrossRef]
- Nguyen, T.Q.; Kisiala, A.B.; Hai, N.N.; Narine, S.; Emery, R.N. Phytohormone dynamics impact fatty acid and oil accumulation during soybean seed maturation. Seed Sci. Res. 2021, 31, 278–291. [Google Scholar] [CrossRef]
- Tayade, R.; Imran, M.; Ghimire, A.; Khan, W.; Nabi, R.B.S.; Kim, Y. Molecular, genetic, and genomic basis of seed size and yield characteristics in soybean. Front. Plant Sci. 2023, 14, 1195210. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, M.A.; Lucas, M.M.; Pueyo, J.J. Adaptive mechanisms make lupin a choice crop for acidic soils affected by aluminum toxicity. Front. Plant Sci. 2022, 12, 810692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiao, Y.; Liu, Z.; Zhu, Y.-X. ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat. Commun. 2015, 6, 6003. [Google Scholar] [CrossRef] [PubMed]
- Wilmowicz, E.; Kućko, A.; Golińska, P.; Burchardt, S.; Przywieczerski, T.; Świdziński, M.; Brzozowska, P.; Kapuścińska, D. Abscisic acid and ethylene in the control of nodule-specific response on drought in yellow lupine. Environ. Exp. Bot. 2020, 169, 103900. [Google Scholar] [CrossRef]
- Cervantes-Pérez, S.A.; Thibivilliers, S.; Laffont, C.; Farmer, A.D.; Frugier, F.; Libault, M. Cell-specific pathways recruited for symbiotic nodulation in the Medicago truncatula legume. Mol. Plant 2022, 15, 1868–1888. [Google Scholar] [CrossRef]
- Andrio, E.; Marino, D.; Marmeys, A.; de Segonzac, M.D.; Damiani, I.; Genre, A.; Huguet, S.; Frendo, P.; Puppo, A.; Pauly, N. Hydrogen peroxide-regulated genes in the Medicago truncatula–Sinorhizobium meliloti symbiosis. New Phytol. 2013, 198, 179–189. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. The Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef]
- Costa, S.R.; Chin, S.; Mathesius, U. Infection of Medicago truncatula by the root-knot nematode Meloidogyne javanica does not require early nodulation genes. Front. Plant Sci. 2020, 11, 1050. [Google Scholar] [CrossRef]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, J.; Chen, L.; Wu, T.; Jiang, B.; Xu, C.; Cai, Y.; Dong, J.; Han, T.; Sun, S. CRISPR/Cas9-Mediated Targeted Mutagenesis of GmEOD1 Enhances Seed Size of Soybean. Agronomy 2023, 13, 2359. [Google Scholar] [CrossRef]
- Ceulemans, E.; Ibrahim, H.M.; De Coninck, B.; Goossens, A. Pathogen effectors: Exploiting the promiscuity of plant signaling hubs. Trends Plant Sci. 2021, 26, 780–795. [Google Scholar] [CrossRef]
- Moellers, T.C.; Singh, A.; Zhang, J.; Brungardt, J.; Kabbage, M.; Mueller, D.S.; Grau, C.R.; Ranjan, A.; Smith, D.L.; Chowda-Reddy, R. Main and epistatic loci studies in soybean for Sclerotinia sclerotiorum resistance reveal multiple modes of resistance in multi-environments. Sci. Rep. 2017, 7, 3554. [Google Scholar] [CrossRef] [PubMed]
- Kofsky, J.; Zhang, H.; Song, B.-H. Novel resistance strategies to soybean cyst nematode (SCN) in wild soybean. Sci. Rep. 2021, 11, 7967. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Shen, D.; Xiong, Q.; Bao, B.; Zhang, W.; Dai, T.; Zhao, Y.; Borriss, R.; Fan, B. The plant-beneficial rhizobacterium Bacillus velezensis FZB42 controls the soybean pathogen Phytophthora sojae due to bacilysin production. Appl. Environ. Microbiol. 2021, 87, e01601-21. [Google Scholar] [CrossRef] [PubMed]
- Nuhse, T.S.; Stensballe, A.; Jensen, O.N.; Peck, S.C. Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 2004, 16, 2394–2405. [Google Scholar] [CrossRef]
- Chongtham, S.K.; Devi, E.L.; Samantara, K.; Yasin, J.K.; Wani, S.H.; Mukherjee, S.; Razzaq, A.; Bhupenchandra, I.; Jat, A.L.; Singh, L.K.; et al. Orphan legumes: Harnessing their potential for food, nutritional and health security through genetic approaches. Planta 2022, 256, 24. [Google Scholar] [CrossRef]
- Song, H.; Taylor, D.C.; Zhang, M. Bioengineering of soybean oil and its impact on agronomic traits. Int. J. Mol. Sci. 2023, 24, 2256. [Google Scholar] [CrossRef]
- Hu, H.; Zhao, J.; Thomas, W.J.W.; Batley, J.; Edwards, D. The role of pangenomics in orphan crop improvement. Nat. Commun. 2025, 16, 118. [Google Scholar] [CrossRef]
- Kokkini, M.; Gazoulis, I.; Danaskos, M.; Kontogeorgou, V.; Kanatas, P.; Travlos, I. Enhancing ecosystem services in agriculture: The special role of legume intercropping. Front. Sustain. Food Syst. 2025, 9, 1547879. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, J.; Beillouin, D.; Lambers, H.; Yang, Y.; Smith, P.; Zeng, Z.; Olesen, J.E.; Zang, H. Global systematic review with meta-analysis reveals yield advantage of legume-based rotations and its drivers. Nat. Commun. 2022, 13, 4926. [Google Scholar] [CrossRef]
- Hendre, P.S.; Muthemba, S.; Kariba, R.; Muchugi, A.; Fu, Y.; Chang, Y.; Song, B.; Liu, H.; Liu, M.; Liao, X. African Orphan Crops Consortium (AOCC): Status of developing genomic resources for African orphan crops. Planta 2019, 250, 989–1003. [Google Scholar] [CrossRef]
- Monfort, M.; Buitink, J.; Roeber, F.; Nogué, F. Genome editing, an opportunity to revive soybean cultivation in Europe. Plant J. 2025, 121, e17266. [Google Scholar] [CrossRef]
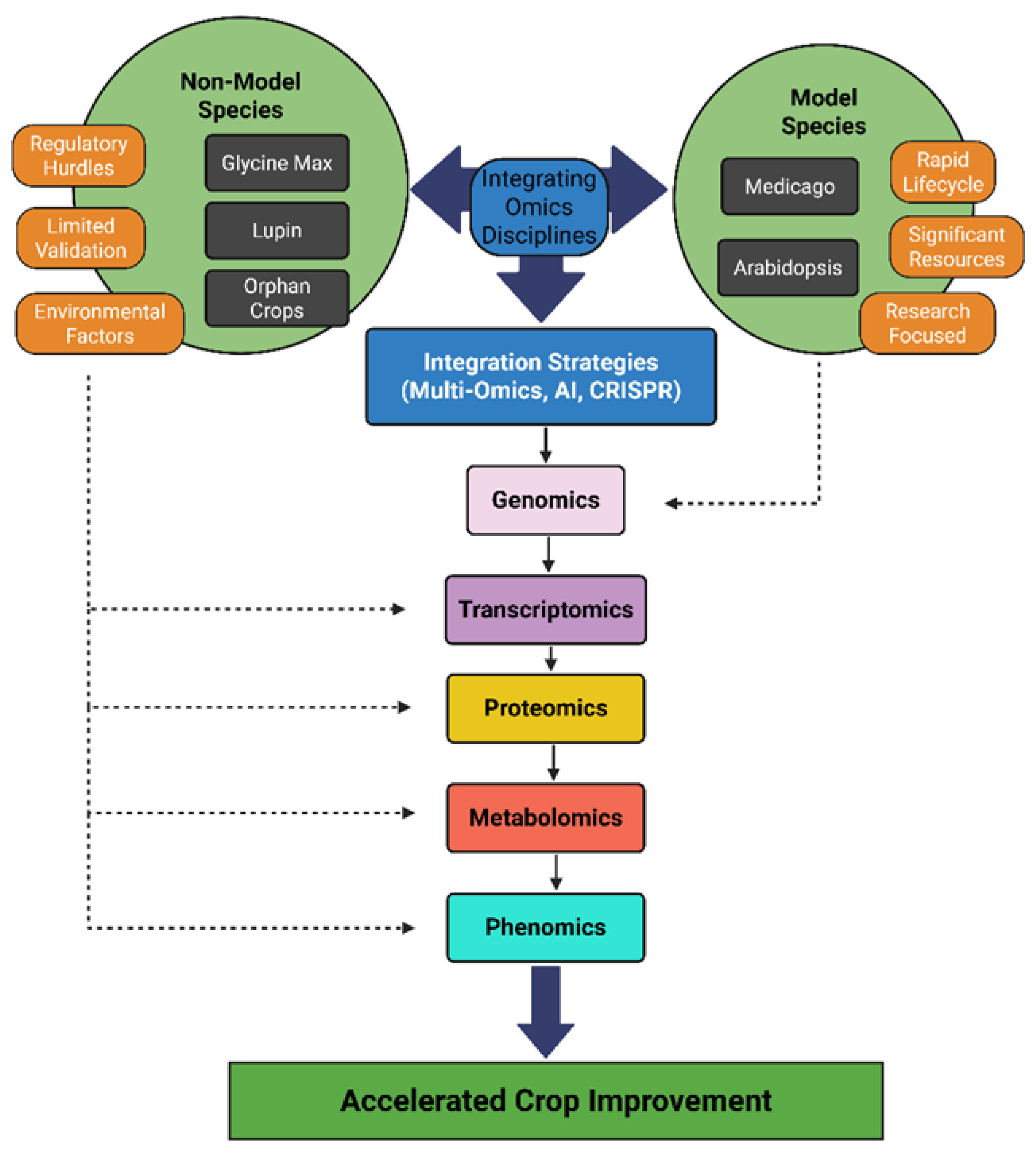
| Species | Model Status | Genome Resources | Key Strengths | Key Challenges |
|---|---|---|---|---|
| Arabidopsis thaliana | Model | Extensive, curated | Comprehensive annotation, mutant libraries | Not a legume; lacks nodule formation |
| Medicago truncatula | Model legume | Moderate to high | Syntenic with legumes, N-fixation model | Fewer large-scale resources than Arabidopsis |
| Glycine max (soybean) | Non-model | Rich but complex | Genomic data, gene editing progress | Paleopolyploid, complicates annotation |
| Lupinus albus | Non-model | Emerging | Cluster root adaptation, nutrient-use traits | Poor transformation systems, limited mutants |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zalzalah, N.; Bruggink, J.; Elian, M.; Lackey, S.; Wozny, J.C.; Haidar, S.; Cober, E.R.; Xing, T.; Samanfar, B. Molecular Network Analysis in Model and Non-Model Legumes: Challenges in Omics Data Interpretation Across Species, with a Focus on Glycine max, Lupinus albus and Medicago truncatula. Plants 2025, 14, 3586. https://doi.org/10.3390/plants14233586
Zalzalah N, Bruggink J, Elian M, Lackey S, Wozny JC, Haidar S, Cober ER, Xing T, Samanfar B. Molecular Network Analysis in Model and Non-Model Legumes: Challenges in Omics Data Interpretation Across Species, with a Focus on Glycine max, Lupinus albus and Medicago truncatula. Plants. 2025; 14(23):3586. https://doi.org/10.3390/plants14233586
Chicago/Turabian StyleZalzalah, Nayla, Jakob Bruggink, Mohamad Elian, Simon Lackey, Julia C. Wozny, Siwar Haidar, Elroy R. Cober, Tim Xing, and Bahram Samanfar. 2025. "Molecular Network Analysis in Model and Non-Model Legumes: Challenges in Omics Data Interpretation Across Species, with a Focus on Glycine max, Lupinus albus and Medicago truncatula" Plants 14, no. 23: 3586. https://doi.org/10.3390/plants14233586
APA StyleZalzalah, N., Bruggink, J., Elian, M., Lackey, S., Wozny, J. C., Haidar, S., Cober, E. R., Xing, T., & Samanfar, B. (2025). Molecular Network Analysis in Model and Non-Model Legumes: Challenges in Omics Data Interpretation Across Species, with a Focus on Glycine max, Lupinus albus and Medicago truncatula. Plants, 14(23), 3586. https://doi.org/10.3390/plants14233586









