Genome-Wide Development and Characterization of 169 gSSR Markers in the Invasive Plant Xanthium strumarium L.
Abstract
1. Introduction
2. Results
2.1. Analysis of SSR Repetition Frequency and Length
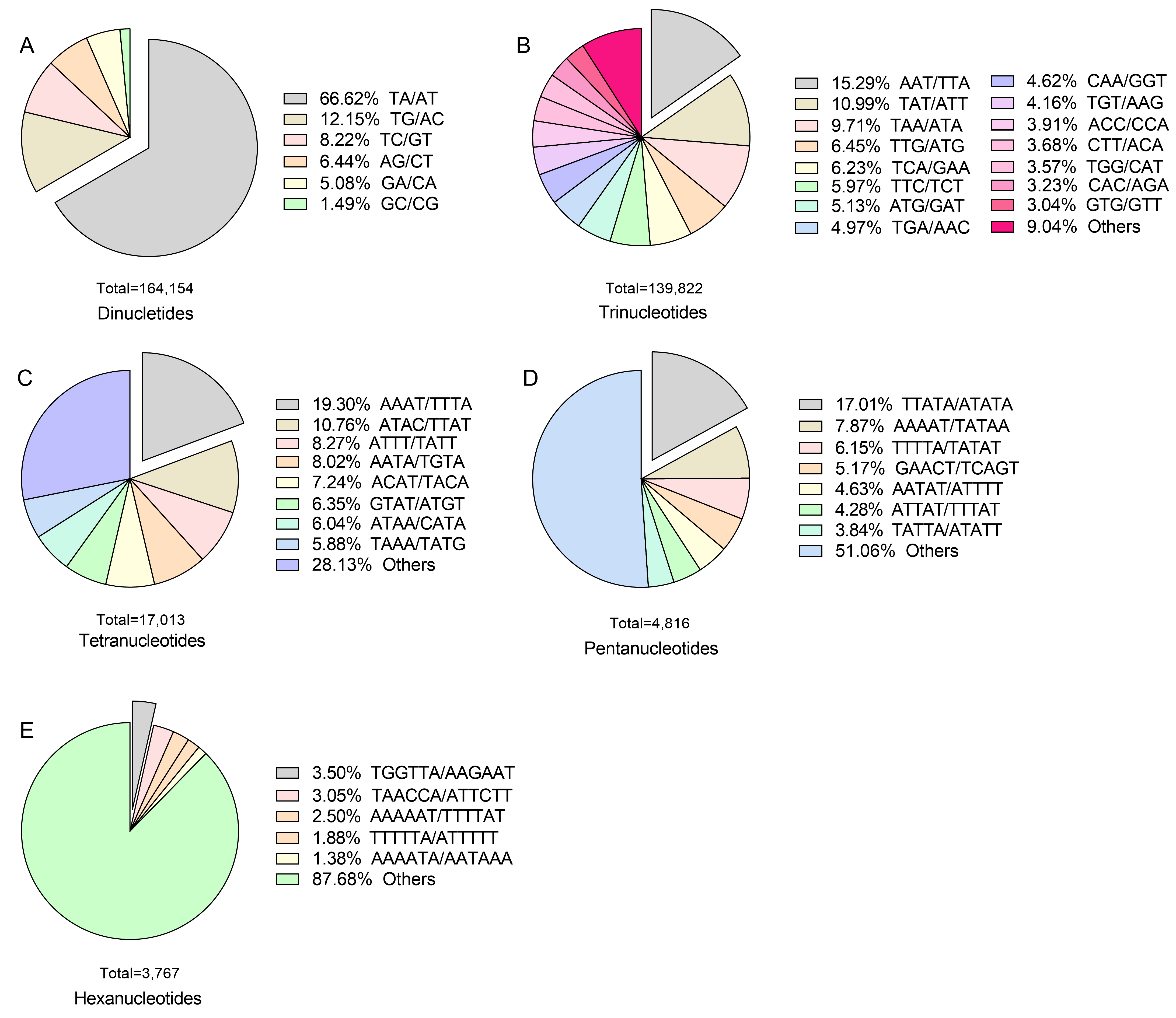
2.2. Characteristics of SSRs Predicted in the Genome of X. strumarium
2.3. Identification and Characterization of 169 gSSR Markers with Polymorphism Across 18 Chromosomes
2.4. gSSR Marker Assay and Their Informativeness
3. Discussion
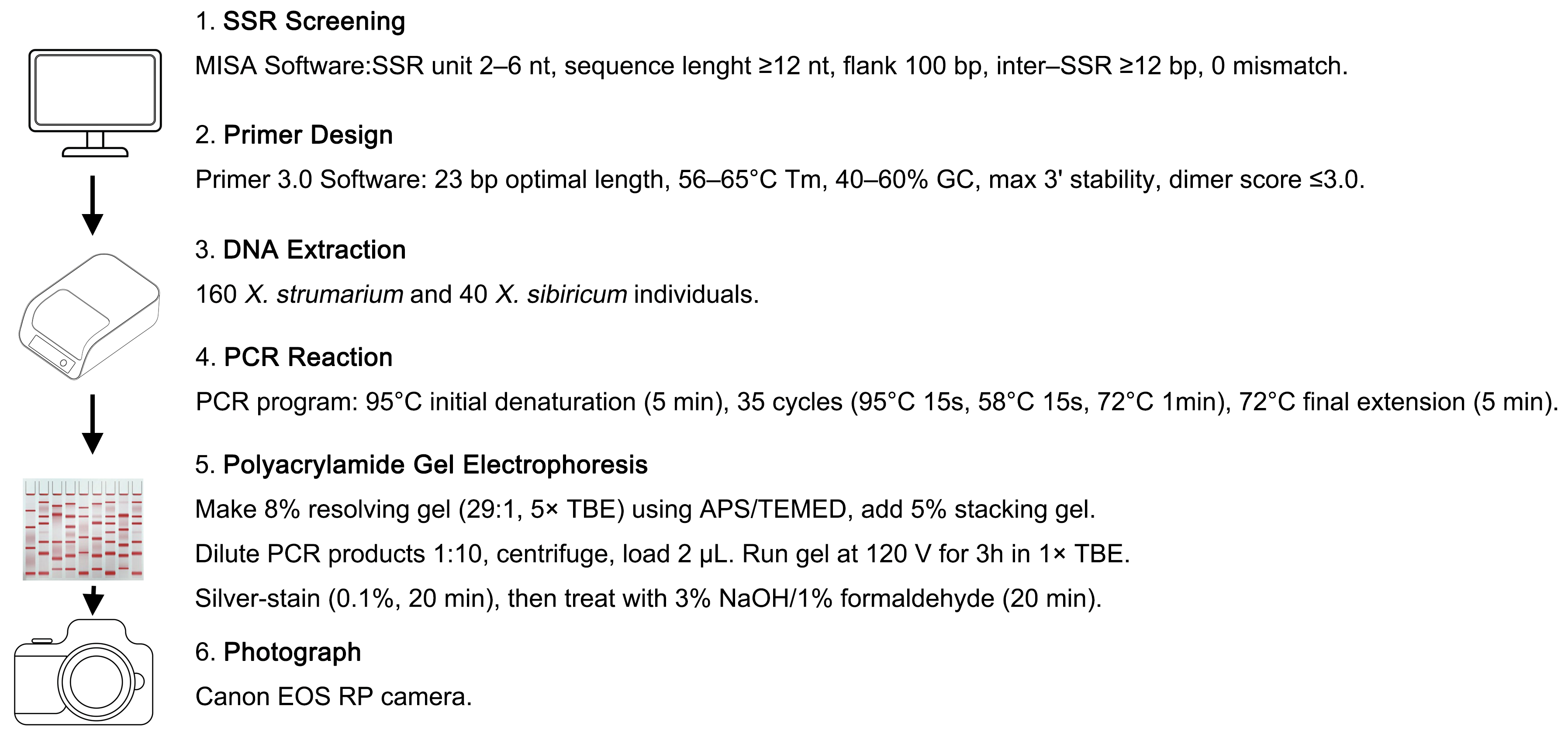
4. Materials and Methods
4.1. Plant Materials and gDNA Extraction
4.2. SSR Screening
4.3. Primer Design and PCR Reaction
4.4. Polyacrylamide Gel Electrophoresis (PAGE) Protocol
4.5. Analysis of gSSR Markers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, J.S.; Ma, M. Genetic diversity and genetic differentiation of invasive weed Xanthium italicum in China. C. R. Biol. 2020, 343, 63–72. [Google Scholar] [CrossRef]
- Sun, S.M.; Chen, J.X.; Feng, W.W.; Zhang, C.; Huang, K.; Guan, M.; Sun, J.K.; Liu, M.C.; Feng, Y.L. Plant Strategies for Nitrogen Acquisition and Their Effects on Exotic Plant Invasions. Biodivers. Sci. 2021, 29, 72–80. [Google Scholar] [CrossRef]
- Luo, J.J.; Gao, Y.M.; Feng, W.W.; Liu, M.C.; Qu, B.; Zhang, C.; Feng, Y.L. Stronger Ability to Absorb Nitrate and Associated Transporters in the Invasive Plant Xanthium strumarium Compared with Its Native Congener. Environ. Exp. Bot. 2022, 198, 104851. [Google Scholar] [CrossRef]
- Zhang, X.; Bai, S.; Min, H.; Cui, Y.; Sun, Y.; Feng, Y. Evolutionary dynamics of nitrate uptake, assimilation, and signalling in plants: Adapting to a changing environment. Physiol. Plant. 2025, 177, e70069. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Liu, H.; Zhang, Q.; Gao, N.; Min, H.; Sun, Y.; Zhang, X.; Feng, Y. Genome-wide identification of cytochrome b5 gene family reveals their potential roles in nitrate response in Xanthium strumarium. Plant. Physiol. Biochem. 2025, 229, 110544. [Google Scholar] [CrossRef]
- Han, M.; Zhang, H.; Liu, M.; Tang, J.; Guo, X.; Ren, W.; Zhao, Y.; Yang, Q.; Guo, B.; Han, Q.; et al. Increased dependence on nitrogen-fixation of a native legume in competition with an invasive plant. Plant. Divers. 2024, 46, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Löve, D.; Dansereau, P. Biosystematic studies on Xanthium: Taxonomic appraisal and ecological status. Can. J. Bot. 1959, 37, 173–208. [Google Scholar] [CrossRef]
- Raman, G.; Park, K.T.; Kim, J.H.; Park, S.J. Correction to: Characteristics of the completed chloroplast genome sequence of Xanthium Spinosum: Comparative analyses, identification of mutational hotspots and phylogenetic implications. BMC Genom. 2020, 22, 97. [Google Scholar] [CrossRef]
- Srivastava, S.; Avvaru, A.K.; Sowpati, D.T.; Mishra, R.K. Patterns of microsatellite distribution across eukaryotic genomes. BMC Genom. 2019, 20, 153. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, J.; Chen, S.; Qi, X.; Peng, H.; Li, P.; Song, A.; Guan, Z.; Fang, W.; Liao, Y. Next-Generation Sequencing of the Chrysanthemum Nankingense (Asteraceae) Transcriptome Permits Large-Scale Unigene Assembly and SSR Marker Discovery. PLoS ONE 2013, 8, e62293. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, S.; Chen, S.; Wei, X.; Shang, H.; Zhang, Q.; Zhang, J. Genome-wide development of SSR molecular markers for modern sugarcane cultivars. Front. Plant Sci. 2025, 16, 1573967. [Google Scholar] [CrossRef]
- Hamm, T.P.; Boggess, S.L.; Kandel, J.S.; Staton, M.E.; Huff, M.L.; Hadziabdic, D.; Shoemaker, D.; Adamczyk, J.J., Jr.; Nowicki, M.; Trigiano, R.N. Development and Characterization of 20 Genomic SSR Markers for Ornamental Cultivars of Weigela. Plants 2022, 11, 1444. [Google Scholar] [CrossRef] [PubMed]
- Hemasai, B.; Kumbha, D.K.; Modem, V.N.; Gannavarapu, S.K.; Bommaka, R.R.; Mallapuram, S.; Chintala, S.; Sreevalli, M.D.; Ramireddy, E.; Vemireddy, L.R. Development of miRNA-SSR and target-SSR markers from yield-associate genes and their applicability in the assessment of genetic diversity and association mapping in rice (Oryza sativa L.). Mol. Breed. 2024, 44, 30. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhao, W.; Li, Z.; Kang, D.; Ai, P.; Ding, H.; Wang, Z. Development and validation of SSR markers related to flower color based on full-length transcriptome sequencing in Chrysanthemum. Sci. Rep. 2022, 12, 22310. [Google Scholar] [CrossRef]
- Park, H.; Sa, K.J.; Hyun, D.Y.; Lee, S.; Lee, J.K. Identifying SSR Markers Related to Seed Fatty Acid Content in Perilla Crop (Perilla frutescens L.). Plants 2021, 10, 1404. [Google Scholar] [CrossRef]
- Chen, M.; Nie, G.; Li, X.; Yang, L.; Cai, Y.; Zhang, Y. Development of EST-SSR markers based on transcriptome sequencing for germplasm evaluation of 65 Lilies (Lilium). Mol. Biol. Rep. 2023, 50, 3259–3269. [Google Scholar] [CrossRef]
- Liu, B.; Wu, H.F.; Cao, Y.Z.; Yang, X.M.; Sui, S.Z. Establishment of Novel Simple Sequence Repeat (SSR) Markers from Chimonanthus praecox Transcriptome Data and Their Application in the Identification of Varieties. Plants 2024, 13, 2131. [Google Scholar] [CrossRef] [PubMed]
- Squirrell, J.; Hollingsworth, P.M.; Woodhead, M.; Russell, J.; Lowe, A.J.; Gibby, M.; Powell, W. How much effort is required to isolate nuclear microsatellites from plants? Mol. Ecol. 2003, 12, 1339–1348. [Google Scholar] [CrossRef]
- Sun, N.; Chen, J.; Wang, Y.; Hussain, I.; Lei, N.; Ma, X.; Li, W.; Liu, K.; Yu, H.; Zhao, K.; et al. Development and utility of SSR markers based on Brassica sp. whole-genome in triangle of U. Front. Plant Sci. 2024, 14, 1259736. [Google Scholar] [CrossRef]
- Jiang, H.; Waseem, M.; Wang, Y.; Basharat, S.; Zhang, X.; Li, Y.; Liu, P. Development of simple sequence repeat markers for sugarcane from data mining of expressed sequence tags. Front. Plant Sci. 2023, 14, 1199210. [Google Scholar] [CrossRef]
- Zhang, Z.; Xie, W.; Zhao, Y.; Zhang, J.; Wang, N.; Ntakirutimana, F.; Yan, J.; Wang, Y. EST-SSR marker development based on RNA-sequencing of E. sibiricus and its application for phylogenetic relationships analysis of seventeen Elymus species. BMC Plant Biol. 2019, 19, 235. [Google Scholar] [CrossRef]
- Li, J.; Zhang, C.; Chen, S.; Jiang, K.; Guan, H.; Liu, W. Characterization and Application of EST-SSR Markers Developed from Transcriptome Sequences in Elymus breviaristatus (Poaceae: Triticeae). Genes 2023, 14, 302. [Google Scholar] [CrossRef]
- Blair, M.W.; Buendía, H.F.; Giraldo, M.C.; Métais, I.; Peltier, D. Characterization of AT-Rich Microsatellites in Common Bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2008, 118, 91–103. [Google Scholar] [CrossRef]
- Lin, Y.; Wang, H.; Wu, Y.; Gao, S.; Schaefer, H.F., 3rd. Proton-transfer in hydrogenated guanine-cytosine trimer neutral species, cations, and anions embedded in B-form DNA. Phys. Chem. Chem. Phys. 2014, 16, 6717–6725. [Google Scholar] [CrossRef]
- Subramanian, H.; Gatenby, R.A. Evolutionary advantage of anti-parallel strand orientation of duplex DNA. Sci. Rep. 2020, 10, 9883. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Huang, S.; Fu, D.; Yu, J.; Wang, X.; Hua, W.; Liu, S.; Liu, G.; Wang, H. Evolutionary Dynamics of Microsatellite Distribution in Plants: Insight from the Comparison of Sequenced Brassica, Arabidopsis and Other Angiosperm Species. PLoS ONE 2013, 8, e59988. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Pan, C.; Diao, Y.; You, Y.; Yang, C.; Hu, Z. Development of microsatellite markers by transcriptome sequencing in two species of Amorphophallus (Araceae). BMC Genom. 2013, 19, 490. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, J.; Han, R.; Zhang, F.; Mao, A.; Luo, J.; Dong, B.; Liu, H.; Tang, H.; Zhang, J.; et al. Target SSR-Seq: A Novel SSR Genotyping Technology Associate With Perfect SSRs in Genetic Analysis of Cucumber Varieties. Front. Plant Sci. 2019, 10, 531. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, S.; Gao, Q.; Liu, F.; Wang, J.; Wang, X. Genetic diversity and population structure analysis in a large collection of Vicia amoena in China with newly developed SSR markers. BMC Plant. Biol. 2021, 21, 544. [Google Scholar] [CrossRef]
- Xiao, N.; Wang, H.; Yao, W.; Zhang, M.; Ming, R.; Zhang, J. Development and Evaluation of SSR Markers Based on Large Scale Full-Length Transcriptome Sequencing in Sugarcane. Trop. Plant. Biol. 2020, 13, 343–352. [Google Scholar] [CrossRef]
- Meyer, L.; Causse, R.; Pernin, F.; Scalone, R.; Bailly, G.; Chauvel, B.; Délye, C.; Le Corre, V. New gSSR and EST-SSR markers reveal high genetic diversity in the invasive plant Ambrosia artemisiifolia L. and can be transferred to other invasive Ambrosia species. PLoS ONE 2017, 12, e0176197. [Google Scholar] [CrossRef]
- Samarina, L.S.; Malyarovskaya, V.I.; Reim, S.; Yakushina, L.G.; Koninskaya, N.G.; Klemeshova, K.V.; Shkhalakhova, R.M.; Matskiv, A.O.; Shurkina, E.S.; Gabueva, T.Y.; et al. Transferability of ISSR, SCoT and SSR Markers for Chrysanthemum × Morifolium Ramat and Genetic Relationships Among Commercial Russian Cultivars. Plants 2021, 10, 1302. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Deng, Z. EST-SSR markers for gerbera (Gerbera hybrida). Mol. Breed. 2010, 26, 125–132. [Google Scholar] [CrossRef]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Toonen, R.J. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecol. Lett. 2006, 9, 615–629. [Google Scholar] [CrossRef]
- Doyle, J.J.; Jane, L.D. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Thiel, T.; Michalek, W.; Varshney, R.K.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Sun, H.; Li, L.; Lou, Y.; Li, R.; Qi, L.; Gao, Z. Transcriptome-based investigation of cirrus development and identifying microsatellite markers in rattan (Daemonorops jenkinsiana). Sci. Rep. 2017, 7, 46107. [Google Scholar] [CrossRef]
- Bonthala, B.; Abdin, M.Z.; Arya, L.; Pandey, C.D.; Sharma, V.; Yadav, P.; Verma, M. Genome-wide SSR markers in bottle gourd: Development, characterization, utilization in assessment of genetic diversity of National Genebank of India and synteny with other related cucurbits. J. Appl. Genet. 2022, 63, 237–263. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [CrossRef] [PubMed]
- Haider, N.; Nabulsi, I.; MirAli, N. Comparison of the efficiency of A–PAGE and SDS–PAGE, ISSRs and RAPDs in resolving genetic relationships among Triticum and Aegilops species. Genet. Resour. Crop Evol. 2010, 57, 1023–1039. [Google Scholar] [CrossRef]
- Sen, A.; Alikamanoglu, S. Analysis of drought-tolerant sugar beet (Beta vulgaris L.) mutants induced with gamma radiation using SDS-PAGE and ISSR markers. Mutat. Res. 2012, 738–739, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
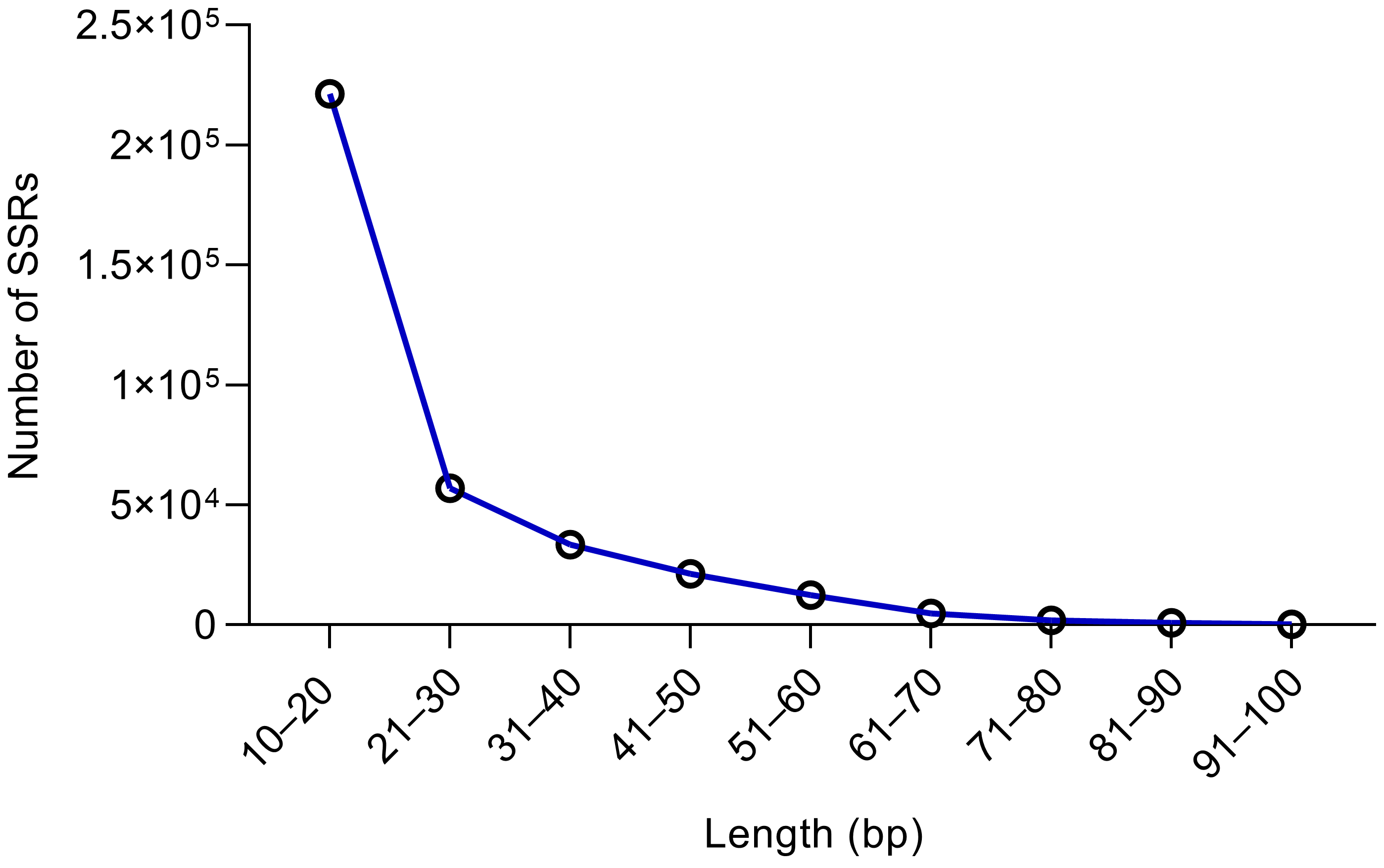

| Length (bp) | Number | Percentage |
|---|---|---|
| 10–20 | 221,425 | 49.11% |
| 21–30 | 57,037 | 12.65% |
| 31–40 | 33,418 | 7.41% |
| 41–50 | 21,287 | 4.72% |
| 51–60 | 12,557 | 2.79% |
| 61–70 | 4858 | 1.08% |
| 71–80 | 2034 | 0.45% |
| 81–90 | 833 | 0.18% |
| 91–100 | 288 | 0.06% |
| 100+ | 97,110 | 21.54% |
| (a) | ||||||||||||||||||||||
| No. of Repeats | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ||||||||||||
| No. | ||||||||||||||||||||||
| SSR Motif Types | ||||||||||||||||||||||
| Dinucletides | 0 | 0 | 36,580 | 21,928 | 16,152 | 11,598 | 8174 | 6122 | 5020 | 4492 | ||||||||||||
| Trinucleotides | 88,734 | 24,411 | 9725 | 4213 | 2087 | 1334 | 970 | 831 | 770 | 703 | ||||||||||||
| Tetranucleotides | 12,482 | 3011 | 823 | 308 | 157 | 82 | 47 | 35 | 17 | 16 | ||||||||||||
| Pentanucleotides | 3463 | 713 | 205 | 98 | 71 | 58 | 40 | 34 | 33 | 23 | ||||||||||||
| Hexanucleotides | 2609 | 620 | 226 | 107 | 60 | 35 | 20 | 19 | 12 | 11 | ||||||||||||
| Total | 107,288 | 28,755 | 47,559 | 26,654 | 18,527 | 13,107 | 9251 | 7041 | 5852 | 5245 | ||||||||||||
| Percentage | 32.55% | 8.72% | 14.43% | 8.09% | 5.62% | 3.98% | 2.81% | 2.14% | 1.78% | 1.59% | ||||||||||||
| (b) | ||||||||||||||||||||||
| No. of Repeats | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 20+ | Total | Percentage | Average Physical Distance | |||||||||||
| No. | ||||||||||||||||||||||
| SSR Motif Types | ||||||||||||||||||||||
| Dinucletides | 4250 | 4159 | 4051 | 3974 | 3809 | 3823 | 3749 | 26,273 | 16,4154 | 49.81% | 10.17 kb | |||||||||||
| Trinucleotides | 641 | 636 | 594 | 531 | 515 | 461 | 454 | 2212 | 139,822 | 42.43% | 12.13 kb | |||||||||||
| Tetranucleotides | 11 | 5 | 6 | 4 | 3 | 1 | 0 | 5 | 17,013 | 5.16% | 80.38 kb | |||||||||||
| Pentanucleotides | 21 | 10 | 8 | 13 | 9 | 4 | 6 | 7 | 4816 | 1.46% | 332.88 kb | |||||||||||
| Hexanucleotides | 9 | 12 | 3 | 2 | 1 | 0 | 2 | 19 | 3767 | 1.14% | 442.9 kb | |||||||||||
| Total | 4932 | 4822 | 4662 | 4524 | 4337 | 4289 | 4211 | 28,516 | 329,572 | 100% | 6.91 kb | |||||||||||
| Percentage | 1.50% | 1.46% | 1.41% | 1.37% | 1.32% | 1.30% | 1.28% | 8.65% | 100 | / | / | |||||||||||
| (a) | ||||||||||||||||
| Locus | INVZ-191 | INVZ-456 | INVZ-705 | INVZ-864 | INVZ-1104 | INVZ-1274 | INVZ-1501 | INVZ-1652 | INVZ-1925 | INVZ-2051 | ||||||
| Types | ||||||||||||||||
| No. of Different Alleles (Na) | 1.200 ± 0.302 | 1.700 ± 0.483 | 2.400 ± 0.369 | 2.900 ± 0.920 | 1.500 ± 0.506 | 3.200 ± 1.207 | 2.400 ± 0.769 | 2.400 ± 0.500 | 2.100 ± 1.282 | 1.500 ± 0.608 | ||||||
| No. of Effective Alleles (Ne) | 1.066 ± 0.120 | 1.282 ± 0.303 | 2.185 ± 0.403 | 2.395 ± 0.460 | 1.350 ± 0.351 | 2.261 ± 0.341 | 1.933 ± 0.326 | 2.101 ± 0.272 | 1.992 ± 1.164 | 1.465 ± 0.596 | ||||||
| Shannon’s Information Index (I) | 0.076 ± 0.056 | 0.250 ± 0.225 | 0.767 ± 0.196 | 0.890 ± 0.212 | 0.265 ± 0.252 | 0.885 ± 0.193 | 0.669 ± 0.210 | 0.763 ± 0.136 | 0.702 ± 0.414 | 0.474 ± 0.235 | ||||||
| Observed Heterozygosity (Ho) | 0.000 ± 0.001 | 0.242 ± 0.128 | 0.800 ± 0.221 | 1.000 ± 0.001 | 0.326 ± 0.146 | 0.950 ± 0.113 | 0.816 ± 0.261 | 0.924 ± 0.139 | 0.700 ± 0.346 | 0.590 ± 0.364 | ||||||
| Expected Heterozygosity (He) | 0.046 ± 0.035 | 0.156 ± 0.154 | 0.507 ± 0.119 | 0.561 ± 0.065 | 0.181 ± 0.174 | 0.544 ± 0.054 | 0.445 ± 0.132 | 0.510 ± 0.063 | 0.424 ± 0.223 | 0.340 ± 0.169 | ||||||
| Polymorphism Information Content (PIC) | 0.215 ± 0.117 | 0.343 ± 0.115 | 0.801 ± 0.066 | 0.778 ± 0.078 | 0.839 ± 0.138 | 0.853 ± 0.045 | 0.612 ± 0.148 | 0.691 ± 0.065 | 0.852 ± 0.195 | 0.774 ± 0.162 | ||||||
| (b) | ||||||||||||||||
| Locus | INVZ-2330 | INVZ-2478 | INVZ-2645 | INVZ-2906 | INVZ-3183 | INVZ-3458 | INVZ-3563 | INVZ-3764 | Average | |||||||
| Types | ||||||||||||||||
| No. of Different Alleles (Na) | 1.200 ± 0.812 | 1.8 ± 0.452 | 2.300 ± 0.483 | 2.000 ± 0.001 | 1.200 ± 0.739 | 2.600 ± 1.129 | 1.700 ± 0.830 | 2.000 ± 0.001 | 2.006 | |||||||
| No. of Effective Alleles (Ne) | 1.077 ± 0.744 | 1.8 ± 0.452 | 2.090 ± 0.202 | 2.000 ± 0.001 | 1.200 ± 0.739 | 1.851 ± 0.557 | 1.104 ± 0.328 | 2.000 ± 0.001 | 1.730 | |||||||
| Shannon’s Information Index (I) | 0.332 ± 0.288 | 0.624 ± 0.157 | 0.756 ± 0.116 | 0.693 ± 0.001 | 0.416 ± 0.256 | 0.712 ± 0.220 | 0.239 ± 0.202 | 0.693 ± 0.001 | 0.567 | |||||||
| Observed Heterozygosity (Ho) | 0.410 ± 0.364 | 0.900 ± 0.226 | 0.945 ± 0.112 | 1.000 ± 0.001 | 0.600 ± 0.370 | 0.714 ± 0.385 | 0.170 ± 0.151 | 1.000 ± 0.001 | 0.672 | |||||||
| Expected Heterozygosity (He) | 0.222 ± 0.193 | 0.450 ± 0.113 | 0.515 ± 0.042 | 0.500 ± 0.001 | 0.300 ± 0.185 | 0.448 ± 0.127 | 0.138 ± 0.120 | 0.500 ± 0.001 | 0.377 | |||||||
| Polymorphism Information Content (PIC) | 0.678 ± 0.170 | 0.491 ± 0.158 | 0.623 ± 0.063 | 0.644 ± 0.043 | 0.375 ± 0.203 | 0.812 ± 0.153 | 0.161 ± 0.090 | 0.614 ± 0.053 | 0.620 | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, J.; Bai, Q.; Mao, Y.; Min, H.; Zhang, C.; Sun, Y.; Zhang, X.; Feng, Y. Genome-Wide Development and Characterization of 169 gSSR Markers in the Invasive Plant Xanthium strumarium L. Plants 2025, 14, 3522. https://doi.org/10.3390/plants14223522
Yin J, Bai Q, Mao Y, Min H, Zhang C, Sun Y, Zhang X, Feng Y. Genome-Wide Development and Characterization of 169 gSSR Markers in the Invasive Plant Xanthium strumarium L. Plants. 2025; 14(22):3522. https://doi.org/10.3390/plants14223522
Chicago/Turabian StyleYin, Junshuang, Qingyao Bai, Yiting Mao, Hui Min, Chunsha Zhang, Yibo Sun, Xiaojia Zhang, and Yulong Feng. 2025. "Genome-Wide Development and Characterization of 169 gSSR Markers in the Invasive Plant Xanthium strumarium L." Plants 14, no. 22: 3522. https://doi.org/10.3390/plants14223522
APA StyleYin, J., Bai, Q., Mao, Y., Min, H., Zhang, C., Sun, Y., Zhang, X., & Feng, Y. (2025). Genome-Wide Development and Characterization of 169 gSSR Markers in the Invasive Plant Xanthium strumarium L. Plants, 14(22), 3522. https://doi.org/10.3390/plants14223522






