Genome-Wide Identification, Characterization, and Expression Analysis of Trehalose Metabolism Genes in Tea Plant (Camellia sinensis) Reveals Their Roles in Response to Heat Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Property Analysis of Trehalose Metabolism Genes in Tea Plant
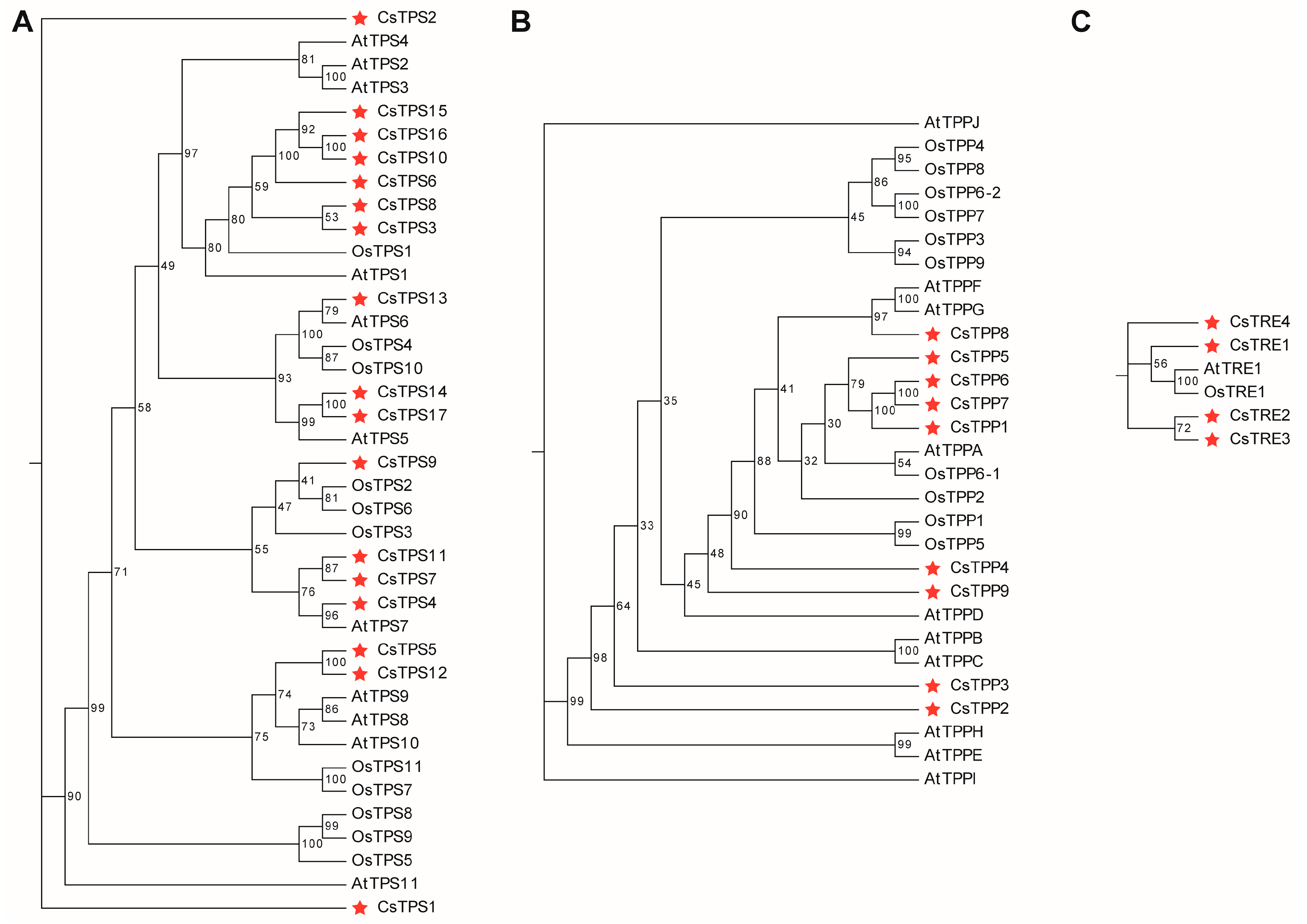
2.2. The Phylogenetic Analysis of Trehalose Metabolism Proteins
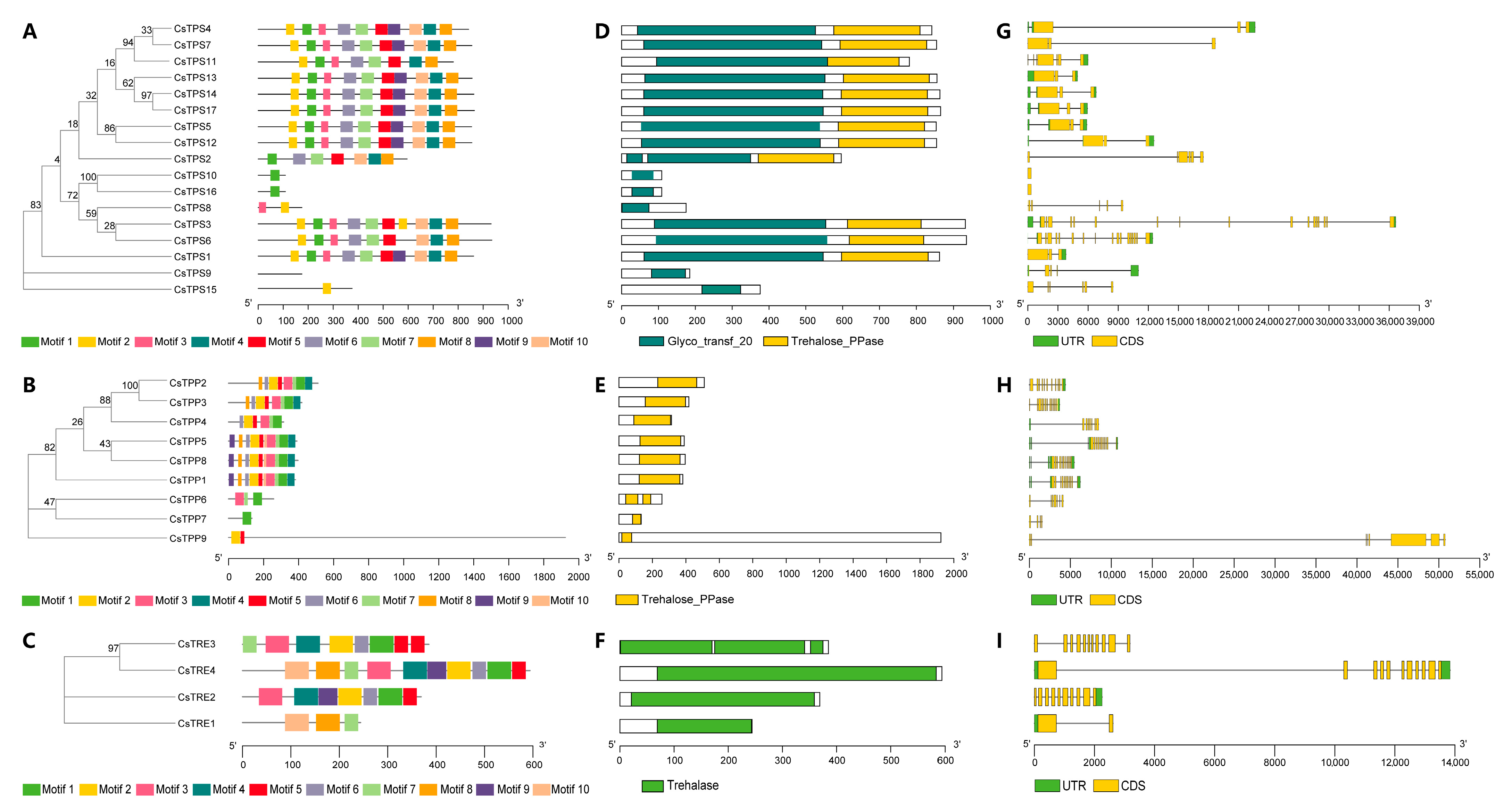
2.3. Phylogenetic Relationships, Conserved Motifs, and Functional Domains Analyses of CsTPS, CsTPP, and CsTRE Proteins, and Gene Structure Analysis of the CsTPS, CsTPP, and CsTRE Gene Families in Tea Plants
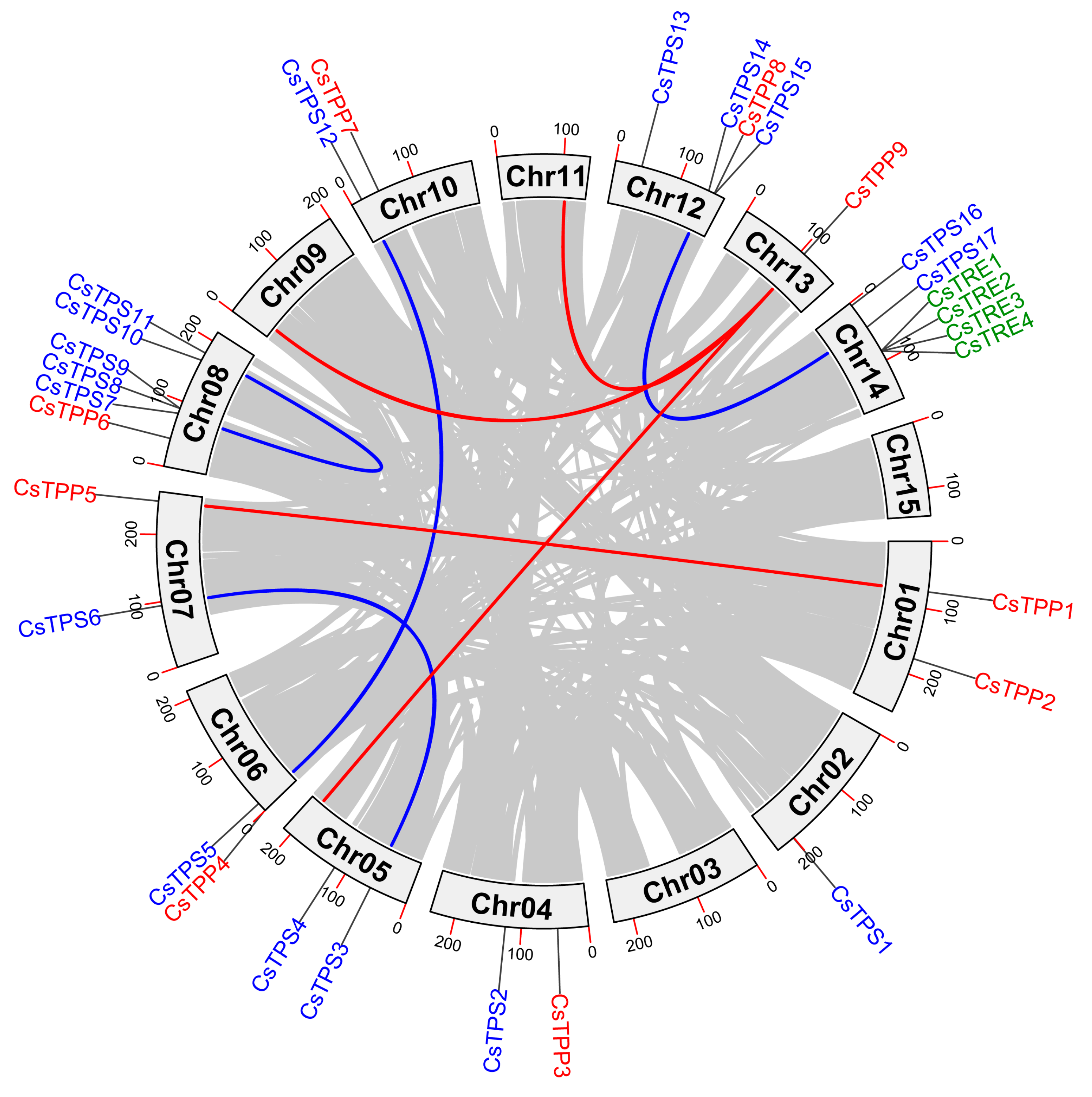
2.4. Collinearity Analysis of CsTPS, CsTPP, and CsTRE Genes
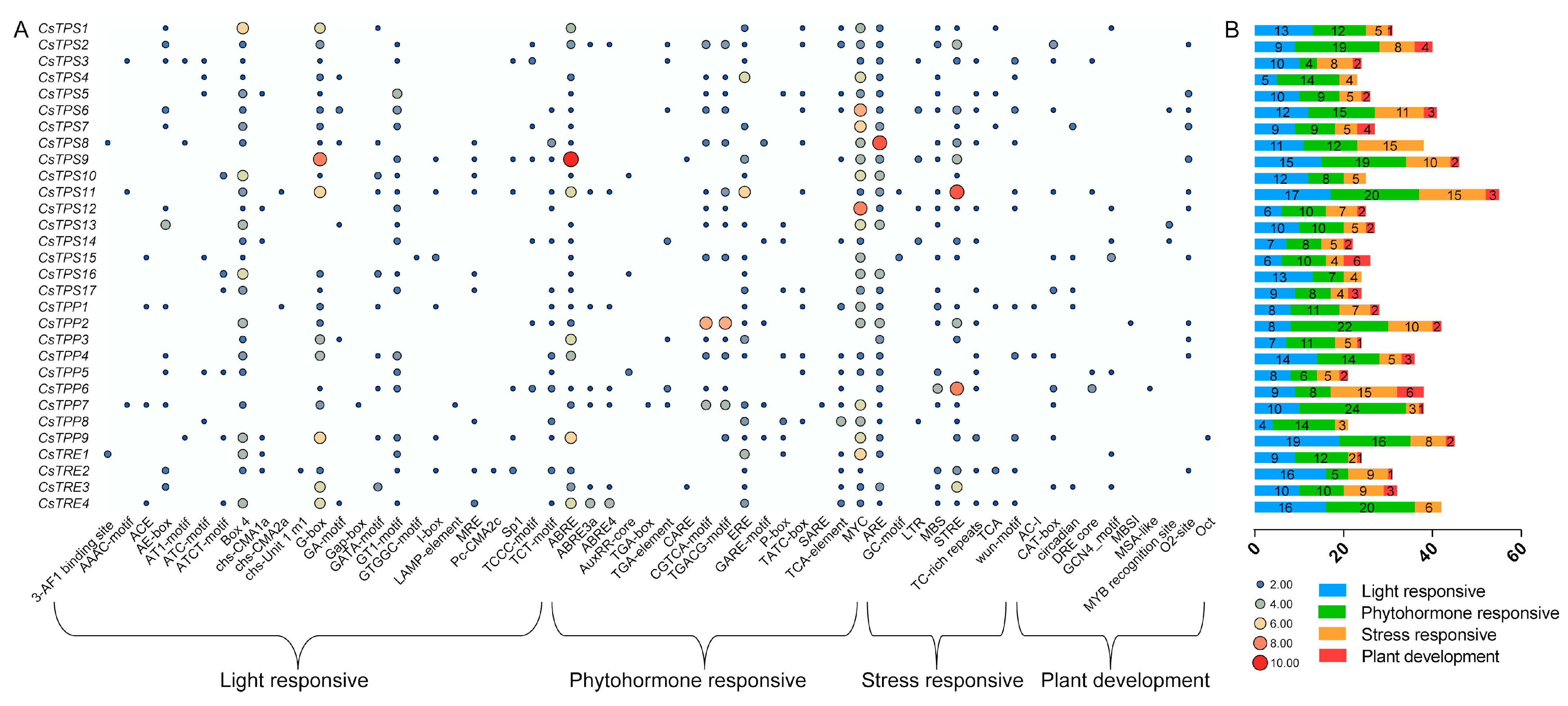
2.5. Cis-Acting Element Analysis of CsTPS, CsTPP, and CsTRE Genes
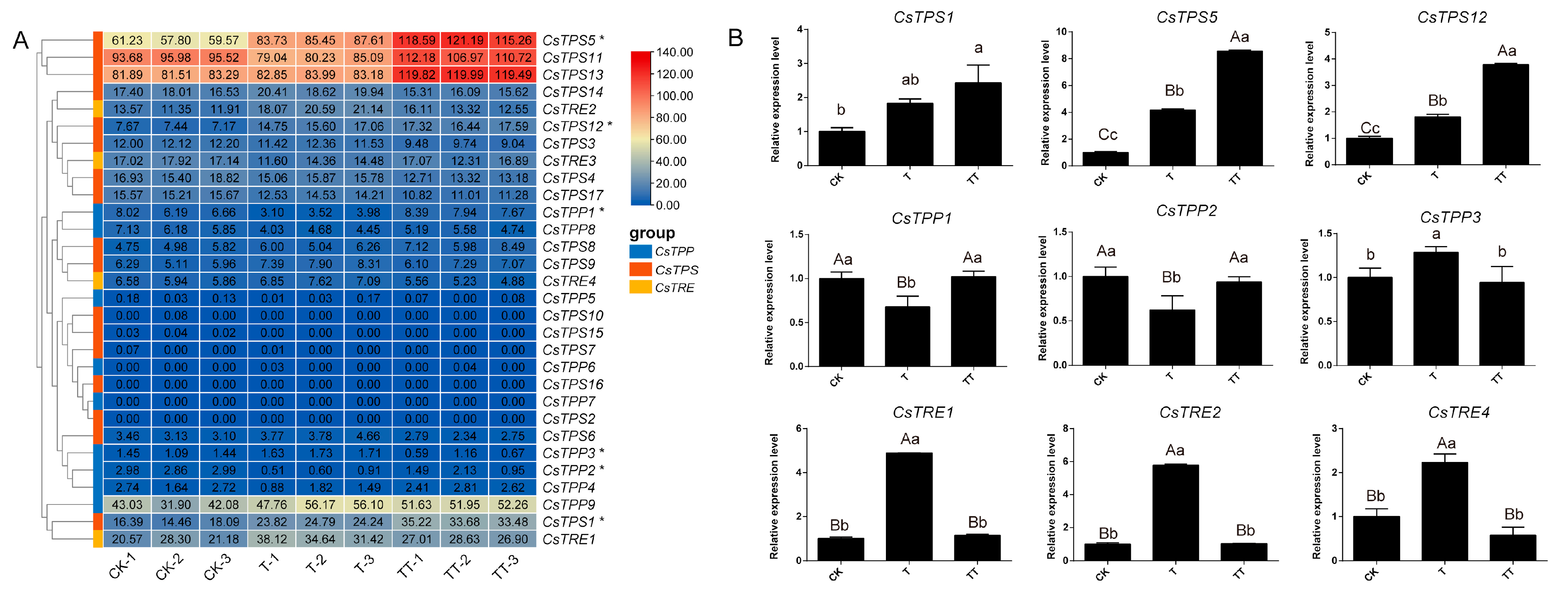
2.6. Expression Patterns of CsTPS, CsTPP, and CsTRE Genes Under Heat Stress with Exogenous Trehalose Treatment
2.7. Analysis of CsTPS1, CsTPS5, CsTPS12, and CsTPP1 Response to Heat Stress
3. Discussion
3.1. Structural Characteristics and Molecular Evolution of Trehalose Metabolism Proteins in Tea Plants
3.2. Trehalose Metabolism Genes Play a Crucial Role in Tea Plants’ Response to Heat Stress
4. Materials and Methods
4.1. Plant Materials and Heat Stress Treatment
4.2. Identification and Characterization of Trehalose Metabolism Genes in Tea Plant
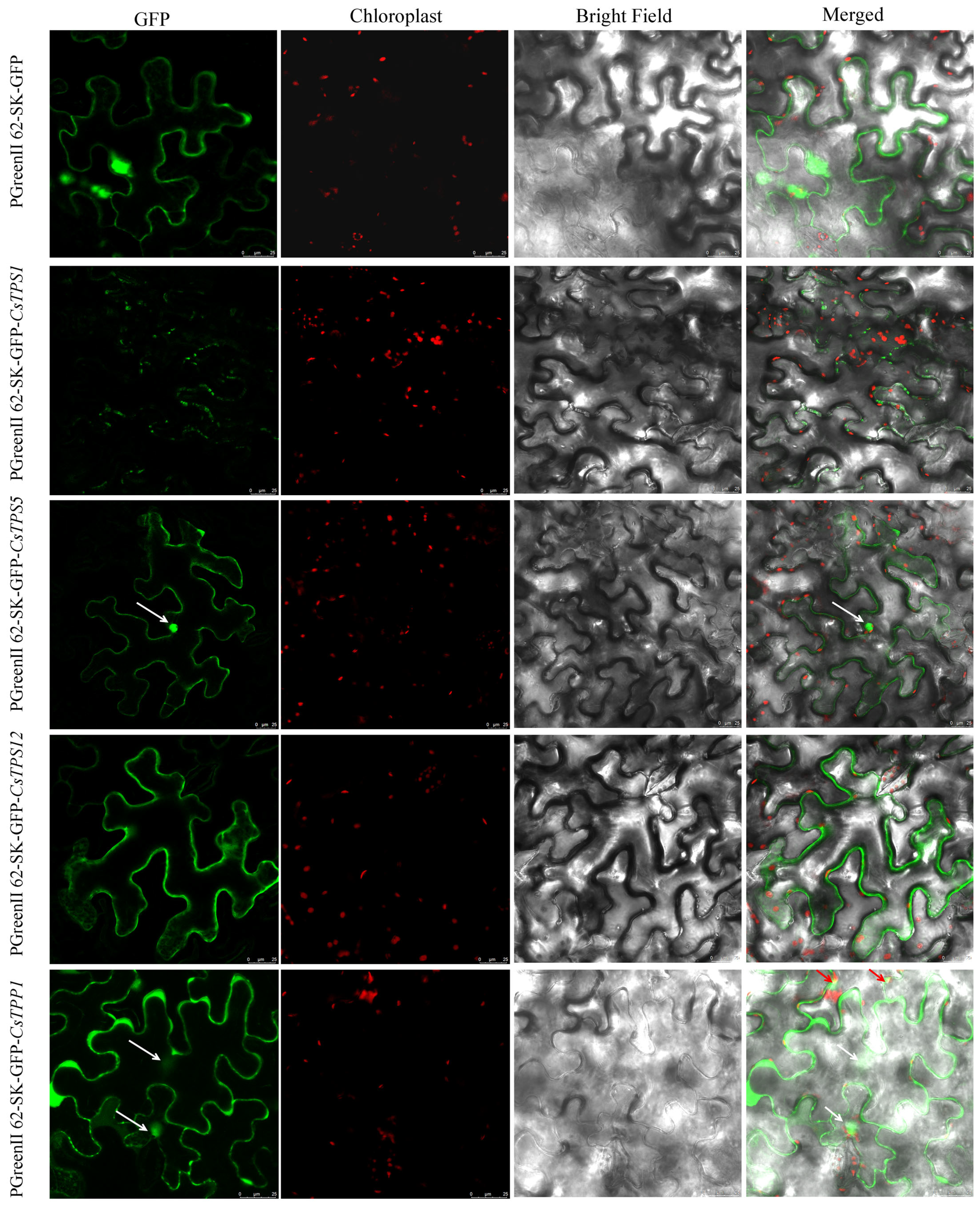
4.3. Subcellular Localization Assay
4.4. Phylogenetic Analysis of Trehalose Metabolism Genes in Tea Plant
4.5. Conserved Motifs, Functional Domains of Trehalose Metabolism Proteins, and Gene Structure Analysis of Trehalose Metabolism Genes in Tea Plants
4.6. Collinearity Analysis of Trehalose Metabolism Genes in Tea Plant
4.7. Cis-Acting Elements Analysis of Trehalose Metabolism Genes in Tea Plant
4.8. Expression Pattern Analysis of Trehalose Metabolism Genes in Tea Plant
4.9. Quantitative Real-Time PCR (RT-qPCR) Analysis
4.10. Heat Stress Tolerance Assays of CsTPS1, CsTPS5, CsTPS12 and CsTPP1 in Yeast
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Djalovic, I.; Kundu, S.; Bahuguna, R.N.; Pareek, A.; Raza, A.; Singla-Pareek, S.L.; Prasad, P.V.V.; Varshney, R.K. Maize and Heat Stress: Physiological, Genetic, and Molecular Insights. Plant Genome 2024, 17, e20378. [Google Scholar] [CrossRef]
- Parankusam, S.; Bhatnagar-Mathur, P.; Sharma, K.K. Heat Responsive Proteome Changes Reveal Molecular Mechanisms Underlying Heat Tolerance in Chickpea. Environ. Exp. Bot. 2017, 141, 132–144. [Google Scholar] [CrossRef]
- Sharma, N.; Thakur, M.; Suryakumar, P.; Mukherjee, P.; Raza, A.; Prakash, C.S.; Anand, A. ‘Breathing Out’ Under Heat Stress—Respiratory Control of Crop Yield Under High Temperature. Agronomy 2022, 12, 806. [Google Scholar] [CrossRef]
- Medina, E.; Kim, S.-H.; Yun, M.; Choi, W.-G. Recapitulation of the Function and Role of ROS Generated in Response to Heat Stress in Plants. Plants 2021, 10, 371. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b Act as Repressors of the Expression of Heat-Inducible Hsfs But Positively Regulate the Acquired Thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Niu, C.Y.; Yang, C.R.; Jinn, T.L. The Heat-Stress Factor HSFA6b Connects ABA Signaling and ABA-Mediated Heat Responses. Plant Physiol. 2016, 172, 1182–1199. [Google Scholar] [CrossRef]
- Liu, H.C.; Liao, H.T.; Charng, Y.Y. The Role of Class A1 Heat Shock Factors (HSFA1s) in Response to Heat and Other Stresses in Arabidopsis. Plant Cell Environ. 2011, 34, 738–751. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.M.; Lin, H.X.; Chong, K. Crop Improvement Through Temperature Resilience. Annu. Rev. Plant Biol. 2019, 70, 753–780. [Google Scholar] [CrossRef]
- Rivero, R.M.; Mittler, R.; Blumwald, E.; Zandalinas, S.I. Developing Climate-Resilient Crops: Improving Plant Tolerance to Stress Combination. Plant J. 2022, 109, 373–389. [Google Scholar] [CrossRef]
- Bae, H.; Herman, E.; Bailey, B.; Bae, H.J.; Sicher, R. Exogenous Trehalose Alters Arabidopsis Transcripts Involved in Cell Wall Modification, Abiotic Stress, Nitrogen Metabolism, and Plant Defense. Physiol. Plant. 2005, 125, 114–126. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, Salt, and Temperature Stress-Induced Metabolic Rearrangements and Regulatory Networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Asthir, B. Mechanisms of Heat Tolerance in Crop Plants. Biol. Plant. 2015, 59, 620–628. [Google Scholar] [CrossRef]
- Hassan, M.U.; Nawaz, M.; Shah, A.N.; Raza, A.; Barbanti, L.; Skalicky, M.; Hashem, M.; Brestic, M.; Pandey, S.; Alamri, S.; et al. Trehalose: A Key Player in Plant Growth Regulation and Tolerance to Abiotic Stresses. J. Plant Growth Regul. 2023, 42, 4935–4957. [Google Scholar] [CrossRef]
- Luo, Y.; Xie, Y.; Li, W.; Wei, M.; Dai, T.; Li, Z.; Wang, B. Physiological and Transcriptomic Analyses Reveal Exogenous Trehalose Is Involved in the Responses of Wheat Roots to High Temperature Stress. Plants 2021, 10, 2644. [Google Scholar] [CrossRef]
- Raza, A.; Bhardwaj, S.; Rahman, M.A.; García-Caparrós, P.; Habib, M.; Saeed, F.; Charagh, S.; Foyer, C.H.; Siddique, K.H.M.; Varshney, R.K. Trehalose: A Sugar Molecule Involved in Temperature Stress Management in Plants. Crop J. 2024, 12, 1–16. [Google Scholar] [CrossRef]
- Zhao, D.Q.; Li, T.T.; Hao, Z.J.; Cheng, M.L.; Tao, J. Exogenous Trehalose Confers High Temperature Stress Tolerance to Herbaceous Peony by Enhancing Antioxidant Systems, Activating Photosynthesis, and Protecting Cell Structure. Cell Stress Chaperones 2019, 24, 247–257. [Google Scholar] [CrossRef]
- Luo, Y.; Li, F.; Wang, G.P.; Yang, X.H.; Wang, W. Exogenously-Supplied Trehalose Protects Thylakoid Membranes of Winter Wheat from Heat-Induced Damage. Biol. Plant. 2010, 54, 495–501. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, W.; Fan, Y.Z.; Gao, Y.M.; Wang, D. Exogenously-Supplied Trehalose Provides Better Protection for D1 Protein in Winter Wheat Under Heat Stress. Russ. J. Plant Physiol. 2018, 65, 115–122. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Ali, S.; Babar, M.A. Crosstalk Amongst Phytohormones from Planta and PGPR Under Biotic and Abiotic Stresses. Plant Growth Regul. 2020, 90, 189–203. [Google Scholar] [CrossRef]
- Onwe, R.O.; Onwosi, C.O.; Ezugworie, F.N.; Ekwealor, C.C.; Okonkwo, C.C. Microbial Trehalose Boosts the Ecological Fitness of Biocontrol Agents, the Viability of Probiotics During Long-Term Storage and Plants Tolerance to Environmental-Driven Abiotic Stress. Sci. Total Environ. 2022, 806, 150432. [Google Scholar] [CrossRef]
- Kumar, S.; Kaushal, N.; Nayyar, H.; Gaur, P. Abscisic Acid Induces Heat Tolerance in Chickpea (Cicer arietinum L.) Seedlings by Facilitated Accumulation of Osmoprotectants. Acta Physiol. Plant. 2012, 34, 1651–1658. [Google Scholar] [CrossRef]
- Lunn, J.E.; Delorge, I.; Figueroa, C.M.; Van Dijck, P.; Stitt, M. Trehalose Metabolism in Plants. Plant J. 2014, 79, 544–567. [Google Scholar] [CrossRef] [PubMed]
- Morgutti, S.; Negrini, N.; Pucciariello, C.; Sacchi, G.A. Role of Trehalose and Regulation of Its Levels as a Signal Molecule in Abiotic Stresses in Plants. Plant Signal. Behav. 2019, 235–255. [Google Scholar]
- Miranda, J.A.; Avonce, N.; Suárez, R.; Thevelein, J.M.; Van Dijck, P.; Iturriaga, G. A Bifunctional TPS-TPP Enzyme from Yeast Confers Tolerance to Multiple and Extreme Abiotic-Stress Conditions in Transgenic Arabidopsis. Planta 2007, 226, 1411–1421. [Google Scholar] [CrossRef]
- Suárez, R.; Calderón, C.; Iturriaga, G. Enhanced Tolerance to Multiple Abiotic Stresses in Transgenic Alfalfa Accumulating Trehalose. Crop Sci. 2009, 49, 1791–1799. [Google Scholar] [CrossRef]
- Lyu, J.I.; Park, J.H.; Kim, J.K.; Bae, C.H.; Jeong, W.J.; Min, S.R.; Liu, J.R. Enhanced Tolerance to Heat Stress in Transgenic Tomato Seeds and Seedlings Overexpressing a Trehalose-6-Phosphate Synthase/Phosphatase Fusion Gene. Plant Biotechnol. Rep. 2018, 12, 399–408. [Google Scholar] [CrossRef]
- Tang, C.; Zhai, Z.; Zhong, Y.; Chen, P.; Zhou, F.; Zeng, D.; Xiang, S.; Song, K.; Guo, H.; Jin, W. Cloning and CRISPR/Cas9-mediated targeted mutagenesis of NtTRE in Nicotiana tabacum. Life Sci. J. 2020, 17, 20–23. [Google Scholar]
- Zheng, S.; Liu, C.; Zhou, Z.; Xu, L.; Lai, Z. Physiological and Transcriptome Analyses Reveal the Protective Effect of Exogenous Trehalose in Response to Heat Stress in Tea Plant (Camellia sinensis). Plants 2024, 13, 1339. [Google Scholar] [CrossRef]
- Zheng, S.; Liu, C.; Zhou, Z.; Xu, L.; Ruan, B.; Chen, X. Genome-Wide Identification and Characterization of Circular RNAs for Exogenous Trehalose-Mediated Heat Stress Responses in Tea Plants (Camellia sinensis). Front. Plant Sci. 2024, 15, 1481169. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Shi, L.; Gong, D.; Zhang, S.; Zhao, Q.; Zhan, D.; Vasseur, L.; Wang, Y.; Yu, J.; et al. Haplotype-Resolved Genome Assembly Provides Insights into Evolutionary History of the Tea Plant Camellia sinensis. Nat. Genet. 2021, 53, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Eastmond, P.J.; Graham, I.A. Trehalose Metabolism: A Regulatory Role for Trehalose-6-Phosphate? Curr. Opin. Plant Biol. 2003, 6, 231–235. [Google Scholar] [CrossRef]
- Zhang, H.; Hong, Y.; Huang, L.; Liu, S.; Tian, L.; Dai, Y.; Cao, Z.; Huang, L.; Li, D.; Song, F. Virus-Induced Gene Silencing-Based Functional Analyses Revealed the Involvement of Several Putative Trehalose-6-Phosphate Synthase/Phosphatase Genes in Disease Resistance Against Botrytis cinerea and Pseudomonas syringae pv. Tomato DC3000 in Tomato. Front. Plant Sci. 2016, 7, 1176. [Google Scholar] [CrossRef]
- Zang, B.; Li, H.; Li, W.; Deng, X.W.; Wang, X. Analysis of Trehalose-6-Phosphate Synthase (TPS) Gene Family Suggests the Formation of TPS Complexes in Rice. Plant Mol. Biol. 2011, 76, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.L.; Liu, Y.J.; Wang, C.L.; Zeng, Q.Y. Molecular Evolution of Trehalose-6-Phosphate Synthase (TPS) Gene Family in Populus, Arabidopsis and Rice. PLoS ONE 2012, 7, e42438. [Google Scholar] [CrossRef]
- Yuan, P.; Zhou, G.; Yu, M.; Hammond, J.P.; Liu, H.; Hong, D.; Cai, H.; Ding, G.; Wang, S.; Xu, F.; et al. Trehalose-6-Phosphate Synthase 8 Increases Photosynthesis and Seed Yield in Brassica napus. Plant J. 2024, 118, 437–456. [Google Scholar] [CrossRef] [PubMed]
- Simola, M.; Hänninen, A.L.; Stranius, S.M.; Makarow, M. Trehalose Is Required for Conformational Repair of Heat-Denatured Proteins in the Yeast Endoplasmic Reticulum But Not for Maintenance of Membrane Traffic Functions After Severe Heat Stress. Mol. Microbiol. 2000, 37, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Sun, Y.; Chai, J.; Liu, L.; Li, J.; Guo, R.; Guo, C. Genome-Wide Identification of the Trehalose-6-Phosphate Synthase Gene Family in Alfalfa (Medicago sativa L.) Reveals Involvement of MsTPS16 in Tolerance to Saline-Alkali Stress. Gene 2025, 963, 149609. [Google Scholar] [CrossRef]
- Lin, M.; Jia, R.; Li, J.; Zhang, M.; Chen, H.; Zhang, D.; Zhang, J.; Chen, X. Evolution and Expression Patterns of the Trehalose-6-Phosphate Synthase Gene Family in Drumstick Tree (Moringa oleifera Lam.). Planta 2018, 248, 999–1015. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiong, T.; Li, K.; Huang, K.; Liao, C.; Liu, G. Evolution and Amplification of the Trehalose-6-Phosphate Synthase Gene Family in Theaceae. BMC Genom. 2025, 26, 273. [Google Scholar] [CrossRef] [PubMed]
- Vandesteene, L.; Ramon, M.; Le Roy, K.; Van Dijck, P.; Rolland, F. A Single Active Trehalose-6-P Synthase (TPS) and a Family of Putative Regulatory TPS-Like Proteins in Arabidopsis. Mol. Plant 2010, 3, 406–419. [Google Scholar] [CrossRef]
- Jiang, D.; Chen, W.; Gao, J.; Yang, F.; Zhuang, C.J.B.P.R. Overexpression of the Trehalose-6-Phosphate Phosphatase OsTPP3 Increases Drought Tolerance in Rice. Plant Biotechnol. Rep. 2019, 13, 285–292. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Li, F.; Liu, H.; Yang, W.; Chong, K.; Xu, Y. OsMAPK3 Phosphorylates OsbHLH002/OsICE1 and Inhibits Its Ubiquitination to Activate OsTPP1 and Enhances Rice Chilling Tolerance. Dev. Cell 2017, 43, 731–743. [Google Scholar] [CrossRef]
- Yuan, G.; Liu, J.; An, G.; Li, W.; Si, W.; Sun, D.; Zhu, Y. Genome-Wide Identification and Characterization of the Trehalose-6-Phosphate Synthetase (TPS) Gene Family in Watermelon (Citrullus lanatus) and Their Transcriptional Responses to Salt Stress. Int. J. Mol. Sci. 2022, 23, 276. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Wu, X.; Huang, C.; Qiu, Z.; Wang, L.; Zhang, R.; Zhang, J. Trehalose Induced by Reactive Oxygen Species Relieved the Radial Growth Defects of Pleurotus ostreatus Under Heat Stress. Appl. Microbiol. Biotechnol. 2019, 103, 5379–5390. [Google Scholar] [CrossRef]
- Chen, W.K.; Zhang, Y.Y.; Lu, J.; Xu, N.J.; Sun, X. Effects of Trehalose on the Physiological Parameters and Gene Expression of High-Temperature Stressed Gracilariopsis lemaneiformis. J. Fish. China 2024, 48, 069605. [Google Scholar]
- Raza, A.; Su, W.; Jia, Z.; Luo, D.; Zhang, Y.; Gao, A.; Hussain, M.A.; Mehmood, S.S.; Cheng, Y.; Lv, Y. Mechanistic 48. Insights into Trehalose-Mediated Cold Stress Tolerance in Rapeseed (Brassica napus L.) Seedlings. Front. Plant Sci. 2022, 13, 857980. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, Z.; Liu, J.; Chen, M.; Pan, R.; Hu, W.; Guan, Y.; Hu, J. Seed Priming with Spermidine and Trehalose Enhances Chilling Tolerance of Rice via Different Mechanisms. J. Plant Growth Regul. 2020, 39, 669–679. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.J.V.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential Expression of RNA-Seq Data at the Gene Level–The DESeq Package; European Molecular Biology Laboratory (EMBL): Heidelberg, Germany, 2012; pp. 1–23; With a Bootstrap Value of 1000. [Google Scholar]


| Genome ID | Gene Name | Location on Chromosome | Amino Acid (aa) | Isoelectric Point | Molecular Weight | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Predicted Subcellular Location | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Start | End | |||||||||
| CsTGY02G0002941 | CsTPS1 | Chr02 | 200,749,218 | 200,753,015 | 862 | 5.84 | 97,862 | 47.91 | 84.19 | −0.269 | chloroplast |
| CsTGY04G0001703 | CsTPS2 | Chr04 | 123,640,109 | 123,657,586 | 596 | 7.03 | 66,845 | 39.54 | 87.75 | −0.19 | nucleus |
| CsTGY05G0000915 | CsTPS3 | Chr05 | 57,599,302 | 57,635,952 | 932 | 7.18 | 104,904 | 43.99 | 85.46 | −0.357 | peroxisome |
| CsTGY05G0001584 | CsTPS4 | Chr05 | 118,346,295 | 118,368,930 | 842 | 5.54 | 95,441 | 48 | 83.47 | −0.292 | nucleus |
| CsTGY06G0000336 | CsTPS5 | Chr06 | 14,036,327 | 14,042,199 | 854 | 6.06 | 96,663 | 47.8 | 90.13 | −0.192 | cytoplasm |
| CsTGY07G0000650 | CsTPS6 | Chr07 | 94,274,222 | 94,286,663 | 935 | 6.57 | 104,640 | 39.85 | 85.29 | −0.343 | mitochondrion |
| CsTGY08G0000966 | CsTPS7 | Chr08 | 81,319,196 | 81,337,863 | 855 | 6.32 | 96,540 | 45 | 87.33 | −0.183 | chloroplast |
| CsTGY08G0001044 | CsTPS8 | Chr08 | 88,871,360 | 88,880,830 | 175 | 9.84 | 19,913 | 43.84 | 98.17 | −0.329 | cytoplasm |
| CsTGY08G0001045 | CsTPS9 | Chr08 | 88,893,568 | 88,904,580 | 185 | 6.44 | 21,360 | 63.25 | 84.86 | −0.465 | chloroplast |
| CsTGY08G0001896 | CsTPS10 | Chr08 | 166,945,078 | 166,945,407 | 109 | 8.23 | 12,687 | 49.69 | 76.06 | 0.236 | chloroplast |
| CsTGY08G0002084 | CsTPS11 | Chr08 | 179,049,653 | 179,055,642 | 781 | 5.67 | 87,823 | 45.4 | 82.84 | −0.273 | nucleus |
| CsTGY10G0000343 | CsTPS12 | Chr10 | 16,604,285 | 16,616,852 | 855 | 5.77 | 96,447 | 49.52 | 89.71 | −0.149 | cytoplasm |
| CsTGY12G0000461 | CsTPS13 | Chr12 | 39,489,896 | 39,494,837 | 856 | 5.65 | 97,350 | 41.47 | 91.51 | −0.207 | nucleus |
| CsTGY12G0001545 | CsTPS14 | Chr12 | 144,915,194 | 144,922,004 | 863 | 5.59 | 97,244 | 44.3 | 92.11 | −0.164 | chloroplast |
| CsTGY12G0001771 | CsTPS15 | Chr12 | 154,839,943 | 154,848,421 | 376 | 9.24 | 41,499 | 40.56 | 87.95 | −0.031 | chloroplast |
| CsTGY14G0000106 | CsTPS16 | Chr14 | 4,531,582 | 4,531,911 | 109 | 8.65 | 12,647 | 48.42 | 76.06 | 0.188 | chloroplast |
| CsTGY14G0000925 | CsTPS17 | Chr14 | 43,647,873 | 43,653,809 | 865 | 5.7 | 97,429 | 50.75 | 91.82 | −0.187 | cytoplasm |
| CsTGY01G0001161 | CsTPP1 | Chr01 | 75,066,031 | 75,072,244 | 382 | 8.98 | 42,683 | 38.31 | 82.88 | −0.388 | nucleus |
| CsTGY01G0002203 | CsTPP2 | Chr01 | 176,281,234 | 176,285,628 | 510 | 9.35 | 57,780 | 38.93 | 80.27 | −0.414 | cytoplasm |
| CsTGY04G0000901 | CsTPP3 | Chr04 | 46,012,012 | 46,015,699 | 418 | 9.02 | 46,615 | 29.76 | 85.38 | −0.376 | nucleus |
| CsTGY06G0000007 | CsTPP4 | Chr06 | 381,529 | 389,971 | 314 | 6.22 | 36,052 | 36.95 | 75.45 | −0.545 | mitochondrion |
| CsTGY07G0002823 | CsTPP5 | Chr07 | 249,344,114 | 249,354,882 | 390 | 6.33 | 43,897 | 35.02 | 84.21 | −0.339 | nucleus |
| CsTGY08G0000603 | CsTPP6 | Chr08 | 42,227,240 | 42,231,370 | 257 | 5.57 | 29,498 | 30.59 | 117.04 | 0.14 | nucleus |
| CsTGY10G0000716 | CsTPP7 | Chr10 | 44,849,190 | 44,850,744 | 133 | 4.72 | 15,070 | 24.06 | 106.09 | −0.165 | cytoplasm |
| CsTGY12G0001751 | CsTPP8 | Chr12 | 154,099,590 | 154,105,064 | 396 | 7.56 | 44,474 | 35.38 | 84.87 | −0.354 | nucleus |
| CsTGY13G0001345 | CsTPP9 | Chr13 | 106,148,166 | 106,198,927 | 1923 | 4.99 | 220,197 | 51.61 | 85.67 | −0.705 | cytoplasm |
| CsTGY14G0001620 | CsTRE1 | Chr14 | 84,599,742 | 84,602,355 | 244 | 6.91 | 27,556 | 27.41 | 94.59 | −0.037 | chloroplast |
| CsTGY14G0001623 | CsTRE2 | Chr14 | 84,614,556 | 84,616,810 | 369 | 5.44 | 41,729 | 39.05 | 77.75 | −0.214 | nucleus |
| CsTGY14G0001626 | CsTRE3 | Chr14 | 84,879,286 | 84,882,465 | 385 | 5.05 | 43,282 | 39.87 | 82.62 | −0.192 | extracellular |
| CsTGY14G0001627 | CsTRE4 | Chr14 | 85,103,603 | 85,117,441 | 594 | 5.49 | 66,878 | 36.26 | 84.39 | −0.169 | extracellular |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S.; Chen, X.; Zhou, Z.; Lin, R.; Jiang, H.; Xu, L.; Su, J. Genome-Wide Identification, Characterization, and Expression Analysis of Trehalose Metabolism Genes in Tea Plant (Camellia sinensis) Reveals Their Roles in Response to Heat Stress. Plants 2025, 14, 3309. https://doi.org/10.3390/plants14213309
Zheng S, Chen X, Zhou Z, Lin R, Jiang H, Xu L, Su J. Genome-Wide Identification, Characterization, and Expression Analysis of Trehalose Metabolism Genes in Tea Plant (Camellia sinensis) Reveals Their Roles in Response to Heat Stress. Plants. 2025; 14(21):3309. https://doi.org/10.3390/plants14213309
Chicago/Turabian StyleZheng, Shizhong, Xiaohui Chen, Ziwei Zhou, Rongzhao Lin, Huangxin Jiang, Liyi Xu, and Jingjing Su. 2025. "Genome-Wide Identification, Characterization, and Expression Analysis of Trehalose Metabolism Genes in Tea Plant (Camellia sinensis) Reveals Their Roles in Response to Heat Stress" Plants 14, no. 21: 3309. https://doi.org/10.3390/plants14213309
APA StyleZheng, S., Chen, X., Zhou, Z., Lin, R., Jiang, H., Xu, L., & Su, J. (2025). Genome-Wide Identification, Characterization, and Expression Analysis of Trehalose Metabolism Genes in Tea Plant (Camellia sinensis) Reveals Their Roles in Response to Heat Stress. Plants, 14(21), 3309. https://doi.org/10.3390/plants14213309







