Comparison of the Genetic Basis of Yield Traits Between Main and Ratoon Rice in an Eight-Way MAGIC Population
Abstract
1. Introduction
2. Results
2.1. Parental Performance of the MAGIC Population in Main and Ratoon Seasons
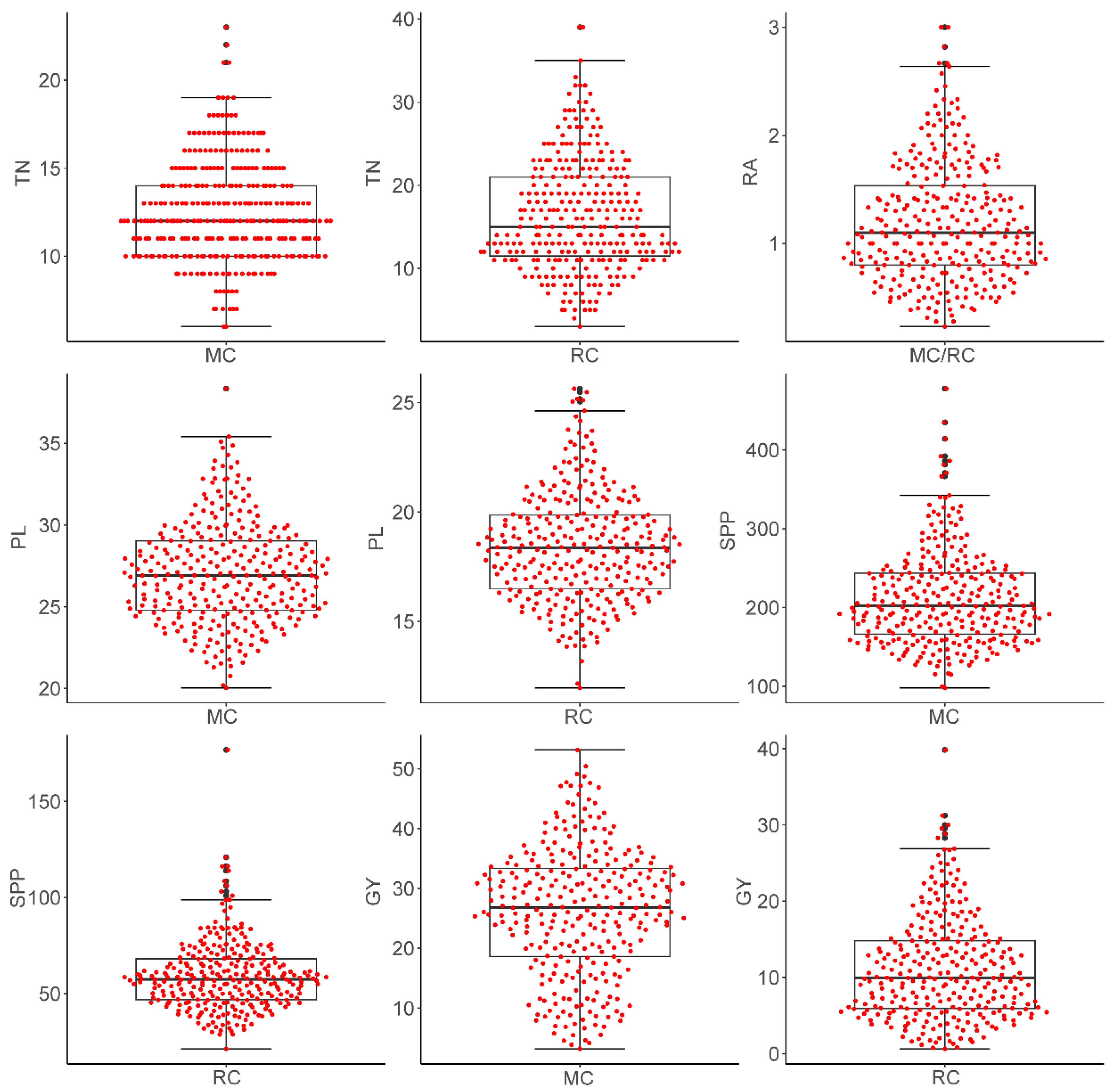
2.2. Yield Performance of the MAGIC Population in Main and Ratoon Crops
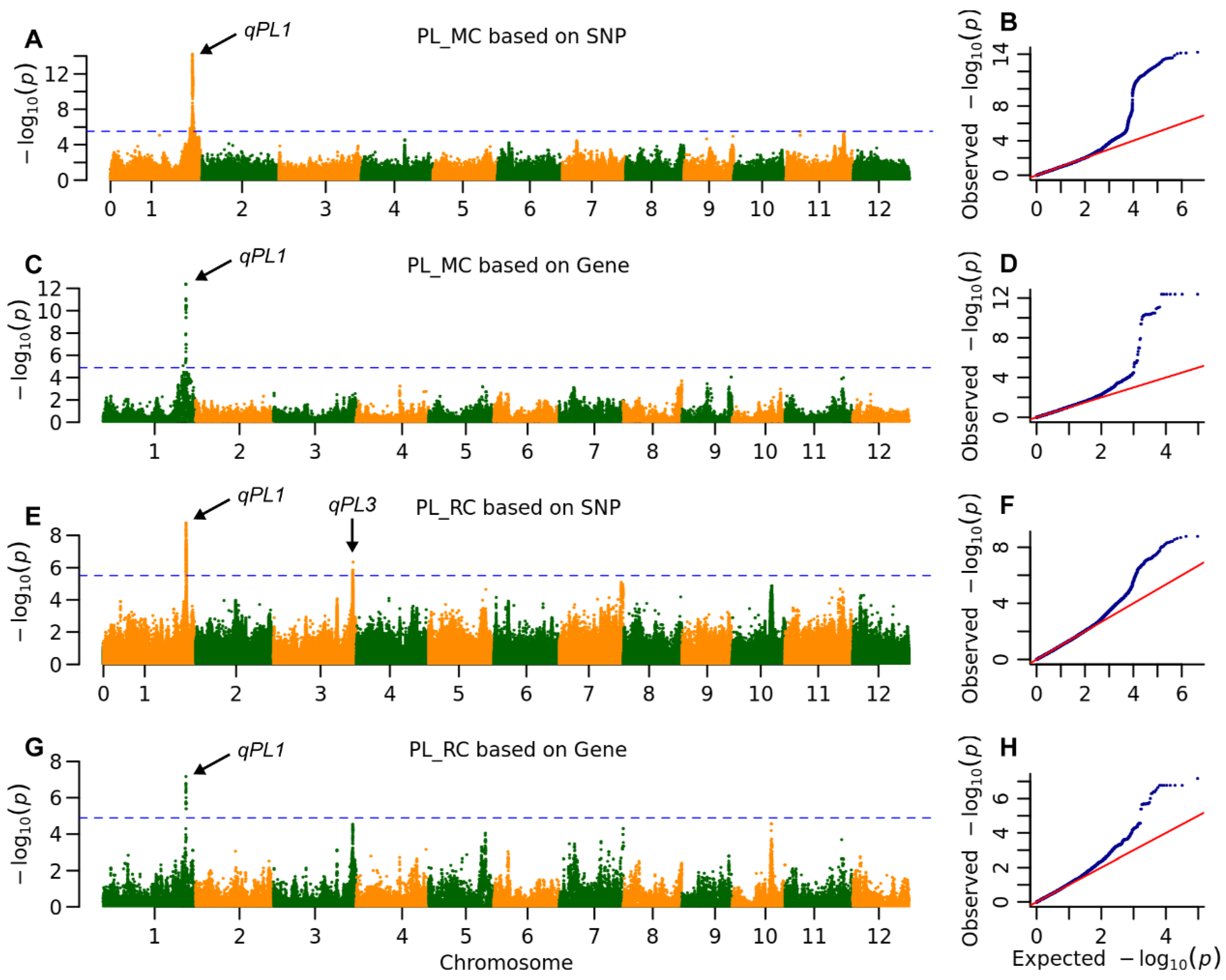
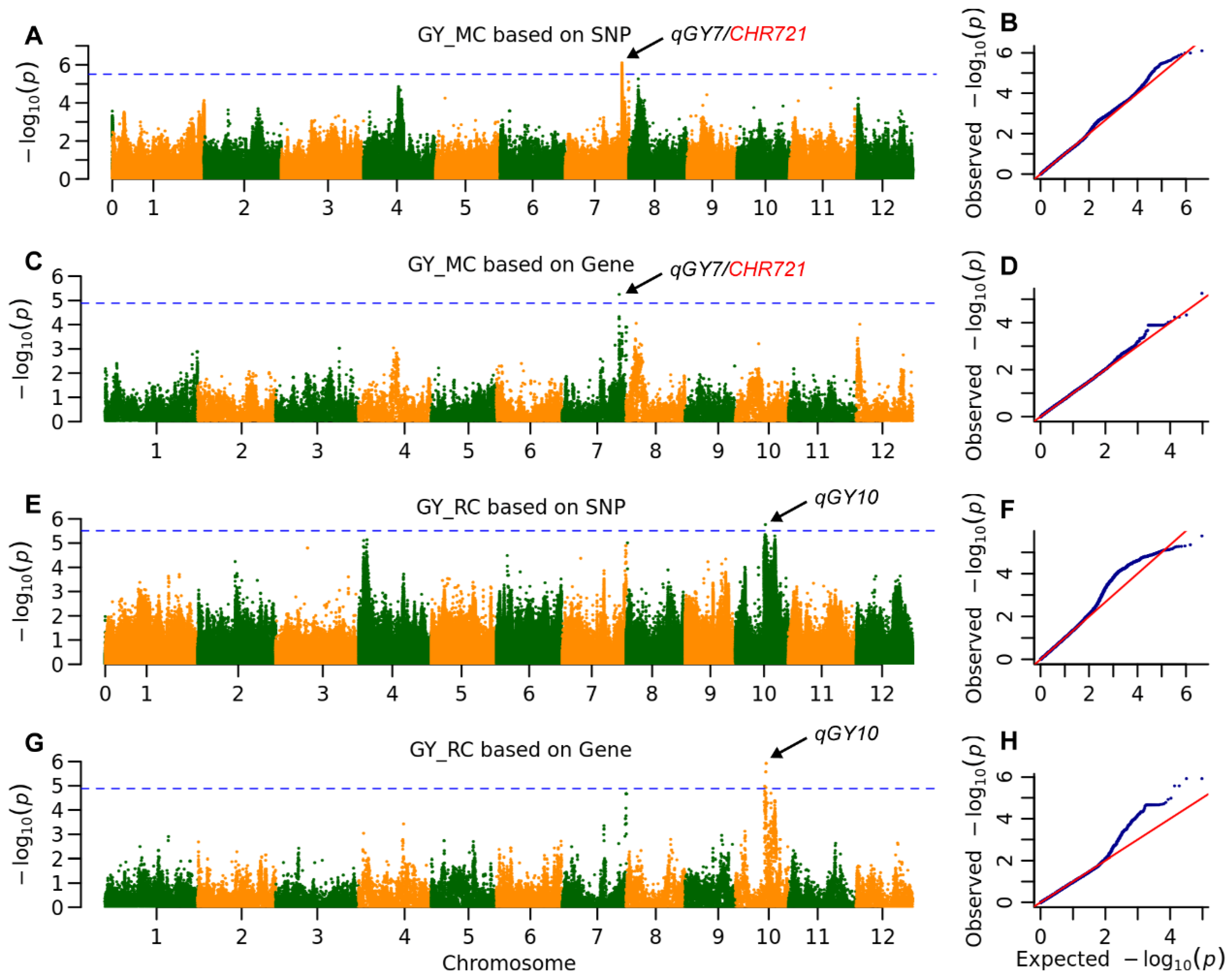
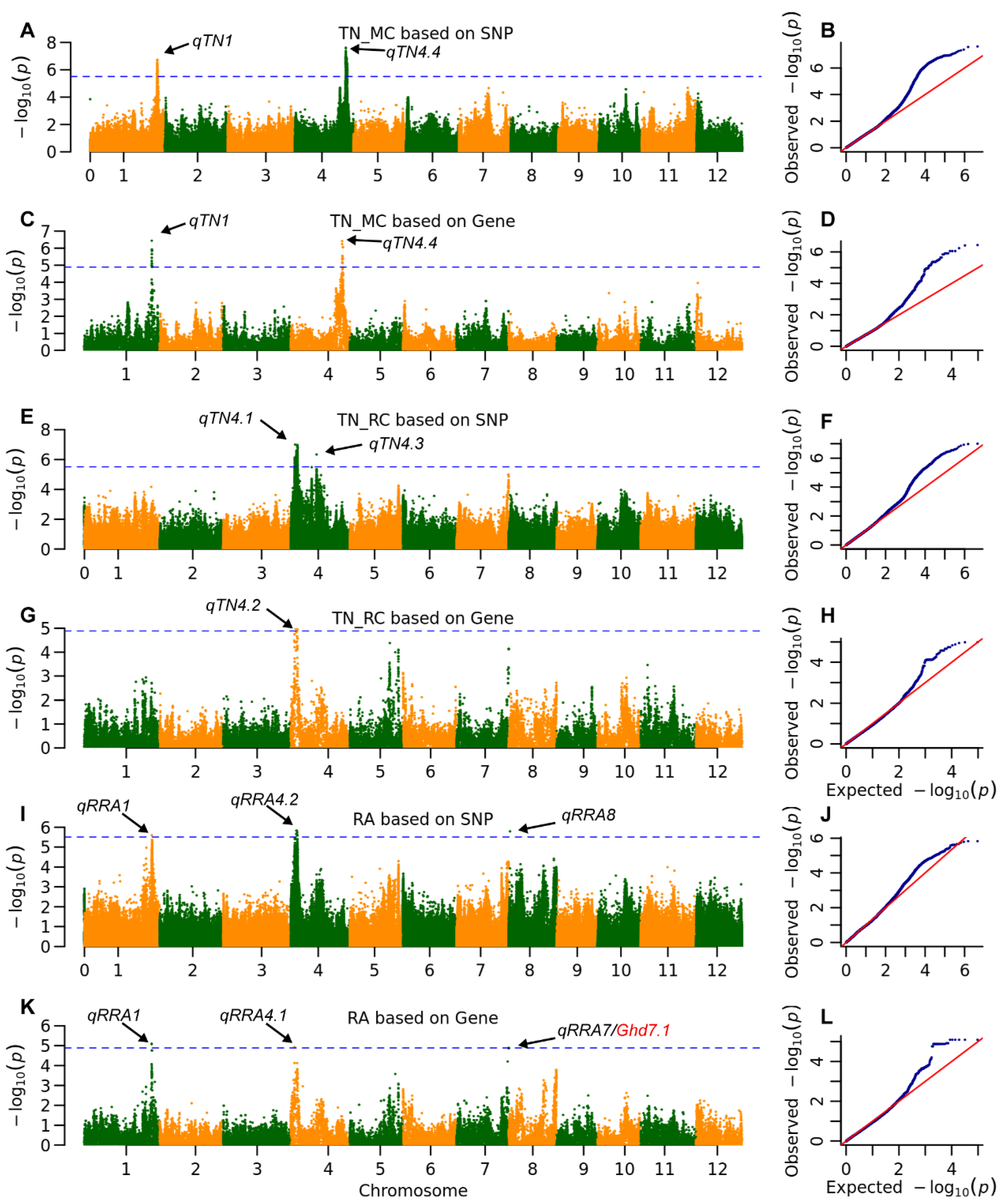
2.3. The Genetic Architecture of Yield and Ratooning Ability Across Cropping Seasons
2.4. Genetic Effects of Multiple Parental Alleles at Key QTLs Impacting Ratoon Crop Performance
3. Discussion
3.1. Diverse Genetic Basis of Grain Yield Between Main and Ratoon Crops
3.2. Genetic Insights into Tiller Number and Ratooning Ability
3.3. Hormones Might Be the Major Endogenous Factor Regulating Ratooning Ability in Ratoon Crop
3.4. The Challenge of QTL Mapping for Ratooning Ability in Ratoon Crop
4. Materials and Methods
4.1. Development of a MAGIC Population
4.2. Field Management and Phenotype Measurement
4.3. Next-Generation Sequencing and Genotyping
4.4. Genome-Wide Association Studies
4.5. Genetic Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munda, G.C.; Das, A.; Patel, D.P. Evaluation of transplanted and ratoon crop for double cropping of rice (Oryza sativa L.) under organic input management in mid altitude sub-tropical Meghalaya. Curr. Sci. 2009, 96, 1620–1627. [Google Scholar]
- Wang, W.; He, A.; Jiang, G.; Sun, H.; Jiang, M.; Man, J.; Ling, X.; Cui, K.; Huang, J.; Peng, S.; et al. Chapter Four—Ratoon Rice Technology: A Green and Resource-Efficient Way for Rice Production. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2020; Volume 159, pp. 135–167. ISBN 0065-2113. [Google Scholar] [CrossRef]
- Peng, S.; Zheng, C.; Yu, X. Progress and challenges of rice ratooning technology in China. Crop Environ. 2023, 2, 5–11. [Google Scholar] [CrossRef]
- Faruq, G.; Taha, R.; Prodhan, Z. Rice Ratoon Crop: A Sustainable Rice Production System for Tropical Hill Agriculture. Sustainability 2014, 6, 5785–5800. [Google Scholar] [CrossRef]
- Hu, H.; Gao, R.; He, L.; Liang, F.; Li, Z.; Xu, J.; Yang, L.; Wang, C.; Liu, Z.; Xu, J.; et al. Genetic Dissection of Rice Ratooning Ability Using an Introgression Line Population and Substitution Mapping of a Pleiotropic Quantitative Trait Locus qRA5. Plants 2022, 11, 1134. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Z.; Wang, B.; Zhang, Z.; Lin, Y.; Cheng, J. Enhancing the annual yield via nitrogen fertilizer application optimization in the direct seeding ratoon rice system. Front. Plant Sci. 2024, 15, 1366718. [Google Scholar] [CrossRef]
- Ji, S.; Luo, X.; Ahn, S.N. Mapping QTL for ratooning ability in advanced backcross lines from an Oryza sativa × O. rufipogon cross. Korean J. Agric. Sci. 2014, 41, 1–7. [Google Scholar] [CrossRef]
- He, N.; Huang, F.; Yang, D. Fine Mapping and Cloning of a qRA2 Affect the Ratooning Ability in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2023, 24, 967. [Google Scholar] [CrossRef]
- Yao, Y.; Xiang, D.; Wu, N.; Wang, Y.; Chen, Y.; Yuan, Y.; Ye, Y.; Hu, D.; Zheng, C.; Yan, Y.; et al. Control of rice ratooning ability by a nucleoredoxin that inhibits histidine kinase dimerization to attenuate cytokinin signaling in axillary buds. Mol. Plant 2023, 16, 1911–1926. [Google Scholar] [CrossRef]
- Yang, H.J.; Zheng, J.S.; Jiang, Z.W.; Li, Y.Z.; Zhuo, C.Y.; Zhang, S.S. The sink structure character of super high-yielding ratooning rice. Fujian J. Agric. Sci. 2005, 20, 65–68. [Google Scholar]
- Banoc, D.M.; Sevillano, R.; Asio, V.B. Ratooning response of lowland rice (Oryza sativa L.) varieties to cutting height of ratoon crop. SVU-Int. J. Agric. Sci. 2022, 4, 99–110. [Google Scholar] [CrossRef]
- Xu, F.; Xiong, H.; Zhu, Y.; Zhang, L. Effect of the amount of N application for bud development on the ratooning ability and in association with ratio of source to sink in mid-season hybrid rice. Southwest China J. Agric. Sci. 2008, 21, 688–693. [Google Scholar]
- Yu, S.B.; Li, J.X.; Xu, C.G.; Tan, Y.F.; Gao, Y.J.; Li, X.H.; Zhang, Q.; Maroof, M.A.S. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 1997, 94, 9226–9231. [Google Scholar] [CrossRef]
- Tan, C.; Han, Z.; Yu, H.; Zhan, W.; Xie, W.; Chen, X.; Zhao, H.; Zhou, F.; Xing, Y. QTL scanning for rice yield using a whole genome SNP array. J. Genet. Genom. 2013, 40, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Tung, C.W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef]
- Yano, K.; Yamamoto, E.; Aya, K.; Takeuchi, H.; Lo, P.; Hu, L.; Yamasaki, M.; Yoshida, S.; Kitano, H.; Hirano, K.; et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 2016, 48, 927–934. [Google Scholar] [CrossRef]
- Han, Z.; Zhang, B.; Zhao, H.; Ayaad, M.; Xing, Y. Genome-Wide Association Studies Reveal that Diverse Heading Date Genes Respond to Short and Long Day Lengths between Indica and Japonica Rice. Front. Plant Sci. 2016, 7, 1270. [Google Scholar] [CrossRef]
- Huang, B.E.; George, A.W.; Forrest, K.L.; Kilian, A.; Hayden, M.J.; Morell, M.K.; Cavanagh, C.R. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol. J. 2012, 10, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yang, Y.; Li, D.; Yu, Z.; Zhang, B.; Zhou, X.; Xiong, L.; Zhang, J.; Xing, Y. Powerful QTL mapping and favorable allele mining in an all-in-one population: A case study of heading date. Natl. Sci. Rev. 2024, 11, nwae222. [Google Scholar] [CrossRef]
- Wei, X.; Chen, M.; Zhang, Q.; Gong, J.; Liu, J.; Yong, K.; Wang, Q.; Fan, J.; Chen, S.; Hua, H.; et al. Genomic investigation of 18,421 lines reveals the genetic architecture of rice. Science 2024, 385, eadm8762. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Q.; Zhu, G.; Zhang, F.; Fang, X.; Ren, H.; Jiang, J.; Yang, F.; Zhang, D.; Chen, F. CHR721, interacting with OsRPA1a, is essential for both male and female reproductive development in rice. Plant Mol. Biol. 2020, 103, 473–487. [Google Scholar] [CrossRef]
- Yan, W.; Liu, H.; Zhou, X.; Li, Q.; Zhang, J.; Lu, L.; Liu, T.; Liu, H.; Zhang, C.; Zhang, Z.; et al. Natural variation in Ghd7. 1 plays an important role in grain yield and adaptation in rice. Cell Res. 2013, 23, 969–971. [Google Scholar] [CrossRef]
- Gao, H.; Jin, M.; Zheng, X.M.; Chen, J.; Yuan, D.; Xin, Y.; Wang, M.; Huang, D.; Zhang, Z.; Zhou, K.; et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 16337–16342. [Google Scholar] [CrossRef]
- Zou, X.; Qin, Z.; Zhang, C.; Liu, B.; Liu, J.; Zhang, C.; Lin, C.; Li, H.; Zhao, T. Over-expression of an S-domain receptor-like kinase extracellular domain improves panicle architecture and grain yield in rice. J. Exp. Bot. 2015, 66, 7197–7209. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Sun, F.; Wang, Q.; Chen, M.; Huang, Y.; Feng, Y.-Q.; Luo, X.; Yang, J. Rice Ethylene-Response AP2/ERF Factor OsEATB Restricts Internode Elongation by Down-Regulating a Gibberellin Biosynthetic Gene. Plant Physiol. 2011, 157, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.U. The Role of Pumilio RNA Binding Protein in Plants. Biomolecules 2021, 11, 1851. [Google Scholar] [CrossRef]
- Huang, R.; Liu, M.; Gong, G.; Wu, P.; Patra, B.; Yuan, L.; Qin, H.; Wang, X.; Wang, G.; Liao, H.; et al. The Pumilio RNA-binding protein APUM24 regulates seed maturation by fine-tuning the BPM-WRI1 module in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 1240–1259. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.; Peng, S.; He, Q.; Yang, H.; Yang, C.; Visperas, R.M.; Cassman, K.G. Comparison of high-yield rice in tropical and subtropical environments. Field Crops Res. 1998, 57, 71–84. [Google Scholar] [CrossRef]
- Santos, A.B.; Fageria, N.K.; Prabhu, A.S. Rice Ratooning Management Practices for Higher Yields. Commun. Soil. Sci. Plant Anal. 2003, 34, 881–918. [Google Scholar] [CrossRef]
- Xu, F. The ratoon rice system with high yield and high efficiency in China: Progress, trend of theory and technology. Field Crops Res. 2021, 272, 108282. [Google Scholar] [CrossRef]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Minakuchi, K.; Kameoka, H.; Yasuno, N.; Umehara, M.; Luo, L.; Kobayashi, K.; Hanada, A.; Ueno, K.; Asami, T.; Yamaguchi, S.; et al. FINE CULM1 (FC1) Works Downstream of Strigolactones to Inhibit the Outgrowth of Axillary Buds in Rice. Plant Cell Physiol. 2010, 51, 1127–1135. [Google Scholar] [CrossRef]
- Dun, E.A.; De Saint Germain, A.; Rameau, C.; Beveridge, C.A. Antagonistic Action of Strigolactone and Cytokinin in Bud Outgrowth Control. Plant Physiol. 2012, 158, 487–498. [Google Scholar] [CrossRef]
- Shinohara, N.; Taylor, C.; Leyser, O. Strigolactone Can Promote or Inhibit Shoot Branching by Triggering Rapid Depletion of the Auxin Efflux Protein PIN1 from the Plasma Membrane. PLoS Biol. 2013, 11, e1001474. [Google Scholar] [CrossRef]
- Liao, Z.; Yu, H.; Duan, J.; Yuan, K.; Yu, C.; Meng, X.; Kou, L.; Chen, M.; Jing, Y.; Liu, G.; et al. SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 2019, 10, 2738. [Google Scholar] [CrossRef]
- Harrell, D.L.; Bond, J.A.; Blanche, S. Evaluation of main-crop stubble height on ratoon rice growth and development. Field Crops Res. 2009, 114, 396–403. [Google Scholar] [CrossRef]
- Jones, D.B. Rice Ratoon Response to Main Crop Harvest Cutting Height. Agron. J. 1993, 85, 1139–1142. [Google Scholar] [CrossRef]
- Jung, Y.; Han, D. BWA-MEME: BWA-MEM emulated with a machine learning approach. Bioinformatics 2022, 38, 2404–2413. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Lippert, C.; Listgarten, J.; Liu, Y.; Kadie, C.M.; Davidson, R.I.; Heckerman, D. FaST linear mixed models for genome-wide association studies. Nat. Methods 2011, 8, 833–835. [Google Scholar] [CrossRef]
- Endelman, J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef]
- Wray, N.; Visscher, P. Estimating trait heritability. Nat. Educ. 2008, 1, 29. [Google Scholar]
| Trait | Crop | QTL | SNP-Based GWAS | Gene-Based GWAS | Nearby Gene | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pos | p | PV (%) | Peak Gene | Pos (bp) | p | PV (%) | ||||
| PL | MC | qPL1 | 36,766,995 | 5.8 × 10−15 | 19.5 | MH01t0732300 | 36,790,627 | 4.3 × 10−13 | 24.0 | |
| RC | qPL1 | 36,746,675 | 1.7 × 10−9 | 11.1 | MH01t0733100 | 36,818,061 | 3.8 × 10−9 | 10.7 | ||
| qPL3 | 35,566,823 | 4.5 × 10−7 | 5.8 | |||||||
| SPP | MC | qSPP1 | 36,912,397 | 2.3 × 10−8 | 10.9 | |||||
| qSPP2 | 2,890,022 | 1.8 × 10−6 | 7.0 | |||||||
| qSPP4 | 28,118,380 | 1.9 × 10−7 | 11.2 | MH04t0625100 | 28,096,106 | 1.3 × 10−5 | 8.5 | |||
| GY | MC | qGY7 | 25,360,255 | 7.8 × 10−7 | 6.7 | MH07t0500600 | 25,371,872 | 5.6 × 10−6 | 6.7 | 3.1 kb to CHR721 |
| RC | qGY10 | 13,346,352 | 1.7 × 10−6 | 6.8 | MH10t0253200 | 13,694,183 | 1.2 × 10−6 | 5.1 | ||
| Trait | Crop | QTL | SNP-Based GWAS | Gene-Based GWAS | Nearby Gene | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pos | p | PV (%) | Peak Gene | Position (bp) | p | PV (%) | ||||
| TN | MC | qTN1 | 36,895,057 | 1.9 × 10−7 | 7.8 | MH01t0732900 | 36,812,637 | 3.6 × 10−7 | 10.1 | |
| qTN4.4 | 28,100,407 | 2.4 × 10−8 | 10.6 | MH04t0625200 | 28,101,376 | 3.8 × 10−7 | 8.9 | |||
| RC | qTN4.1 | 2,333,293 | 9.9 × 10−8 | 11.6 | ||||||
| qTN4.2 | MH04t0067200 | 2,745,892 | 1.0 × 10−5 | 2.1 | ||||||
| qTN4.3 | 14,036,939 | 4.6 × 10−7 | 4.4 | |||||||
| RA | qRRA1 | 36,742,004 | 2.5 × 10−6 | 8.3 | MH01t0731300 | 36,742,004 | 8.1 × 10−6 | 7.4 | ||
| qRRA4.1 | MH04t0054700 | 2,082,223 | 1.2 × 10−5 | 1.3 | ||||||
| qRRA4.2 | 3,268,530 | 1.5 × 10−6 | 7.0 | |||||||
| qRRA7 | MH07t0551600 | 28,514,126 | 1.3 × 10−5 | 5.4 | Ghd7.1 | |||||
| qRRA8 | 664,886 | 1.6 × 10−6 | 5.9 | |||||||
| Hap 1 | Hap 2 | Hap 3 | |
|---|---|---|---|
| G1212C (Lys404Asn) | G | C | G |
| T1440C (Cys480Cys) | T | C | T |
| C1962T (Cys654Cys) | C | T | C |
| C2030T (Ala677Val) | C | T | C |
| T2104C (Ser702Pro) | T | C | C |
| A2201G (As34Gly) | A | G | A |
| G2227T (Ala743Ser) | G | T | G |
| G2365C (Val789Leu) | G | C | G |
| G2385T (Glu795Asp) | G | T | G |
| Frequency | 168 | 95 | 22 |
| Parents | IR34/GC2/Cyp/ YJMS/MH63 | DA5/Pra | ZS97 |
| TN_MC | 13.2 ± 3.1 b* | 11.5 ± 2.2 c | 14.5 ± 3.5 a |
| TN_RC | 15.5 ± 6.1 a | 17.4 ± 7.4 a | 16.6 ± 7.1 a |
| RA | 1.1 ± 0.5 b | 1.4 ± 0.6 a | 1.0 ± 0.4 b |
| PL_MC | 26.3 ± 2.7 b | 28.8 ± 3 a | 24.9 ± 3.2 c |
| PL_RC | 18 ± 2.3 b | 19.5 ± 2.6 a | 17 ± 2.7 b |
| GY_MC | 26.2 ± 10.3 a | 24.4 ± 11.9 a | 27.9 ± 12.5 a |
| GY_RC | 10.4 ± 6.3 a | 12.2 ± 6.9 a | 10.4 ± 6.8 a |
| Hap 1 | Hap 2 | Hap 3 | Hap 4 | |
|---|---|---|---|---|
| Ins 2222–2223 (FS) | 0 | 0 | 0 | 1 |
| Del 2219 (FS) | 0 | 0 | 0 | −1 |
| Ins 2192–2193 (FS) | 0 | 0 | 0 | 2 |
| Del 2190–2191 (FS) | 0 | 0 | 0 | −2 |
| Frequency | 168 | 72 | 19 | 21 |
| Parents | IR34/MH63/ Pra/ZS97 | GC2/YJMS | DA5 | Cyp |
| TN_MC | 12.6 ± 2.7 a* | 12.2 ± 3.3 a | 12.4 ± 2.5 a | 12.9 ± 2.9 a |
| TN_RC | 16.1 ± 6.5 b | 15.4 ± 6.2 b | 17.4 ± 8.9 a,b | 19.7 ± 8.3 a |
| RA | 1.2 ± 0.6 a | 1.1 ± 0.5 a | 1.3 ± 0.7 a | 1.4 ± 0.5 a |
| PL_MC | 27.1 ± 3.1 a | 27.0 ± 3.1 a | 26.8 ± 3.1 a | 28.2 ± 3.5 a |
| PL_RC | 18.4 ± 2.4 b | 18.2 ± 2.8 b | 17.7 ± 2.4 b | 20.1 ± 2.3 a |
| SPP_MC | 217.7 ± 68.3 a | 215.3 ± 63.3 a | 196.8 ± 32.0 a | 218.6 ± 64.7 a |
| SPP_RC | 59.9 ± 18.7 a,b | 55.5 ± 17.6 b | 59.4 ± 14.8 a,b | 68.5 ± 13.4 a |
| GY_MC | 25.6 ± 10.6 a | 25.9 ± 12.2 a | 26.3 ± 10.1 a | 25.7 ± 12.6 a |
| GY_RC | 10.7 ± 6.2 bc | 9.4 ± 5.7 c | 12.7 ± 8.2 b | 18.0 ± 9.1 a |
| Hap 1 | Hap 2 | |
|---|---|---|
| Del 1515_1522; (FS *) | 0 | −8 |
| Frequency | 168 | 124 |
| Parents | GC2/DA5/Cyp/ YJSM/MH63/Pra | IR34/ZS97 |
| TN_RC | 17.7 ± 7.1 a | 14.7 ± 6 b |
| RA | 1.3 ± 0.6 a | 1.0 ± 0.5 b |
| PL_MC | 27.4 ± 3.3 a | 26.7 ± 2.9 b |
| PL_RC | 18.8 ± 2.5 a | 17.9 ± 2.4 b |
| SPP_MC | 225.4 ± 68.1 a | 196.2 ± 54.3 b |
| SPP_RC | 61.3 ± 18.1 a | 56.5 ± 18.3 b |
| GY_MC | 24.2 ± 11 b | 27.4 ± 10.8 a |
| GY_RC | 12.5 ± 7.1 a | 9.0 ± 5.5 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Z.; Sherif, A.; Ayaad, M.; Xing, Y.; Lu, Y. Comparison of the Genetic Basis of Yield Traits Between Main and Ratoon Rice in an Eight-Way MAGIC Population. Plants 2025, 14, 3527. https://doi.org/10.3390/plants14223527
Han Z, Sherif A, Ayaad M, Xing Y, Lu Y. Comparison of the Genetic Basis of Yield Traits Between Main and Ratoon Rice in an Eight-Way MAGIC Population. Plants. 2025; 14(22):3527. https://doi.org/10.3390/plants14223527
Chicago/Turabian StyleHan, Zhongmin, Ahmed Sherif, Mohammed Ayaad, Yongzhong Xing, and Yuncai Lu. 2025. "Comparison of the Genetic Basis of Yield Traits Between Main and Ratoon Rice in an Eight-Way MAGIC Population" Plants 14, no. 22: 3527. https://doi.org/10.3390/plants14223527
APA StyleHan, Z., Sherif, A., Ayaad, M., Xing, Y., & Lu, Y. (2025). Comparison of the Genetic Basis of Yield Traits Between Main and Ratoon Rice in an Eight-Way MAGIC Population. Plants, 14(22), 3527. https://doi.org/10.3390/plants14223527





