Genetic and Molecular Basis for Heat Tolerance in Rice: Strategies for Resilience Under Climate Change
Abstract
1. Introduction
2. Advances in Understanding the Molecular Mechanisms Behind Rice Heat Tolerance
2.1. Heat Sensing and Signal Transduction Mechanisms
2.2. HSF-Mediated Multi-Level Regulatory Mechanisms of Heat Tolerance in Rice
2.3. Regulation of Heat Response by ALBA Proteins via mRNA Stabilization Mechanism
2.4. Epigenetic Regulation, Chromatin Remodeling, and Transcriptional Coregulation Comprise the Heat Response Expression Framework
2.5. Regulation of Heat Tolerance via Membrane Homeostasis and Antioxidant Networks
3. Overview of Key Genes Involved in Rice Heat Tolerance
3.1. TT1- and TT2-Mediated Regulation of Heat Tolerance in Rice
3.2. The TT3 Genetic Module Enhances Heat Tolerance in Rice by Regulating PSII Stability
3.3. QT12, a Regulator of Heat Response Affecting Grain Quality and Yield
3.4. HTS1 and Regulation of Membrane Homeostasis
3.5. Antioxidant-Related Regulators in the OsHTAS Network
3.6. SLG1 and tRNA Thiolation
3.7. Other Key Genes
4. Mechanisms of Crosstalk Between Rice Heat Tolerance and Grain Development
4.1. Long-Distance ABA Transport and Grain Filling
4.2. Regulation of Endosperm Development by MADS-Box Transcription Factors
4.3. Impact of Heat Stress During the Grain-Filling Window
5. Emerging Technologies for Rice Heat Tolerance Research
5.1. Multi-Omics Approaches in Heat Tolerance Research
5.2. Applications of Gene Editing
6. Molecular Breeding Strategies and Integration of Technology
6.1. Marker-Assisted Selection (MAS)
6.2. Gene Editing in Heat Tolerance Breeding
6.3. Gene Pyramiding and Breeding by Design
6.4. Coordinated Improvement of Multiple Traits
7. Discussion
7.1. Fragmented Mechanistic Models
7.2. Disconnect Between Laboratory Simulations and Field Conditions
7.3. Functional Validation and Breeding Translation Lag Behind
7.4. Trait Trade-Offs Limit Breeding Efficiency
7.5. Limitations of This Review
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Intergovernmental Panel on Climate Change. Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2021. [Google Scholar]
- Ju, Y.; Choi, J.; Yun, S.; Mittra, P.K.; Woo, S.; Sakagami, J.I. Stage-dependent heat priming mitigates reproductive heat stress via proteomic regulation in Oryza sativa L. Plant Sci. 2025, 359, 112639. [Google Scholar] [CrossRef]
- Prerostova, S.; Jarošová, J.; Dobrev, P.; Gaudinova, A.; Knirsch, V.; Kobzova, E.; Benczúr, K.; Szalai, G.; Novak, O.; Vankova, R. Cytokinin elevation caused by high light intensity contributes substantially to the increase of thermotolerance of rice plants. Plant Stress 2025, 16, 100904. [Google Scholar] [CrossRef]
- Kompas, T.; Che, T.N.; Grafton, R.Q. Global impacts of heat and water stress on food production and severe food insecurity. Sci. Rep. 2024, 14, 14398. [Google Scholar] [CrossRef]
- Lu, F.; Feng, B.; Chen, L.; Qiu, J.; Wei, X. How Does Rice Cope with High-Temperature Stress During Its Growth and Development, Especially at the Grain-Filling Stage? Agronomy 2025, 15, 623. [Google Scholar] [CrossRef]
- Park, J.R.; Kim, E.G.; Jang, Y.H.; Kim, K.M. Screening and identification of genes affecting grain quality and spikelet fertility during high-temperature treatment in grain filling stage of rice. BMC Plant Biol. 2021, 21, 263. [Google Scholar] [CrossRef]
- Riaz, A.; Thomas, J.; Ali, H.H.; Zaheer, M.S.; Ahmad, N.; Pereira, A. High night temperature stress on rice (Oryza sativa)—insights from phenomics to physiology. A review. Funct. Plant Biol. 2024, 51, fp24057. [Google Scholar] [CrossRef]
- Li, W.; Yang, K.; Hu, C.; Abbas, W.; Zhang, J.; Xu, P.; Cheng, B.; Zhang, J.; Yin, W.; Shalmani, A.; et al. A natural gene on-off system confers field thermotolerance for grain quality and yield in rice. Cell 2025, 188, 3661–3678.e21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, J.-F.; Kan, Y.; Shan, J.-X.; Ye, W.-W.; Dong, N.-Q.; Guo, T.; Xiang, Y.-H.; Yang, Y.-B.; Li, Y.-C.; et al. A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance. Science 2022, 376, 1293–1300. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhong, X.; Liao, J.; Ji, P.; Yang, J.; Cao, Z.; Duan, X.; Xiong, J.; Wang, Y.; Xu, C.; et al. Exogenous abscisic acid improves grain filling capacity under heat stress by enhancing antioxidative defense capability in rice. BMC Plant Biol. 2023, 23, 619. [Google Scholar] [CrossRef]
- Liu, H.; Zeng, B.; Zhao, J.; Yan, S.; Wan, J.; Cao, Z. Genetic Research Progress: Heat Tolerance in Rice. Int. J. Mol. Sci. 2023, 24, 7140. [Google Scholar] [CrossRef]
- Xing, Y.H.; Lu, H.; Zhu, X.; Deng, Y.; Xie, Y.; Luo, Q.; Yu, J. How Rice Responds to Temperature Changes and Defeats Heat Stress. Rice 2024, 17, 73. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wei, Y.; Xia, S.; Xie, W.; Ren, D.; Rao, Y. Improvements in Tolerance to Heat Stress in Rice via Molecular Mechanisms and Rice Varieties. Agriculture 2025, 15, 318. [Google Scholar] [CrossRef]
- Li, X.M.; Chao, D.Y.; Wu, Y.; Huang, X.; Chen, K.; Cui, L.G.; Su, L.; Ye, W.W.; Chen, H.; Chen, H.C.; et al. Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 2015, 47, 827–833. [Google Scholar] [CrossRef]
- Kan, Y.; Mu, X.R.; Zhang, H.; Gao, J.; Shan, J.X.; Ye, W.W.; Lin, H.X. TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis. Nat. Plants 2022, 8, 53–67. [Google Scholar] [CrossRef]
- Ps, S.; Sv, A.M.; Prakash, C.; Mk, R.; Tiwari, R.; Mohapatra, T.; Singh, N.K. High Resolution Mapping of QTLs for Heat Tolerance in Rice Using a 5K SNP Array. Rice 2017, 10, 28. [Google Scholar] [CrossRef]
- Yiwei, F.; Jiayelu, W.; Mingming, W.; Shenghai, Y.; Rongrong, Z.; Jing, Y.; Guofu, Z.; Faming, Y.; Yanting, L.; Xiaoming, Z. Progress on Molecular Mechanism of Heat Tolerance in Rice. Rice Sci. 2024, 31, 673–687. [Google Scholar] [CrossRef]
- Chen, F.; Dong, G.; Wang, F.; Shi, Y.; Zhu, J.; Zhang, Y.; Ruan, B.; Wu, Y.; Feng, X.; Zhao, C.; et al. A β-ketoacyl carrier protein reductase confers heat tolerance via the regulation of fatty acid biosynthesis and stress signaling in rice. New Phytol. 2021, 232, 655–672. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Wei, C.; Liu, X.; Wang, M.; Yu, F.; Xie, Q.; Tu, J. The RING Finger Ubiquitin E3 Ligase OsHTAS Enhances Heat Tolerance by Promoting H2O2-Induced Stomatal Closure in Rice. Plant Physiol. 2016, 170, 429–443. [Google Scholar] [CrossRef]
- Wu, N.; Yao, Y.; Xiang, D.; Du, H.; Geng, Z.; Yang, W.; Li, X.; Xie, T.; Dong, F.; Xiong, L. A MITE variation-associated heat-inducible isoform of a heat-shock factor confers heat tolerance through regulation of JASMONATE ZIM-DOMAIN genes in rice. New Phytol. 2022, 234, 1315–1331. [Google Scholar] [CrossRef] [PubMed]
- Mei, W.; Chen, W.; Wang, Y.; Liu, Z.; Dong, Y.; Zhang, G.; Deng, H.; Liu, X.; Lu, X.; Wang, F.; et al. Exogenous Kinetin Modulates ROS Homeostasis to Affect Heat Tolerance in Rice Seedlings. Int. J. Mol. Sci. 2023, 24, 6252. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.H.; Lyu, Y.S.; Yang, W.; Yang, Z.T.; Lu, S.J.; Liu, J.X. A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol. J. 2020, 18, 1317–1329. [Google Scholar] [CrossRef]
- Gandass, N.; Kajal; Salvi, P. Intrinsically disordered protein, DNA binding with one finger transcription factor (OsDOF27) implicates thermotolerance in yeast and rice. Front. Plant Sci. 2022, 13, 956299. [Google Scholar] [CrossRef]
- Chen, S.; Cao, H.; Huang, B.; Zheng, X.; Liang, K.; Wang, G.L.; Sun, X. The WRKY10-VQ8 module safely and effectively regulates rice thermotolerance. Plant Cell Environ. 2022, 45, 2126–2144. [Google Scholar] [CrossRef]
- El-Kereamy, A.; Bi, Y.M.; Ranathunge, K.; Beatty, P.H.; Good, A.G.; Rothstein, S.J. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 2012, 7, e52030. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Ma, Z.; Kang, Y.; Zhang, B.; Gao, X.; Yu, F.; Yang, P.; Ke, Y. ENHANCED DISEASE SUSCEPTIBILITY 1 promotes hydrogen peroxide scavenging to enhance rice thermotolerance. Plant Physiol. 2023, 192, 3106–3119. [Google Scholar] [CrossRef]
- He, Y.; Zhang, X.; Shi, Y.; Xu, X.; Li, L.; Wu, J.L. PREMATURE SENESCENCE LEAF 50 Promotes Heat Stress Tolerance in Rice (Oryza sativa L.). Rice 2021, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Zhang, X.; Liu, J.; Hou, L.; Liu, H.; Zhao, X. OsProDH Negatively Regulates Thermotolerance in Rice by Modulating Proline Metabolism and Reactive Oxygen Species Scavenging. Rice 2020, 13, 61. [Google Scholar] [CrossRef]
- Liu, J.; Sun, X.; Xu, F.; Zhang, Y.; Zhang, Q.; Miao, R.; Zhang, J.; Liang, J.; Xu, W. Suppression of OsMDHAR4 enhances heat tolerance by mediating H2O2-induced stomatal closure in rice plants. Rice 2018, 11, 38. [Google Scholar] [CrossRef]
- Qin, P.; Zhang, G.; Hu, B.; Wu, J.; Chen, W.; Ren, Z.; Liu, Y.; Xie, J.; Yuan, H.; Tu, B.; et al. Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism. Sci. Adv. 2021, 7, eabc8873. [Google Scholar] [CrossRef] [PubMed]
- She, K.C.; Kusano, H.; Koizumi, K.; Yamakawa, H.; Hakata, M.; Imamura, T.; Fukuda, M.; Naito, N.; Tsurumaki, Y.; Yaeshima, M.; et al. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 2010, 22, 3280–3294. [Google Scholar] [CrossRef]
- Takehara, K.; Murata, K.; Yamaguchi, T.; Yamaguchi, K.; Chaya, G.; Kido, S.; Iwasaki, Y.; Ogiwara, H.; Ebitani, T.; Miura, K. Thermo-responsive allele of sucrose synthase 3 (Sus3) provides high-temperature tolerance during the ripening stage in rice (Oryza sativa L.). Breed. Sci. 2018, 68, 336–342. [Google Scholar] [CrossRef]
- Kusano, H.; Arisu, Y.; Nakajima, J.; Yaeshima, M.; She, K.-C.; Shimada, H. Implications of the gene for F1–ATPase β subunit (AtpB) for the grain quality of rice matured in a high-temperature environment. Plant Biotechnol. 2016, 33, 169–175. [Google Scholar] [CrossRef]
- Yan, Y.; Li, C.; Liu, Z.; Zhuang, J.J.; Kong, J.R.; Yang, Z.K.; Yu, J.; Shah Alam, M.; Ruan, C.C.; Zhang, H.M.; et al. A new demethylase gene, OsDML4, is involved in high temperature-increased grain chalkiness in rice. J. Exp. Bot. 2022, 73, 7273–7284. [Google Scholar] [CrossRef]
- Xu, H.; Li, X.; Zhang, H.; Wang, L.; Zhu, Z.; Gao, J.; Li, C.; Zhu, Y. High temperature inhibits the accumulation of storage materials by inducing alternative splicing of OsbZIP58 during filling stage in rice. Plant Cell Environ. 2020, 43, 1879–1896. [Google Scholar] [CrossRef]
- Tabassum, R.; Dosaka, T.; Ichida, H.; Morita, R.; Ding, Y.; Abe, T.; Katsube-Tanaka, T. FLOURY ENDOSPERM11-2 encodes plastid HSP70-2 involved with the temperature-dependent chalkiness of rice (Oryza sativa L.) grains. Plant J. 2020, 103, 604–616. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, L.; Ou, S.; Wang, R.; Wang, Y.; Chu, C.; Yao, S. Natural variations of SLG1 confer high-temperature tolerance in indica rice. Nat. Commun. 2020, 11, 5441. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 2008, 227, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Gupta, A.; Bansal, C.; Sorin, C.; Crespi, M.; Mathur, S. A conserved HSF:miR169:NF-YA loop involved in tomato and Arabidopsis heat stress tolerance. Plant J. 2022, 112, 7–26. [Google Scholar] [CrossRef]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Doring, P.; Vierling, E.; Scharf, K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef]
- Schramm, F.; Ganguli, A.; Kiehlmann, E.; Englich, G.; Walch, D.; von Koskull-Doring, P. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 2006, 60, 759–772. [Google Scholar] [CrossRef]
- Jin, Q.; Chachar, M.; Ali, A.; Chachar, Z.; Zhang, P.; Riaz, A.; Ahmed, N.; Chachar, S. Epigenetic Regulation for Heat Stress Adaptation in Plants: New Horizons for Crop Improvement under Climate Change. Agronomy 2024, 14, 2105. [Google Scholar] [CrossRef]
- Cortijo, S.; Charoensawan, V.; Brestovitsky, A.; Buning, R.; Ravarani, C.; Rhodes, D.; van Noort, J.; Jaeger, K.E.; Wigge, P.A. Transcriptional Regulation of the Ambient Temperature Response by H2A.Z Nucleosomes and HSF1 Transcription Factors in Arabidopsis. Mol. Plant 2017, 10, 1258–1273. [Google Scholar] [CrossRef]
- Huang, Y.; An, J.; Sircar, S.; Bergis, C.; Lopes, C.D.; He, X.; Da Costa, B.; Tan, F.Q.; Bazin, J.; Antunez-Sanchez, J.; et al. HSFA1a modulates plant heat stress responses and alters the 3D chromatin organization of enhancer-promoter interactions. Nat. Commun. 2023, 14, 469. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zuo, Z.; Yao, P.; Li, X.; Zhang, Q.; Chen, X. Bromodomain-containing proteins interact with a non-canonical RNA polymerase II kinase to maintain gene expression upon heat stress. Nat. Plants 2025, 11, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Islam, M.N.; Sarker, S.; Tuteja, N.; Seraj, Z.I. Overexpression of heterotrimeric G protein beta subunit gene (OsRGB1) confers both heat and salinity stress tolerance in rice. Plant Physiol. Biochem. 2019, 144, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, K.; Wang, G.; Peng, Z.; Wang, T.; Meng, Y.; Huang, J.; Huo, J.; Li, X.; Zhu, X.; et al. The OsEBF1-OsEIL5-OsPP91 module regulates rice heat tolerance via ubiquitination and transcriptional activation. Cell Rep. 2025, 44, 115271. [Google Scholar] [CrossRef]
- Ma, Y.; Zhao, S.; Ma, X.; Dong, G.; Liu, C.; Ding, Y.; Hou, B. A high temperature responsive UDP-glucosyltransferase gene OsUGT72F1 enhances heat tolerance in rice and Arabidopsis. Plant Cell Rep. 2025, 44, 48. [Google Scholar] [CrossRef]
- Yang, B.; Xie, Y.; Liu, Y. A novel NF-Ys-QT12-IRE1 module controlling grain quality and yield thermotolerance in rice. Mol. Plant 2025, 18, 1106–1108. [Google Scholar] [CrossRef]
- Lo, S.F.; Cheng, M.L.; Hsing, Y.C.; Chen, Y.S.; Lee, K.W.; Hong, Y.F.; Hsiao, Y.; Hsiao, A.S.; Chen, P.J.; Wong, L.I.; et al. Rice Big Grain 1 promotes cell division to enhance organ development, stress tolerance and grain yield. Plant Biotechnol. J. 2020, 18, 1969–1983. [Google Scholar] [CrossRef]
- Xiong, J.; Wang, H.; Zhong, Z.; Li, S.; Qin, P. Emerging strategies to improve heat stress tolerance in crops. aBIOTECH 2025, 6, 97–115. [Google Scholar] [CrossRef]
- Zhang, P.; Zhu, W.; He, Y.; Fan, J.; Shi, J.; Fu, R.; Hu, J.; Li, L.; Zhang, D.; Liang, W. THERMOSENSITIVE BARREN PANICLE (TAP) is required for rice panicle and spikelet development at high ambient temperature. New Phytol. 2023, 237, 855–869. [Google Scholar] [CrossRef]
- Chen, C.; Begcy, K.; Liu, K.; Folsom, J.J.; Wang, Z.; Zhang, C.; Walia, H. Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant Physiol. 2016, 171, 606–622. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yin, T.; Zhao, Y.; Wang, X.; Wang, K.; Shen, Y.; Ding, Y.; Tang, S. Effects of High Temperature on Rice Grain Development and Quality Formation Based on Proteomics Comparative Analysis Under Field Warming. Front. Plant Sci. 2021, 12, 746180. [Google Scholar] [CrossRef]
- Yan, H.; Wang, C.; Liu, K.; Tian, X. Detrimental effects of heat stress on grain weight and quality in rice (Oryza sativa L.) are aggravated by decreased relative humidity. PeerJ 2021, 9, e11218. [Google Scholar] [CrossRef] [PubMed]
- Gann, P.J.; Esguerra, M.; Counce, P.A.; Srivastava, V. Genotype-dependent and heat-induced grain chalkiness in rice correlates with the expression patterns of starch biosynthesis genes. Plant Environ. Interact. 2021, 2, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Cai, H.; Liu, K.; An, B.; Wang, R.; Yang, F.; Zeng, C.; Jiao, C.; Xu, Y. DNA Methylation Alterations and Their Association with High Temperature Tolerance in Rice Anthesis. J. Plant Growth Regul. 2023, 42, 780–794. [Google Scholar] [CrossRef]
- Chakraborty, A.; Wylie, S.J. CRISPR/Cas9 for Heat Stress Tolerance in Rice: A Review. Plant Mol. Biol. Report. 2025, 43, 1047–1056. [Google Scholar] [CrossRef]
- Zhang, Y.; Zou, B.; Lu, S.; Ding, Y.; Liu, H.; Hua, J. Expression and promoter analysis of the OsHSP16.9C gene in rice. Biochem. Biophys. Res. Commun. 2016, 479, 260–265. [Google Scholar] [CrossRef]
- Xuan, Q.; Wang, J.; Nie, Y.; Fang, C.; Liang, W. Research Progress and Application of Miniature CRISPR-Cas12 System in Gene Editing. Int. J. Mol. Sci. 2024, 25, 12686. [Google Scholar] [CrossRef]
- Lou, H.; Li, S.; Shi, Z.; Zou, Y.; Zhang, Y.; Huang, X.; Yang, D.; Yang, Y.; Li, Z.; Xu, C. Engineering source-sink relations by prime editing confers heat-stress resilience in tomato and rice. Cell 2025, 188, 530–549.e520. [Google Scholar] [CrossRef]
- Li, X.; Xie, J.; Dong, C.; Zheng, Z.; Shen, R.; Cao, X.; Chen, X.; Wang, M.; Zhu, J.K.; Tian, Y. Efficient and heritable A-to-K base editing in rice and tomato. Hortic. Res. 2024, 11, uhad250. [Google Scholar] [CrossRef]
- Hu, C.; Jiang, J.; Li, Y.; Song, S.; Zou, Y.; Jing, C.; Zhang, Y.; Wang, D.; He, Q.; Dang, X. QTL mapping and identification of candidate genes using a genome-wide association study for heat tolerance at anthesis in rice (Oryza sativa L.). Front. Genet. 2022, 13, 983525. [Google Scholar] [CrossRef]
- Liu, X.; Ji, P.; Liao, J.; Duan, X.; Luo, Z.; Yu, X.; Jiang, C.J.; Xu, C.; Yang, H.; Peng, B.; et al. CRISPR/Cas knockout of the NADPH oxidase gene OsRbohB reduces ROS overaccumulation and enhances heat stress tolerance in rice. Plant Biotechnol. J. 2025, 23, 336–351. [Google Scholar] [CrossRef]
- Li, P.; Jiang, J.; Zhang, G.; Miao, S.; Lu, J.; Qian, Y.; Zhao, X.; Wang, W.; Qiu, X.; Zhang, F.; et al. Integrating GWAS and transcriptomics to identify candidate genes conferring heat tolerance in rice. Front. Plant Sci. 2022, 13, 1102938. [Google Scholar] [CrossRef]
- Pan, Y.H.; Chen, L.; Zhu, X.Y.; Li, J.C.; Rashid, M.A.R.; Chen, C.; Qing, D.J.; Zhou, W.Y.; Yang, X.H.; Gao, L.J.; et al. Utilization of natural alleles for heat adaptability QTLs at the flowering stage in rice. BMC Plant Biol. 2023, 23, 256. [Google Scholar] [CrossRef]
- Grenier, C.; Cao, T.V.; Ospina, Y.; Quintero, C.; Chatel, M.H.; Tohme, J.; Courtois, B.; Ahmadi, N. Accuracy of Genomic Selection in a Rice Synthetic Population Developed for Recurrent Selection Breeding. PLoS ONE 2015, 10, e0136594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, M.; Ye, J.; Xu, Q.; Feng, Y.; Xu, S.; Hu, D.; Wei, X.; Hu, P.; Yang, Y. Integrating genome-wide association study into genomic selection for the prediction of agronomic traits in rice (Oryza sativa L.). Mol. Breed. 2023, 43, 81. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Xu, Y.; Fu, W.; Li, H.; Li, G.; Li, J.; Wang, W.; Tao, L.; Chen, T.; Fu, G. RGA1 Negatively Regulates Thermo-tolerance by Affecting Carbohydrate Metabolism and the Energy Supply in Rice. Rice 2023, 16, 32. [Google Scholar] [CrossRef] [PubMed]
- El-Esawi, M.A.; Alayafi, A.A. Overexpression of Rice Rab7 Gene Improves Drought and Heat Tolerance and Increases Grain Yield in Rice (Oryza sativa L.). Genes 2019, 10, 56. [Google Scholar] [CrossRef]
- Guo, Z.; Zuo, Y.; Wang, S.; Zhang, X.; Wang, Z.; Liu, Y.; Shen, Y. Early signaling enhance heat tolerance in Arabidopsis through modulating jasmonic acid synthesis mediated by HSFA2. Int. J. Biol. Macromol. 2024, 267, 131256. [Google Scholar] [CrossRef]
- Huang, J.; Gao, L.; Luo, S.; Liu, K.; Qing, D.; Pan, Y.; Dai, G.; Deng, G.; Zhu, C. The genetic editing of GS3 via CRISPR/Cas9 accelerates the breeding of three-line hybrid rice with superior yield and grain quality. Mol. Breed. 2022, 42, 22. [Google Scholar] [CrossRef]
- Li, N.; Zhao, Y.; Han, J.; Yang, Q.; Liang, J.; Liu, X.; Wang, Y.; Huang, Z. Impacts of future climate change on rice yield based on crop model simulation-A meta-analysis. Sci. Total Environ. 2024, 949, 175038. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, J.; Qian, Q.; Shang, L. Enhancement of Heat and Drought Stress Tolerance in Rice by Genetic Manipulation: A Systematic Review. Rice 2022, 15, 67. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Ren, Z.; Sun, L.; Zhou, S.; Yuan, W.; Hui, Y.; Ci, D.; Wang, W.; Fan, L.M.; Wu, Z.; et al. ALBA proteins confer thermotolerance through stabilizing HSF messenger RNAs in cytoplasmic granules. Nat. Plants 2022, 8, 778–791. [Google Scholar] [CrossRef] [PubMed]
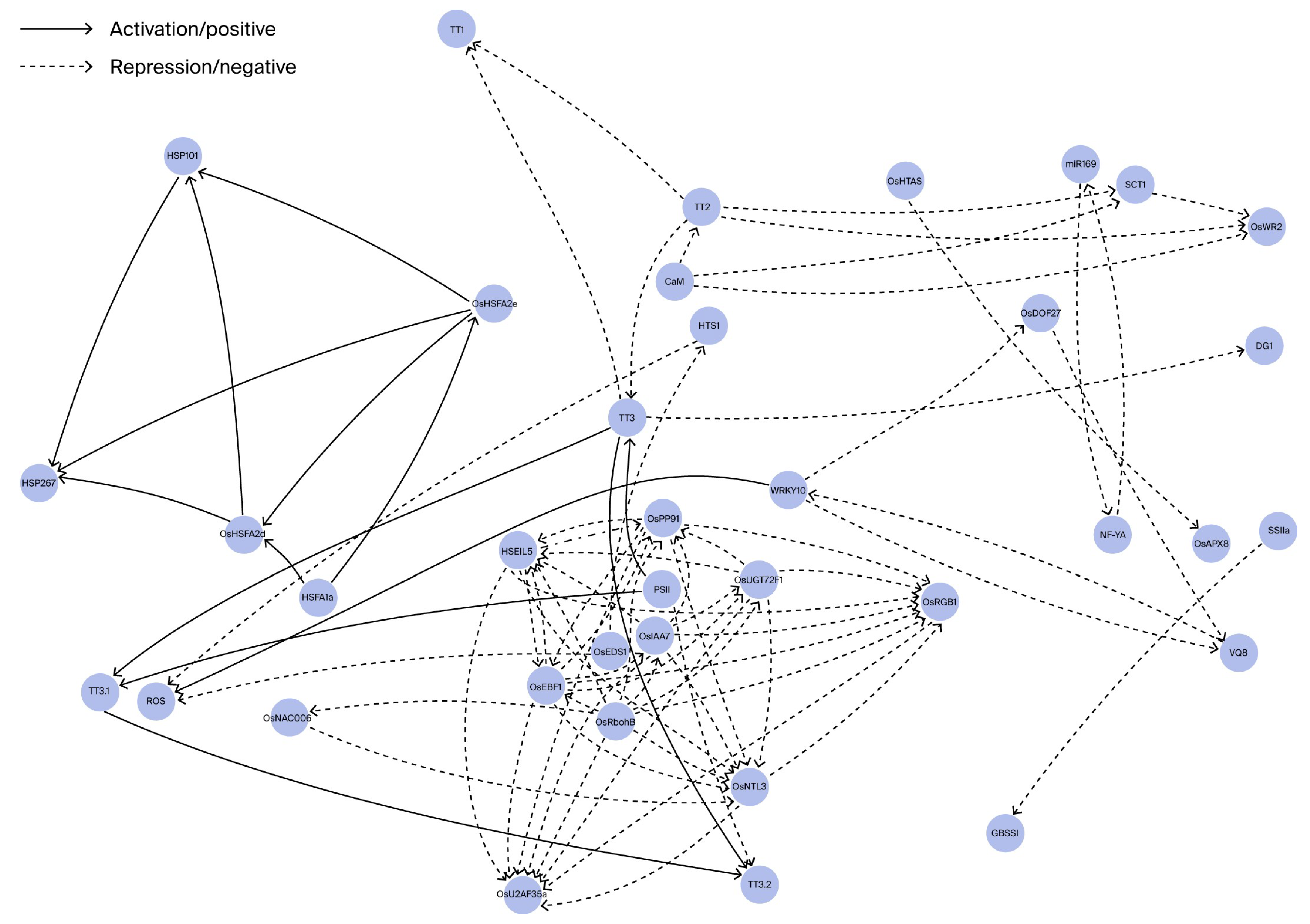
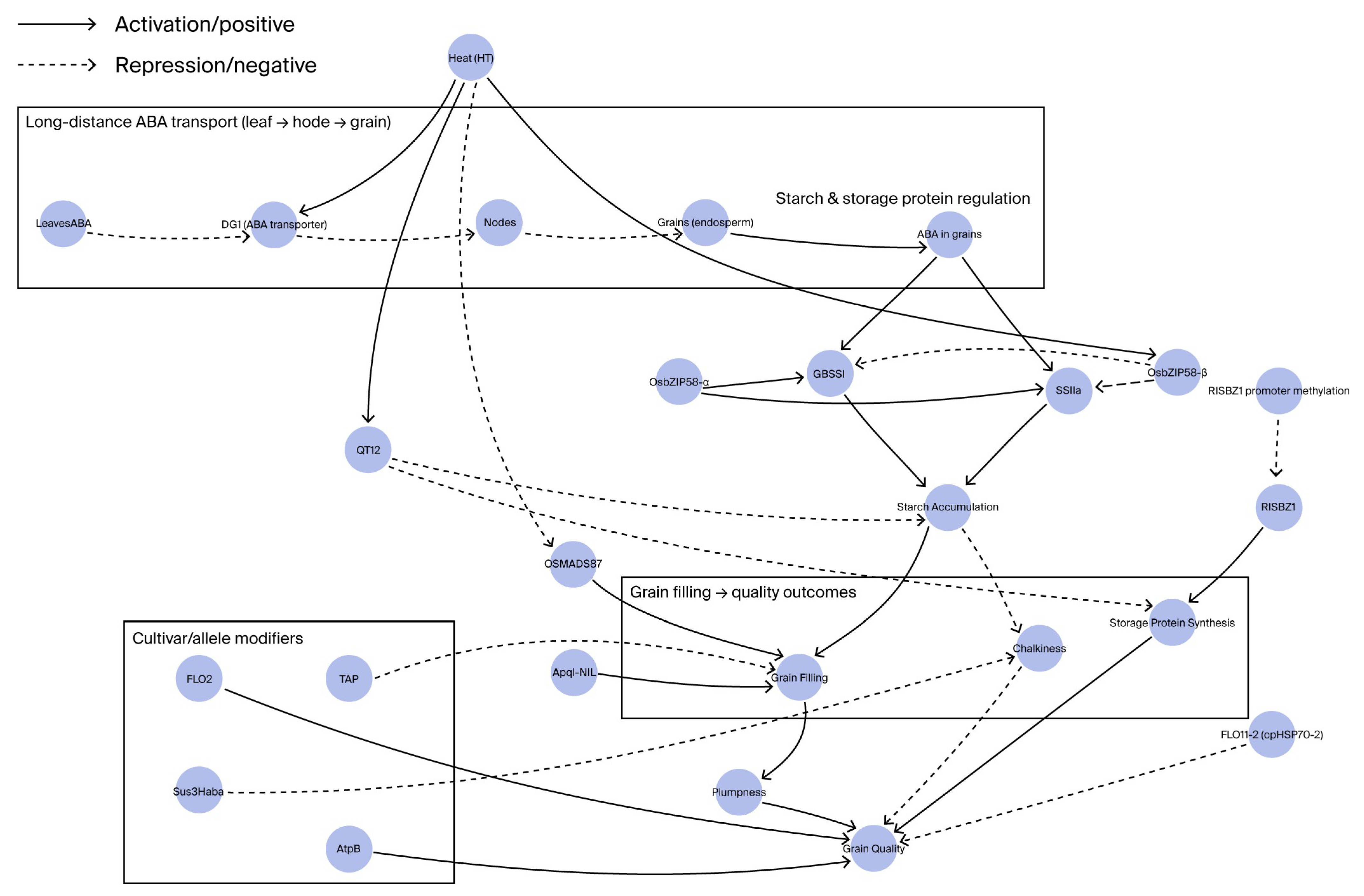
| Category | Gene/Locus | Functional Description | Regulatory Mechanism/Pathway | Heat Tolerance Effect |
|---|---|---|---|---|
| Heat Sensing and Signal Transduction | HTS1 [18] | β-ketoacyl carrier protein reductase in chloroplast thylakoid membrane regulates lipid unsaturation | Maintains membrane stability, suppresses ROS accumulation, and modulates Ca2+ signaling | Loss leads to membrane rupture, increased PCD; normal function enhances heat perception |
| OsHTAS [19] | RING-type E3 ubiquitin ligase | Regulates H2O2 accumulation to promote stomatal closure; interacts with OsAPX8 for ROS scavenging | Overexpression markedly improves survival under heat stress | |
| TT2 [15] | G protein γ subunit | Regulates Ca2+–CaM–SCT1–OsWR2 pathway to maintain wax deposition | Preserves leaf surface barrier and improves heat tolerance | |
| Transcriptional Regulation | OsHSFA2e/ [20] | Heat shock transcription factor; binds HSE to activate stress genes | Induces HSPs (OsHSP70, OsHSP90) and activates OsHsfA2d and stress TFs (OsSNAC1, OsDREB2A, OsLEA3) | Enhances floral organ and whole-plant thermotolerance |
| OsHSFA2d [21] | Core Hsf in heat response | Upregulates HSPs and LEAs and stabilizes proteins and membranes | Improves survival and reproductive stability under heat | |
| OsNTL3–OsbZIP74 [22] | Membrane-tethered NAC TF (OsNTL3) and spliced bZIP TF (OsbZIP74) | Heat/ER stress → OsNTL3 cleavage and nuclear import. OsbZIP74 activates OsNTL3; both form a positive loop to induce UPR/heat-responsive genes | Enhances stress gene expression, reduces ROS, and improves survival under heat | |
| OsDOF27 [23] | Plant-specific DOF transcription factor with intrinsically disordered protein (IDP) features; is nuclear-localized | Heat-inducible; promoter enriched in HSE/ABRE motifs. May self-regulate via DOF sites; activates HSPs and stress-related genes | Overexpression lines show ~2× survival rate | |
| WRKY10–VQ8 [24] | WRKY–VQ interaction module | Suppresses pro-senescence genes and reduces ROS accumulation | Delays cell death and improves survival under heat stress | |
| OsMYB55 [25] | R2R3-MYB transcription factor | R2R3-MYB transcription factor that promotes amino acid biosynthesis and maintains metabolic stability | Reduces yield loss under high temperatures | |
| Protein Homeostasis and Degradation | TT1 [14] | 26S proteasome α2 subunit | Degrades heat-damaged proteins and maintains proteostasis | The elite allele improves heat adaptation significantly |
| TT3.1–TT3.2 [9] | E3 ubiquitin ligase–chloroplast chaperone | Promotes TT3.2 degradation and protects PSII | Enhances photosynthesis and yield under heat | |
| Membrane Lipid and Antioxidant Regulation | OsEDS1 [26] | Positive regulator of thermotolerance; interacts with catalases | Stabilizes and enhances the activity of OsCATB/OsCATC in peroxisomes; maintains H2O2 homeostasis under heat | Overexpression reduces ROS accumulation and improves survival, fertility, grain weight, and yield under heat |
| PSL50 [27] | Premature senescence-related protein | Reduces ROS accumulation and cell death | Mutants display decreased thermotolerance | |
| OsProDH [28] | Proline dehydrogenase catalyzes proline degradation to P5C | Promotes proline catabolism; knockout increases proline accumulation, and overexpression decreases proline | Negative regulator of thermotolerance: knockout reduces ROS and enhances survival; overexpression increases ROS and sensitivity under heat | |
| OsMDHAR4 [29] | Ascorbate metabolism enzyme | Inhibits H2O2-induced stomatal closure | Knockout enhances survival | |
| Grain Development and Quality-Related Genes | QT12 [8] | Grain quality regulator; expression negatively correlated with thermotolerance | NF-YA8–NF-YB9/NF-YC10 module controls QT12 via CCAAT-box. Indica promoter variation disrupts NF-YA8 binding → low QT12 expression; japonica retains binding → high expression | Low QT12 expression maintains starch–protein balance and improves fertility, yield, and grain quality under heat; high QT12 expression increases chalkiness and reduces tolerance |
| DG1 [30] | MATE transporter essential for seed filling; mediates ABA efflux | Promotes ABA long-distance transport and stabilizes grain filling | Maintains grain filling and starch synthesis under heat | |
| FLO2 [31] | Participates in the regulation of grain development under high-temperature stress | Affects grain filling and endosperm development | Improves grain shape, quality, and yield under heat | |
| Sus3Haba [32] | Maintains starch synthesis and reduces the formation of chalky grains | |||
| Apql [32] | Heat response sucrose synthase Sus3 | |||
| AtpB [33] | Responsible for driving ATP synthesis and playing a crucial role in energy supply and grain development | |||
| OsDML4 [34] | Maintains a genome-wide hypomethylation state at high temperatures | |||
| OsbZIP58 [35] | Maintains the normal development of grains by activating starch and storage protein synthesis genes to inhibit the expression of hydrolase genes | |||
| FLO11-2 [36] | Regulate rice grain quality under high temperatures | |||
| tRNA Modification | SLG1 [37] | Is a member of the CTU2 superfamily; regulates tRNA 2-thiolation; and is localized in the nucleus and cytoplasm | Heat-inducible; controls tRNA 2-thiolation levels; loss reduces thiolation and thermotolerance; and overexpression enhances them | Knockout causes heat sensitivity; indica haplotype (Hap2) confers stronger tolerance than japonica (Hap1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Zhou, L.; Zhang, D. Genetic and Molecular Basis for Heat Tolerance in Rice: Strategies for Resilience Under Climate Change. Plants 2025, 14, 3492. https://doi.org/10.3390/plants14223492
Zhang W, Zhou L, Zhang D. Genetic and Molecular Basis for Heat Tolerance in Rice: Strategies for Resilience Under Climate Change. Plants. 2025; 14(22):3492. https://doi.org/10.3390/plants14223492
Chicago/Turabian StyleZhang, Wei, Liang Zhou, and Dewen Zhang. 2025. "Genetic and Molecular Basis for Heat Tolerance in Rice: Strategies for Resilience Under Climate Change" Plants 14, no. 22: 3492. https://doi.org/10.3390/plants14223492
APA StyleZhang, W., Zhou, L., & Zhang, D. (2025). Genetic and Molecular Basis for Heat Tolerance in Rice: Strategies for Resilience Under Climate Change. Plants, 14(22), 3492. https://doi.org/10.3390/plants14223492





