Identification of Auxin, Cytokinin, Transcription Factors, and Other Zygotic Embryogenesis-Related Genes in Persea americana: A Transcriptomic-Based Study †
Abstract
1. Introduction
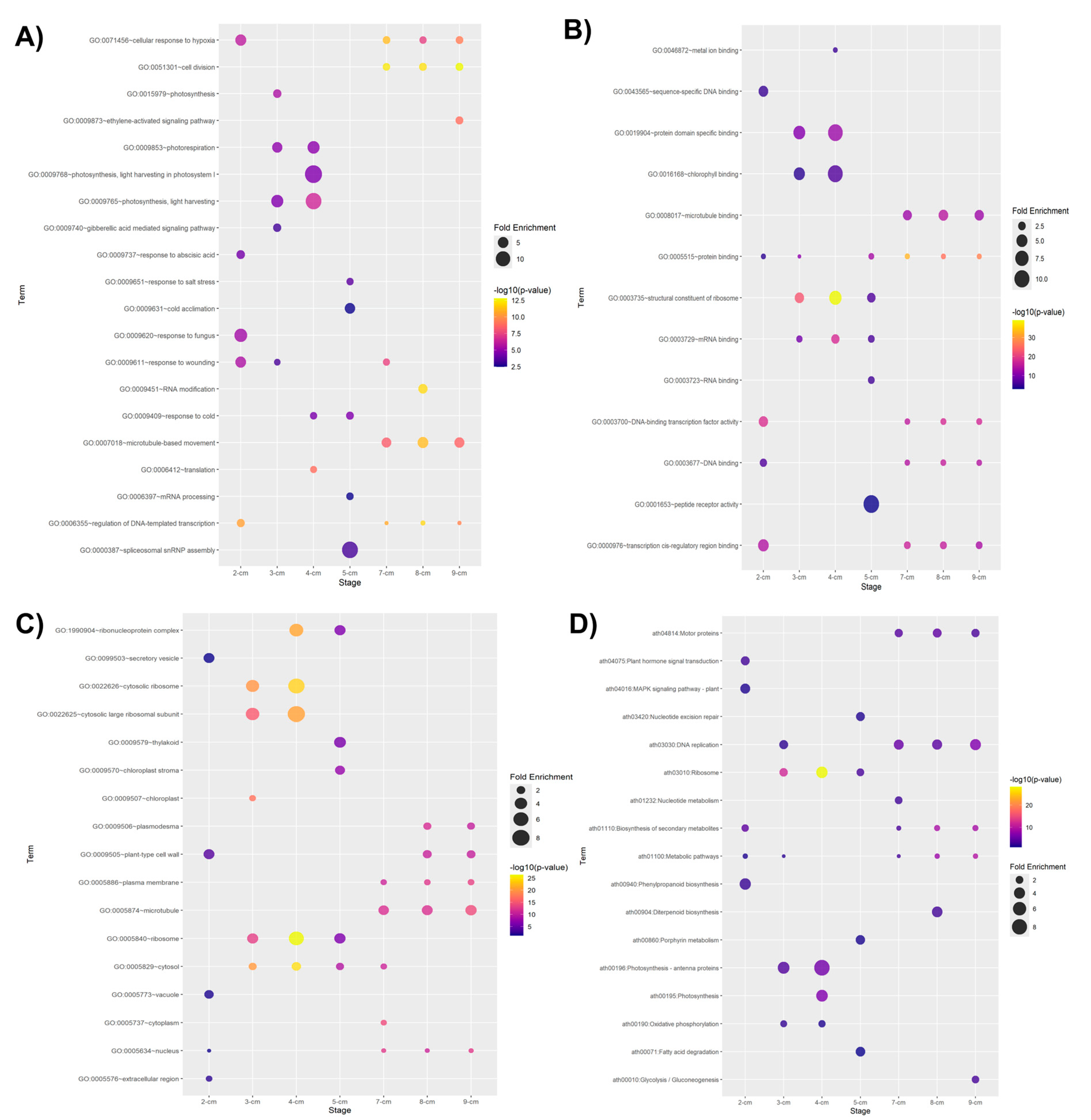
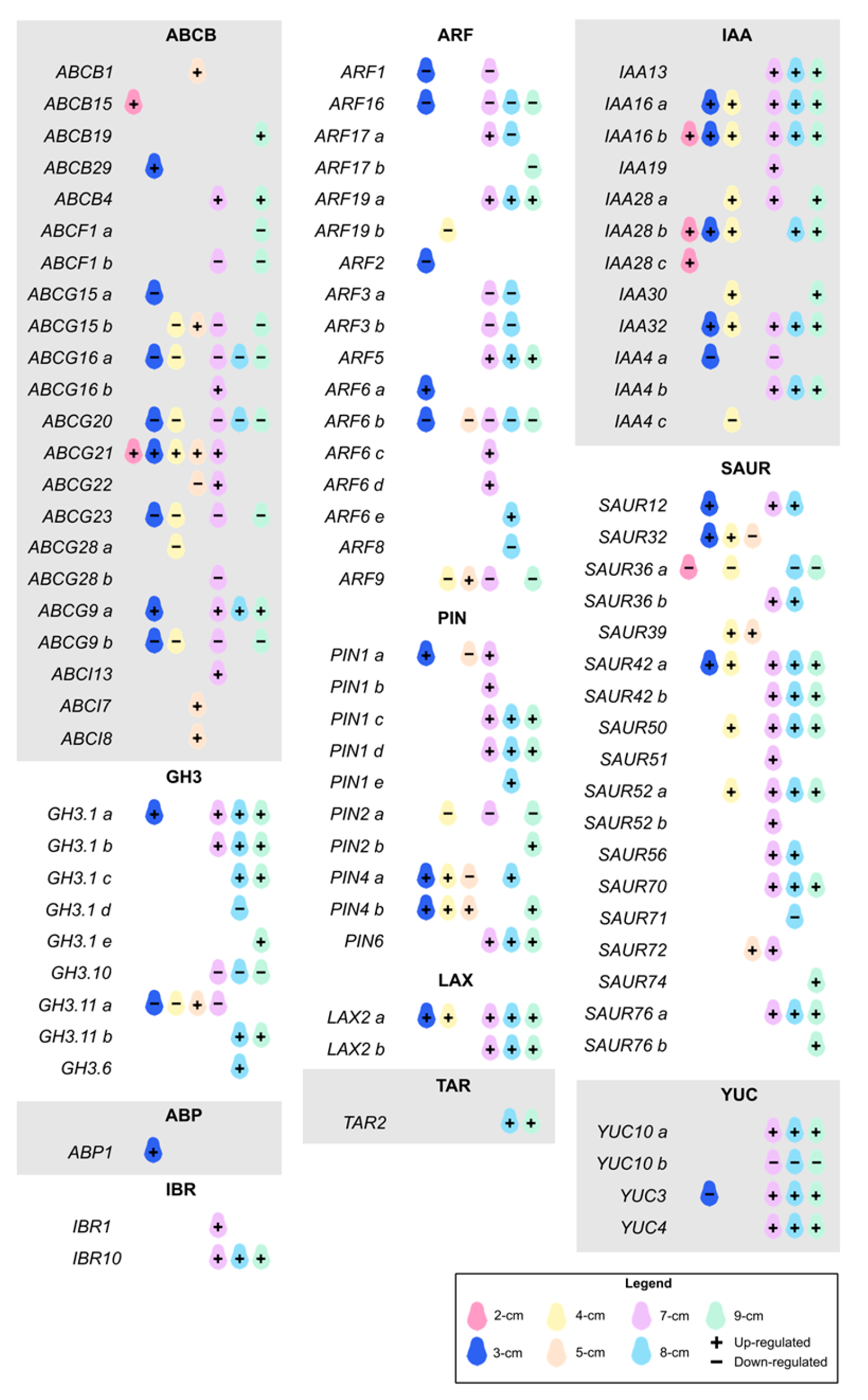
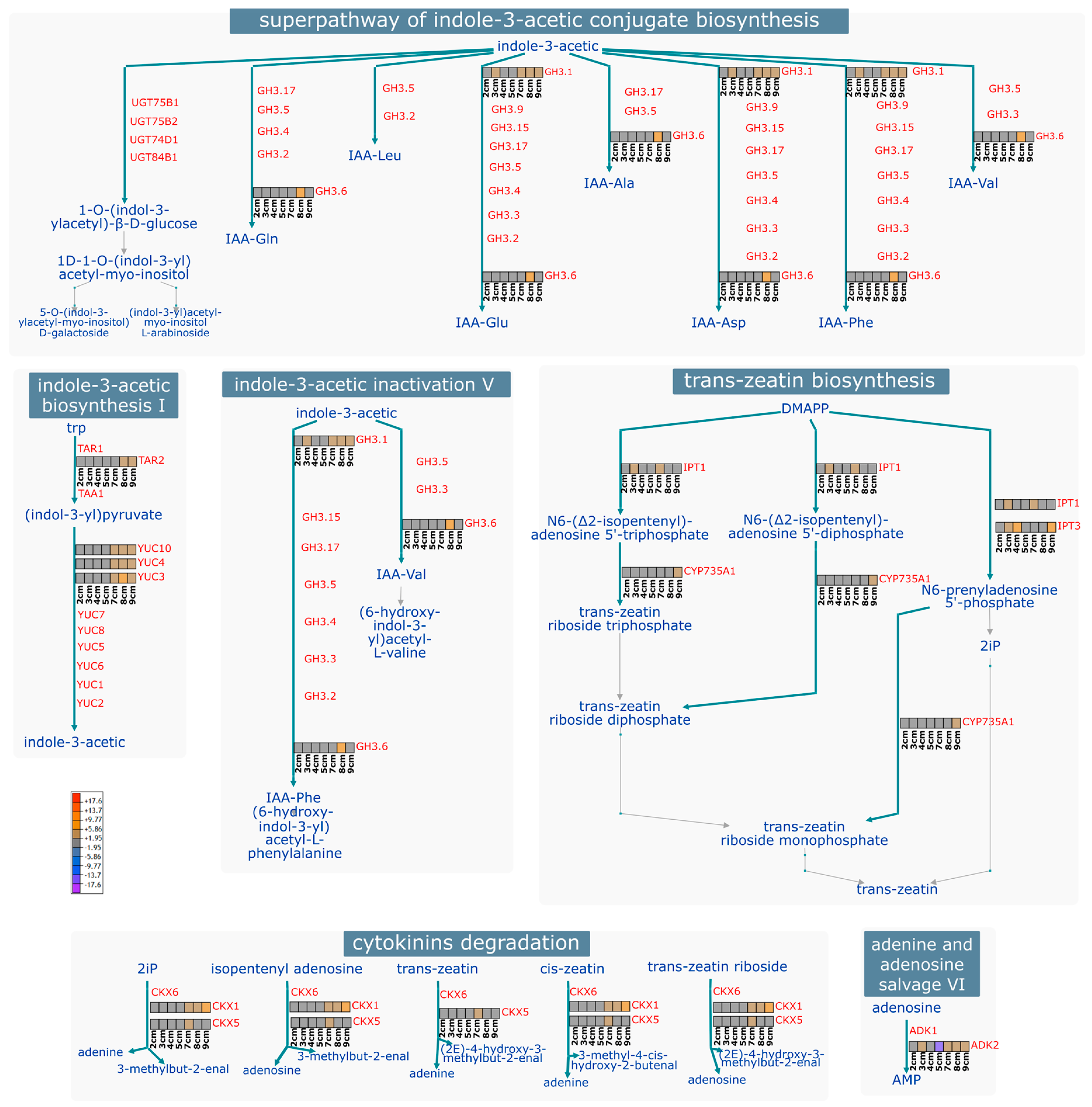
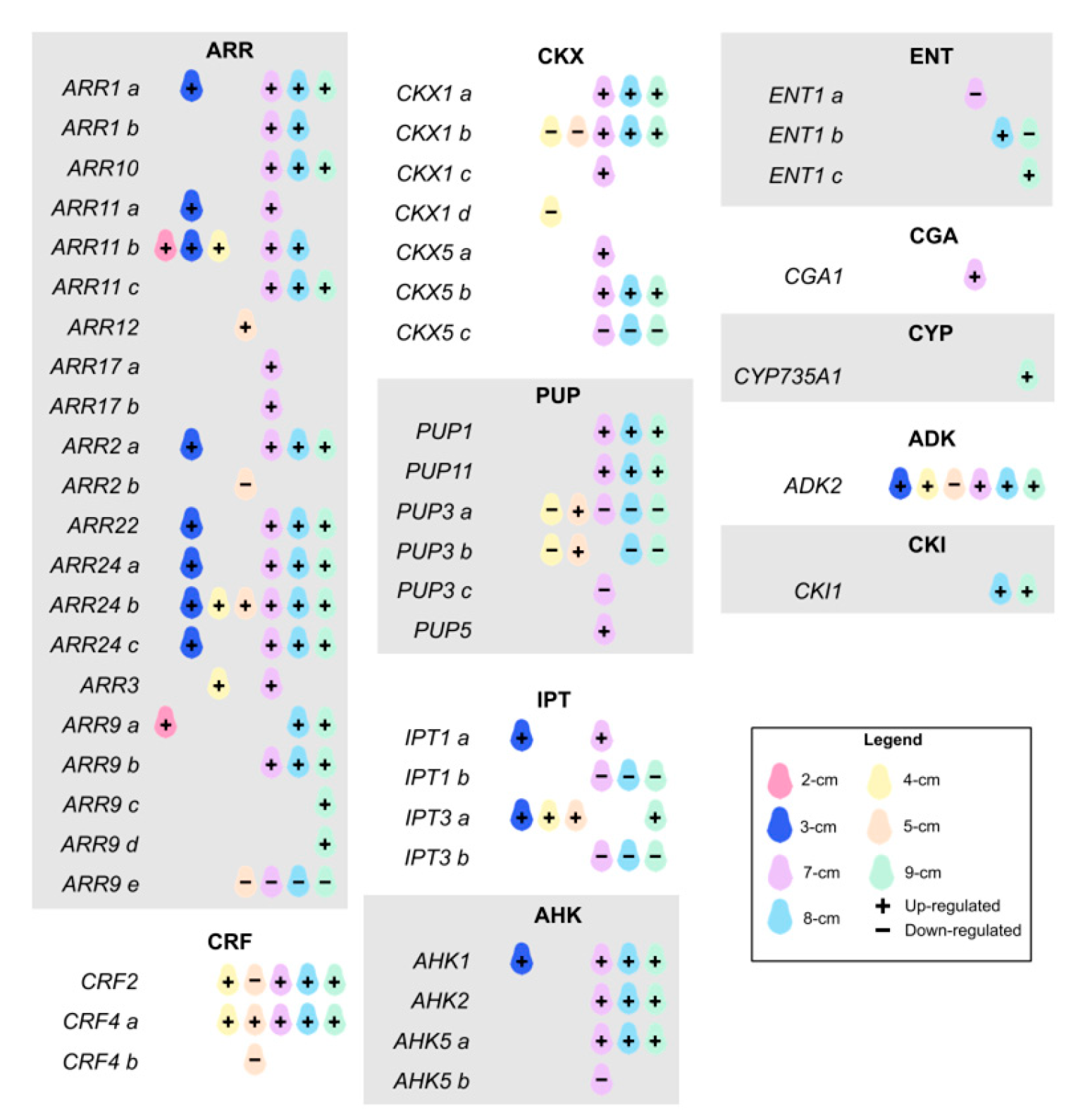
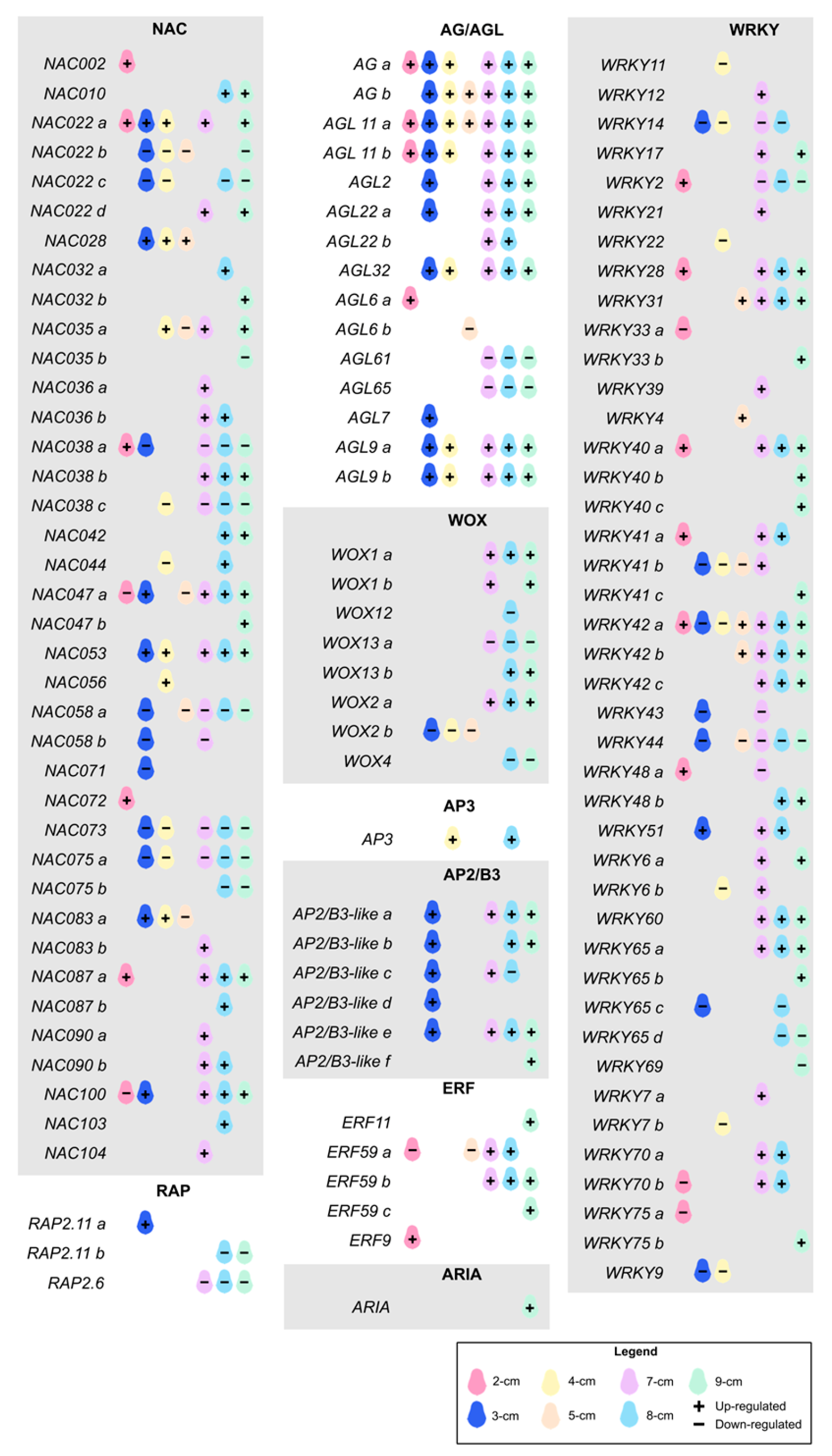
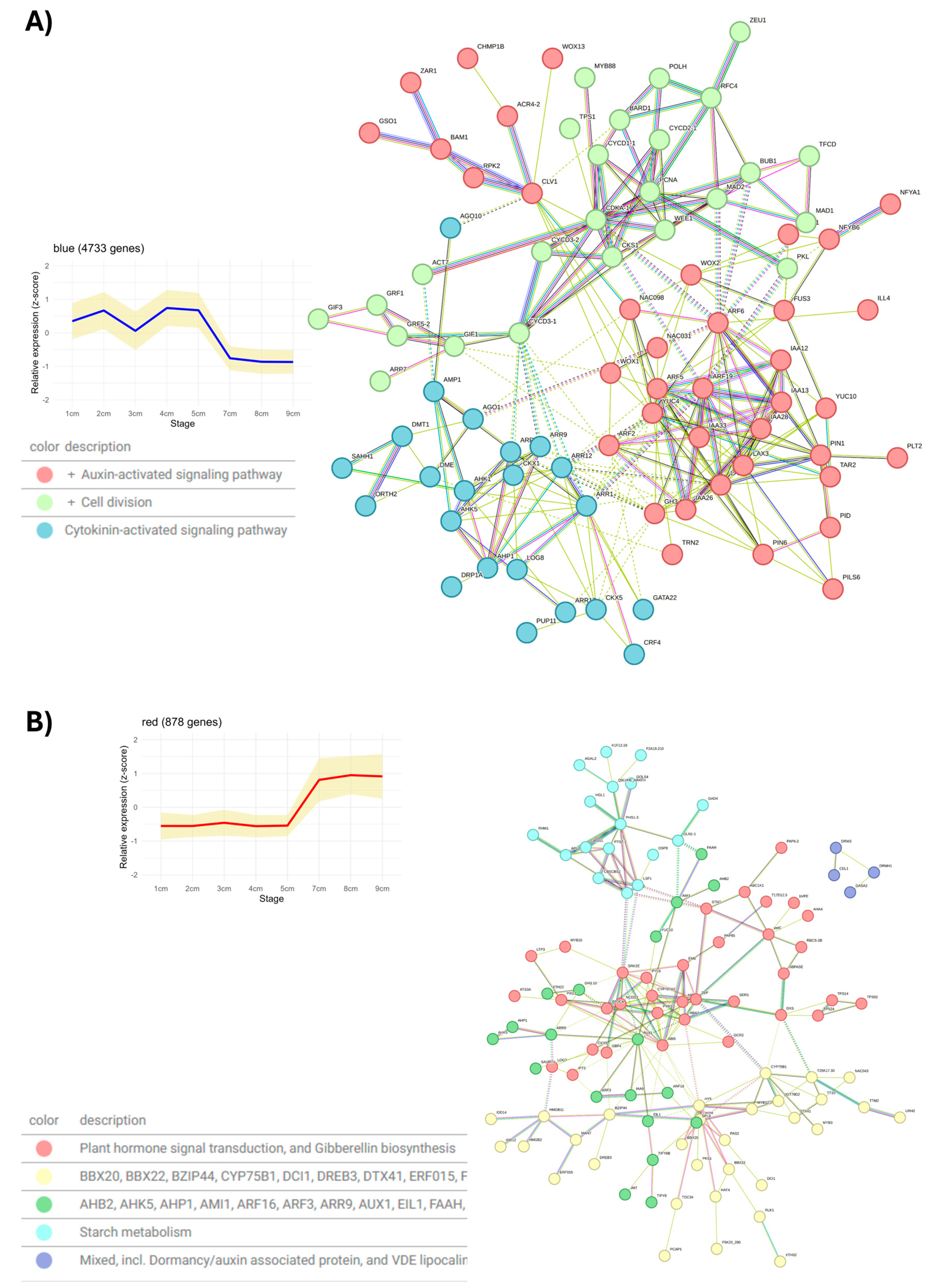
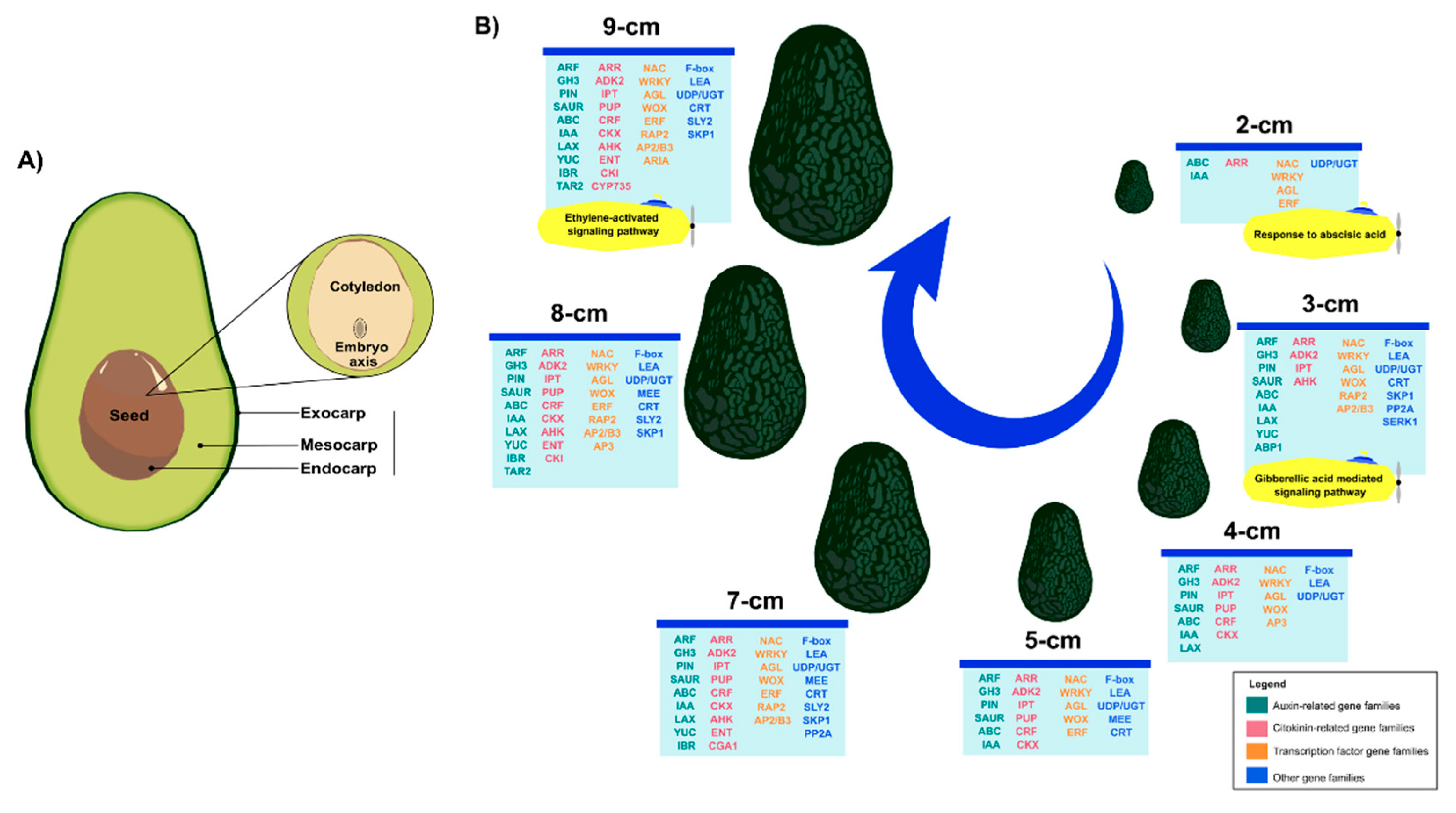
2. Results and Discussion
3. Materials and Methods
3.1. Biological Material and RNA-Seq
3.2. Bioinformatic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Denvir, A.; Arima, E.Y.; González-Rodríguez, A.; Young, K.R. Ecological and human dimensions of avocado expansion in México: Towards supply-chain sustainability. Ambio 2022, 51, 152–166. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/faostat/en/#search/AVOCADO (accessed on 2 August 2025).
- Solares, E.; Morales-Cruz, A.; Balderas, R.F.; Focht, E.; Ashworth, V.E.; Wyant, S.; Minio, A.; Cantu, D.; Arpaia, M.L.; Gaut, B.S. Insights into the domestication of avocado and potential genetic contributors to heterodichogamy. G3 Genes Genomes Genet. 2023, 13, jkac323. [Google Scholar] [CrossRef]
- Furnier, G.R.; Cummings, M.P.; Clegg, M.T. Evolution of the avocados as revealed by DNA restriction fragment variation. J. Hered. 1990, 81, 183–188. [Google Scholar] [CrossRef]
- Fiedler, J.; Bufler, G.; Bangerth, F. Genetic relationships of avocado (Persea americana Mill.) using RAPD markers. Euphytica 1998, 101, 249–255. [Google Scholar] [CrossRef]
- Chen, H.; Morrell, P.L.; Cruz, M.d.l.; Clegg, M.T. Nucleotide diversity and linkage disequilibrium in wild avocado (Persea americana Mill.). J. Hered. 2008, 99, 382–389. [Google Scholar] [CrossRef]
- Liu, Y.; Ge, Y.; Zhan, R.; Lin, X.; Zang, X.; Li, Y.; Yang, Y.; Ma, W. Molecular markers and a quality trait evaluation for assessing the genetic diversity of avocado landraces from China. Agriculture 2020, 10, 102. [Google Scholar] [CrossRef]
- Ibarra-Laclette, E.; Méndez-Bravo, A.; Pérez-Torres, C.A.; Albert, V.A.; Mockaitis, K.; Kilaru, A.; López-Gómez, R.; Cervantes-Luevano, J.I.; Herrera-Estrella, L. Deep sequencing of the Mexican avocado transcriptome, an ancient angiosperm with a high content of fatty acids. BMC Genom. 2015, 16, 599. [Google Scholar] [CrossRef]
- Pochamreddy, M.; Haim, D.; Halon, E.; Keinan, E.; Rai, A.C.; Kamara, I.; Sadka, A.; Irihimovitch, V. Alternate bearing in ‘Hass’ avocado—Fruit load-induced changes in bud auxin homeostasis are associated with flowering repression. J. Exp. Bot. 2024, 75, 5717–5733. [Google Scholar] [CrossRef] [PubMed]
- Winkelmann, T. Somatic versus zygotic embryogenesis: Learning from seeds. In In Vitro Embryogenesis in Higher Plants; Germanà, M.A., Lambardi, M., Eds.; Springer: New York, NY, USA, 2016; pp. 25–46. [Google Scholar] [CrossRef]
- Gaj, M.D. Somatic embryogenesis and plant regeneration in the culture of Arabidopsis thaliana (L.) Heynh. immature zygotic embryo. In Plant Embryo Culture; Thorpe, T.A., Yeung, E.C., Eds.; Springer: New York, NY, USA; London, UK; Dordrecht, The Netherlands; Heidelberg, Germany, 2011; pp. 257–265. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, P.; Peng, X.; Sun, M.-X. Seed development in Arabidopsis: What we have learnt in the past 30 years. Seed Biol. 2023, 2, 6. [Google Scholar] [CrossRef]
- Tian, R.; Paul, P.; Joshi, S.; Perry, S.E. Genetic activity during early plant embryogenesis. Biochem. J. 2020, 477, 3743–3767. [Google Scholar] [CrossRef]
- Harnvanichvech, Y.; Gorelova, V.; Sprakel, J.; Weijers, D. The Arabidopsis embryo as a quantifiable model for studying pattern formation. Quant. Plant Biol. 2021, 2, e3. [Google Scholar] [CrossRef] [PubMed]
- Kao, P.; Nodine, M.D. Transcriptional activation of Arabidopsis zygotes is required for initial cell divisions. Sci. Rep. 2019, 9, 17159. [Google Scholar] [CrossRef]
- Su, Y.H.; Liu, Y.B.; Zhang, X.S. Auxin-cytokinin interaction regulates meristem development. Mol. Plant 2011, 4, 616–625. [Google Scholar] [CrossRef]
- Möller, B.; Weijers, D. Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. 2009, 1, a001545. [Google Scholar] [CrossRef]
- Argueso, C.T.; Ferreira, F.J.; Kieber, J.J. Environmental perception avenues: The interaction of cytokinin and environmental response pathways. Plant Cell Environ. 2009, 32, 1147–1160. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhu, D.; Ma, C.; Cao, H.; Wang, Y.; Xu, Y.; Zhang, W.; Yan, Y. Transcriptome analysis reveals key differentially expressed genes involved in wheat grain development. Crop J. 2016, 4, 92–106. [Google Scholar] [CrossRef]
- Chen, J.; Zeng, B.; Zhang, M.; Xie, S.; Wang, G.; Hauck, A.; Lai, J. Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 2014, 166, 252–264. [Google Scholar] [CrossRef]
- Méndez-Hernández, H.A.; Quintana-Escobar, A.O.; Uc-Chuc, M.Á.; Loyola-Vargas, V.M. Genome-wide analysis, modeling, and identification of amino acid binding motifs suggest the involvement of GH3 genes during somatic embryogenesis of Coffea canephora. Plants 2021, 10, 2034. [Google Scholar] [CrossRef]
- Miransari, M.; Smith, D.L. Plant hormones and seed germination. Environ. Exp. Bot. 2014, 99, 110–121. [Google Scholar] [CrossRef]
- Shackira, A.; Sarath, N.G.; Aswathi, K.R.; Pardha-Saradhi, P.; Puthur, J.T. Green seed photosynthesis: What is it? What do we know about it? Where to go? Plant Physiol. Rep. 2022, 27, 573–579. [Google Scholar] [CrossRef]
- Juárez-Escobar, J.; Monribot-Villanueva, J.L.; Guerrero-Analco, J.A.; Zamora-Briseño, J.A.; Elizalde-Contreras, J.M.; Bautista-Valle, M.V.; Loyola-Vargas, V.M.; Ruíz-May, E. Tissue-specific proteome characterization of avocado seed during postharvest shelf life. J. Proteom. 2021, 235, 104112. [Google Scholar] [CrossRef]
- Rodriguez, L.; Fiedler, L.; Zou, M.; Giannini, C.; Monzer, A.; Vladimirtsev, D.; Randuch, M.; Yu, Y.; Gelová, Z.; Verstraeten, I.; et al. ABP1/ABL3-TMK1 cell-surface auxin signaling directly targets PIN2-mediated auxin fluxes for root gravitropism. bioRxiv 2025. [Google Scholar] [CrossRef]
- Chen, J.G.; Ullah, H.; Young, J.C.; Sussman, M.R.; Jones, A.M. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 2001, 15, 902–911. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef]
- Lorenzo-Manzanarez, J.L.; Enríquez-Valencia, A.J.; Olivares-García, C.A.; Ibarra-Laclette, E.; Velázquez-López, O.; Ruiz-May, E.; Loyola-Vargas, V.M.; Kú-González, A.F.; Arteaga-Vázquez, M.A.; Mata-Rosas, M. Genome-wide analysis of ARF gene family and miR160 in avocado (Persea americana Mill.) and their roles in somatic embryogenesis from zygotic embryos. Planta 2025, 261, 61. [Google Scholar] [CrossRef]
- Wójcikowska, B.; Belaidi, S.; Robert, H.S. Game of thrones among AUXIN RESPONSE FACTORs—Over 30 years of MONOPTEROS research. J. Exp. Bot. 2023, 74, 6904–6921. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Bian, J.; Ren, Z.; Ta, W.; Peng, Y. Plant Aux/IAA gene family: Significance in growth, development and stress responses. Agronomy 2025, 15, 1228. [Google Scholar] [CrossRef]
- Weijers, D.; Benková, E.; Jager, K.E.; Schlereth, A.; Hamann, T.; Kientz, M.; Wilmoth, J.C.; Reed, J.W.; Jurgens, G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005, 24, 1874–1885. [Google Scholar] [CrossRef]
- Luschnig, C.; Friml, J. Over 25 years of decrypting PIN-mediated plant development. Nat. Commun. 2024, 15, 9904. [Google Scholar] [CrossRef]
- Furutani, M.; Vernoux, T.; Traas, J.; Kato, T.; Tasaka, M.; Aida, M. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 2004, 131, 5021–5030. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Forestan, C.; Canova, S.; Traas, J.; Varotto, S. ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol. 2006, 142, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Křeček, P.; Skůpa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zažímalová, E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Vanstraelen, M.; Simon, S.; Yin, K.; Carron-Arthur, A.; Nisar, N.; Tarle, G.; Cuttriss, A.J.; Searle, I.R.; Benková, E. Role of the Arabidopsis PIN6 auxin transporter in auxin homeostasis and auxin-mediated development. PLoS ONE 2013, 8, e70069. [Google Scholar] [CrossRef]
- Hu, Y.; Shani, E. Cytokinin activity—Transport and homeostasis at the whole plant, cell, and subcellular levels. New Phytol. 2023, 239, 1603–1608. [Google Scholar] [CrossRef]
- Le Hir, R.; Sorin, C.; Chakraborti, D.; Moritz, T.; Schaller, H.; Tellier, F.; Robert, S.; Morin, H.; Bako, L.; Bellini, C. ABCG9, ABCG11 and ABCG14 ABC transporters are required for vascular development in Arabidopsis. Plant J. 2013, 76, 811–824. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, X.; Liu, R.; Du, W.; Ma, M.; Han, Y.; Li, H.; Liu, L.; Hou, S. Comparative transcriptomic analysis reveals the regulatory mechanism of the gibberellic acid pathway of Tartary buckwheat (Fagopyrum tataricum (L.) Gaertn.) dwarf mutants. BMC Plant Biol. 2021, 21, 206. [Google Scholar] [CrossRef]
- Titapiwatanakun, B.; Murphy, A.S. Post-transcriptional regulation of auxin transport proteins: Cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J. Exp. Bot. 2009, 60, 1093–1107. [Google Scholar] [CrossRef]
- Wu, G.; Carville, J.S.; Spalding, E.P. ABCB19-mediated polar auxin transport modulates Arabidopsis hypocotyl elongation and the endoreplication variant of the cell cycle. Plant J. 2016, 85, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.R.; Wu, G.; Ljung, K.; Spalding, E.P. Auxin transport into cotyledons and cotyledon growth depend similarly on the ABCB19 Multidrug Resistance-like transporter. Plant J. 2009, 60, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Shang, W.; Li, L.; Song, Y.; Wang, G.; Shi, L.; Shen, Y.; Sun, Y.; He, S.; Wang, Z. Transcriptome landscape analyses of the regulatory network for zygotic embryo development in Paeonia ostii. Int. J. Mol. Sci. 2023, 24, 10715. [Google Scholar] [CrossRef]
- Do, T.H.T.; Martinoia, E.; Lee, Y. Functions of ABC transporters in plant growth and development. Curr. Opin. Plant Biol. 2018, 41, 32–38. [Google Scholar] [CrossRef]
- Swarup, R.; Péret, B. AUX/LAX family of auxin influx carriers—An overview. Front. Plant Sci. 2012, 3, 225. [Google Scholar] [CrossRef]
- Swarup, R.; Bhosale, R. Developmental roles of AUX1/LAX auxin influx carriers in plants. Front. Plant Sci. 2019, 10, 1036. [Google Scholar] [CrossRef] [PubMed]
- Zolman, B.K.; Martinez, N.; Millius, A.; Adham, A.R.; Bartel, B. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 2008, 180, 237–251. [Google Scholar] [CrossRef]
- Strader, L.C.; Culler, A.H.; Cohen, J.D.; Bartel, B. Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 2010, 153, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Hernández, H.A.; Ledezma-Rodríguez, M.; Avilez-Montalvo, R.N.; Juárez-Gómez, Y.L.; Skeete, A.; Avilez-Montalvo, J.; De-la-Peña, C.; Loyola-Vargas, V.M. Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 2019, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yang, G.; Pan, D.; Wang, X.; Han, Q.; Qin, Y.; Li, K.; Huang, G. Analysis of the plant hormone expression profile during somatic embryogenesis induction in teak (Tectona grandis). Front. Plant Sci. 2024, 15, 1429575. [Google Scholar] [CrossRef]
- Guo, R.; Hu, Y.; Aoi, Y.; Hira, H.; Ge, C.; Dai, X.; Kasahara, H.; Zhao, Y. Local conjugation of auxin by the GH3 amido synthetases is required for normal development of roots and flowers in Arabidopsis. Biochem. Biophys. Res. Commun. 2022, 589, 16–22. [Google Scholar] [CrossRef]
- Ren, H.; Gray, W.M. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef]
- Sun, R.; Wang, S.; Ma, D.; Li, Y.; Liu, C. Genome-wide analysis of cotton auxin early response gene families and their roles in somatic embryogenesis. Genes 2019, 10, 730. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, X.; Xue, X.; Liu, M.; Zhang, X.; Xiao, X.; Lai, C.; Zhang, Z.; Lai, Z.; Lin, Y. Genome-wide analysis of the SAUR gene family and function exploration of DlSAUR32 during early longan somatic embryogenesis. Plant Physiol. Biochem. 2023, 195, 362–374. [Google Scholar] [CrossRef]
- Wójcik, A.M.; Wójcikowska, B.; Gaj, M.D. Current perspectives on the auxin-mediated genetic network that controls the induction of somatic embryogenesis in plants. Int. J. Mol. Sci. 2020, 21, 1333. [Google Scholar] [CrossRef]
- Ayil-Gutiérrez, B.A.; Galaz-Ávalos, R.M.; Peña-Cabrera, E.; Loyola-Vargas, V.M. Dynamics of the concentration of IAA and some of its conjugates during the induction of somatic embryogenesis in Coffea canephora. Plant Signal. Behav. 2013, 8, e26998. [Google Scholar] [CrossRef]
- Nishimura, C.; Ohashi, Y.; Sato, S.; Kato, T.; Tabata, S.; Ueguchi, C. Genetic analysis of Arabidopsis histidine kinase genes encoding cytokinin receptors reveals their overlapping biological functions in the regulation of shoot and root growth in Arabidopsis thaliana. Plant Cell 2004, 16, 1365–1377. [Google Scholar] [CrossRef]
- Choi, J.; Hwang, I. Cytokinin: Perception, signal transduction, and role in plant growth and development. J. Plant Biol. 2007, 50, 98–108. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Kieber, J. Cytokinin signaling in plants. In Molecular Biology. The Plant Sciences 2; Howell, S.H., Ed.; Springer: New York, NY, USA, 2014; pp. 269–289. [Google Scholar] [CrossRef]
- Hwang, I.; Sakakibara, H. Cytokinin biosynthesis and perception. Physiol. Plant. 2006, 126, 528–538. [Google Scholar] [CrossRef]
- Deng, Y.; Dong, H.; Mu, J.; Ren, B.; Zheng, B.; Ji, Z.; Yang, W.C.; Liang, Y.; Zuo, J. Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 2010, 22, 1232–1248. [Google Scholar] [CrossRef] [PubMed]
- Hnatuszko-Konka, K.; Gerszberg, A.; Weremczuk-Jezyna, I.; Grzegorczyk-Karolak, I. Cytokinin signaling and de novo shoot organogenesis. Genes 2021, 12, 265. [Google Scholar] [CrossRef]
- Avilez-Montalvo, J.; Quintana-Escobar, A.O.; Méndez-Hernández, H.A.; Uc-Chuc, M.Á.; Brito-Argáez, L.; Aguilar-Hernández, V.; Galaz-Ávalos, R.M.; Loyola-Vargas, V.M. Auxin-cytokinin cross talk in somatic embryogenesis of Coffea canephora. Plants 2022, 11, 2013. [Google Scholar] [CrossRef] [PubMed]
- Raines, T.; Shanks, C.; Cheng, C.-Y.; McPherson, D.; Argueso, C.T.; Kim, H.J.; Franco-Zorrilla, J.M.; López-Vidriero, I.; Solano, R.; Vaňková, R.; et al. The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J. 2016, 85, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Gentile, D.; Serino, G.; Frugis, G. CRF transcription factors in the trade-off between abiotic stress response and plant developmental processes. Front. Genet. 2024, 15, 1377204. [Google Scholar] [CrossRef]
- Miyawaki, K.; Matsumoto-Kitano, M.; Kakimoto, T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: Tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004, 37, 128–138. [Google Scholar] [CrossRef]
- Dello Ioio, R.; Linhares, F.S.; Scacchi, E.; Casamitjana-Martinez, E.; Heidstra, R.; Costantino, P.; Sabatini, S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 2007, 17, 678–682. [Google Scholar] [CrossRef]
- Werner, S.; Bartrina, I.; Novák, O.; Strnad, M.; Werner, T.; Schmülling, T. The cytokinin status of the epidermis regulates aspects of vegetative and reproductive development in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 613488. [Google Scholar] [CrossRef] [PubMed]
- Terceros, G.C.; Resentini, F.; Cucinotta, M.; Manrique, S.; Colombo, L.; Mendes, M.A. The Importance of Cytokinins during Reproductive Development in Arabidopsis and Beyond. Int. J. Mol. Sci. 2020, 21, 8161. [Google Scholar] [CrossRef]
- Bernard, C.; Traub, M.; Kunz, H.-H.; Hach, S.; Trentmann, O.; Möhlmann, T. Equilibrative nucleoside transporter 1 (ENT1) is critical for pollen germination and vegetative growth in Arabidopsis. J. Exp. Bot. 2011, 62, 4627–4637. [Google Scholar] [CrossRef] [PubMed]
- Hirose, N.; Makita, N.; Yamaya, T.; Sakakibara, H. Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiol. 2005, 138, 196–206. [Google Scholar] [CrossRef]
- Bürkle, L.; Cedzich, A.; Döpke, C.; Stransky, H.; Okumoto, S.; Gillissen, B.; Kühn, C.; Frommer, W.B. Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 2003, 34, 13–26. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhang, J.; Yu, G.; Lu, X.; Mei, W.; Deng, H.; Zhang, G.; Chen, G.; Chu, C.; Tong, H.; et al. Endoplasmic reticulum-localized PURINE PERMEASE1 regulates plant height and grain weight by modulating cytokinin distribution in rice. Front. Plant Sci. 2020, 11, 613488. [Google Scholar] [CrossRef] [PubMed]
- Rong, C.; Zhang, R.; Liu, Y.; Chang, Z.; Liu, Z.; Ding, Y.; Ding, C. Purine permease (PUP) family gene PUP11 positively regulates the rice seed setting rate by influencing seed development. Plant Cell Rep. 2024, 43, 112. [Google Scholar] [CrossRef]
- Chiang, Y.-H.; Zubo, Y.O.; Tapken, W.; Kim, H.J.; Lavanway, A.M.; Howard, L.; Pilon, M.; Kieber, J.J.; Schaller, G.E. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012, 160, 332–348. [Google Scholar] [CrossRef]
- Schoor, S.; Farrow, S.; Blaschke, H.; Lee, S.; Perry, G.; von Schwartzenberg, K.; Emery, N.; Moffatt, B. Adenosine Kinase Contributes to Cytokinin Interconversion in Arabidopsis. Plant Physiol. 2011, 157, 659–672. [Google Scholar] [CrossRef]
- Young, L.-S.; Harrison, B.R.; Narayana Murthy, U.M.; Moffatt, B.A.; Gilroy, S.; Masson, P.H. Adenosine kinase modulates root gravitropism and cap morphogenesis in Arabidopsis. Plant Physiol. 2006, 142, 564–573. [Google Scholar] [CrossRef]
- Valencia-Lozano, E.; Cabrera-Ponce, J.L.; Barraza, A.; López-Calleja, A.C.; García-Vázquez, E.; Rivera-Toro, D.M.; de Folter, S.; Alvarez-Venegas, R. Editing of SlWRKY29 by CRISPR-activation promotes somatic embryogenesis in Solanum lycopersicum cv. Micro-Tom. PLoS ONE 2024, 19, e0301169. [Google Scholar] [CrossRef]
- Grunewald, W.; De Smet, I.; De Rybel, B.; Robert, H.S.; van de Cotte, B.; Willemsen, V.; Gheysen, G.; Weijers, D.; Friml, J.; Beeckman, T. Tightly controlled WRKY23 expression mediates Arabidopsis embryo development. EMBO Rep. 2013, 14, 1136–1142. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Li, S.; Ma, M.; Quan, C.; Tian, X.; Tu, J.; Shen, J.; Yi, B.; Fu, T.; Ma, C.; et al. The transcription factor BnaWRKY10 regulates cytokinin dehydrogenase BnaCKX2 to control cytokinin distribution and seed size in Brassica napus. J. Exp. Bot. 2023, 74, 4994–5013. [Google Scholar] [CrossRef]
- Lagacé, M.; Matton, D.P. Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta 2004, 219, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Zhang, X.; Pang, C.; Song, M.; Wei, H.; Fan, S.; Yu, S. Genome-wide analysis of the WRKY gene family in cotton. Mol. Gen. Genom. 2014, 289, 1103–1121. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Zhang, Z.; Laux, T. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell 2011, 20, 264–270. [Google Scholar] [CrossRef]
- Xiong, H.; He, H.; Chang, Y.; Miao, B.; Liu, Z.; Wang, Q.; Dong, F.; Xiong, L. Multiple roles of NAC transcription factors in plant development and stress responses. J. Integr. Plant Biol. 2025, 67, 510–538. [Google Scholar] [CrossRef]
- Li, X.; Sun, M.; Cui, Z.; Jiang, Y.; Yang, L.; Jiang, Y. Transcription factor ZmNAC19 promotes embryo development in Arabidopsis thaliana. Plant Cell Rep. 2024, 43, 244. [Google Scholar] [CrossRef]
- Kunieda, T.; Mitsuda, N.; Ohme-Takagi, M.; Takeda, S.; Aida, M.; Tasaka, M.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. Plant Cell 2008, 20, 2631–2642. [Google Scholar] [CrossRef]
- Paul, P.; Joshi, S.; Tian, R.; Diogo Junior, R.; Chakrabarti, M.; Perry, S.E. The MADS-domain factor AGAMOUS-Like18 promotes somatic embryogenesis. Plant Physiol. 2022, 188, 1617–1631. [Google Scholar] [CrossRef]
- Zheng, Q.; Perry, S. Alterations in the transcriptome of soybean in response to enhanced somatic embryogenesis promoted by orthologs of AGAMOUS-like 15 and AGAMOUS-like 18. Plant Physiol. 2014, 164, 1365–1377. [Google Scholar] [CrossRef]
- El Ouakfaoui, S.; Schnell, J.; Abdeen, A.; Colville, A.; Labbé, H.; Han, S.; Baum, B.; Laberge, S.; Miki, B. Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol. Biol. 2010, 74, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Rashotte, A.M.; Mason, M.G.; Hutchison, C.E.; Ferreira, F.J.; Schaller, G.E.; Kieber, J.J. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 11081–11085. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Dong, H.; Xue, Y.; Su, S.; Wu, Y.; Li, S.; Liu, H.; Li, H.; Han, J.; Shan, X.; et al. Transcriptomic analysis reveals somatic embryogenesis-associated signaling pathways and gene expression regulation in maize (Zea mays L.). Plant Mol. Biol. 2020, 104, 647–663. [Google Scholar] [CrossRef] [PubMed]
- Piyatrakul, P.; Putranto, R.-A.; Martin, F.; Rio, M.; Dessailly, F.; Leclercq, J.; Dufayard, J.-F.; Montoro, P. Some ethylene biosynthesis and AP2/ERF genes reveal a specific pattern of expression during somatic embryogenesis in Hevea brasiliensis. BMC Plant Biol. 2012, 12, 244. [Google Scholar] [CrossRef]
- Mantiri, F.R.; Kurdyukov, S.; Lohar, D.P.; Sharopova, N.; Saeed, N.A.; Wang, X.D.; VandenBosch, K.A.; Rose, R.J. The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiol. 2008, 146, 1622–1636. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, L.; Wu, Y.; Shen, Y.; Wu, X.; Wang, J. Analysis of global gene expression profiles to identify differentially expressed genes critical for embryo development in Brassica rapa. Plant Mol. Biol. 2014, 86, 425–442. [Google Scholar] [CrossRef]
- Zhou, X.; Guo, Y.; Zhao, P.; Sun, M.-x. Comparative analysis of WUSCHEL-related homeobox genes revealed their parent-of-origin and cell type-specific expression pattern during early embryogenesis in tobacco. Front. Plant Sci. 2018, 9, 311. [Google Scholar] [CrossRef]
- Chen, H.; Miao, Y.; Wang, K.; Bayer, M. Zygotic embryogenesis in flowering plants. In Doubled Haploid Technology: Volume 2: Hot Topics, Apiaceae, Brassicaceae, Solanaceae; Seguí-Simarro, J.M., Ed.; Springer: New York, NY, USA, 2021; pp. 73–88. [Google Scholar] [CrossRef]
- Breuninger, H.; Rikirsch, E.; Hermann, M.; Ueda, M.; Laux, T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 2008, 14, 867–876. [Google Scholar] [CrossRef]
- Kao, P.; Schon, M.A.; Mosiolek, M.; Enugutti, B.; Nodine, M.D. Gene expression variation in Arabidopsis embryos at single-nucleus resolution. Development 2021, 148, dev199589. [Google Scholar] [CrossRef]
- Amara, I.; Zaidi, I.; Masmoudi, K.; Ludevid, M.D.; Pagès, M.; Goday, A.; Brini, F. Insights into late embryogenesis abundant (LEA) proteins in plants: From structure to the functions. Am. J. Plant Sci. 2014, 5, 3440–3455. [Google Scholar] [CrossRef]
- Olvera-Carrillo, Y.; Campos, F.; Reyes, J.L.; Garciarrubio, A.; Covarrubias, A.A. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis thaliana. Plant Physiol. 2010, 154, 373–390. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 441–445. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, M.J.; Terrile, M.C.; Correa-Aragunde, N.; Colman, S.L.; Izquierdo-Álvarez, A.; Fiol, D.F.; París, R.; Sánchez-López, N.; Marina, A.; Calderón Villalobos, L.I.A.; et al. Regulation of SCFTIR1/AFBs E3 ligase assembly by S-nitrosylation of Arabidopsis SKP1-like1 impacts on auxin signaling. Redox Biol. 2018, 18, 200–210. [Google Scholar] [CrossRef]
- Kim, H.J.; Kieber, J.J.; Schaller, G.E. The rice F-box protein KISS ME DEADLY2 functions as a negative regulator of cytokinin signalling. Plant Signal. Behav. 2013, 8, e26434. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Toriyama, K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 2011, 62, 437–460. [Google Scholar] [CrossRef]
- Borisjuk, N.; Sitailo, L.; Adler, K.; Malysheva, L.; Tewes, A.; Borisjuk, L.; Manteuffel, R. Calreticulin expression in plant cells: Developmental regulation, tissue specificity and intracellular distribution. Planta 1998, 206, 504–514. [Google Scholar] [CrossRef]
- Jia, X.-Y.; He, L.-H.; Jing, R.-L.; Li, R.-Z. Calreticulin: Conserved protein and diverse functions in plants. Physiol. Plant. 2009, 136, 127–138. [Google Scholar] [CrossRef]
- Quintana-Escobar, A.O.; Bojórquez-Velázquez, E.; Ruiz-May, E.; Loyola-Vargas, V.M. Proteomic approach during the induction of somatic embryogenesis in Coffea canephora. Plants 2023, 12, 4095. [Google Scholar] [CrossRef]
- Kawadza, D.; Dikobe, T.; Chatukuta, P.; Takundwa, M.; Bobo, E.; Sehlabane, K.; Ruzvidzo, O. An Arabidopsis maternal effect embryo arrest protein is an adenylyl cyclase with predicted roles in embryo development and response to abiotic stress. Open Biotechnol. J. 2023, 17, e187407072212060. [Google Scholar] [CrossRef]
- Nolan, K.E.; Kurdyukov, S.; Rose, R.J. Expression of the SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (SERK1) gene is associated with developmental change in the life cycle of the model legume Medicago truncatula. J. Exp. Bot. 2009, 60, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Michniewicz, M.; Zago, M.K.; Abas, L.; Weijers, D.; Schweighofer, A.; Meskiene, I.; Heisler, M.G.; Ohno, C.; Zhang, J.; Huang, F.; et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 2007, 130, 1044–1056. [Google Scholar] [CrossRef] [PubMed]
- Rehman, H.M.; Nawaz, M.A.; Bao, L.; Shah, Z.H.; Lee, J.-M.; Ahmad, M.Q.; Chung, G.; Yang, S.H. Genome-wide analysis of Family-1 UDP-glycosyltransferases in soybean confirms their abundance and varied expression during seed development. J. Plant Physiol. 2016, 206, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.H.; Ma, X.M.; Han, P.; Wang, B.; Sun, Y.G.; Zhang, G.Z.; Li, Y.j.; Hou, B.K. UGT74D1 is a novel auxin gluycosyltransferase from Arabidopsis thaliana. PLoS ONE 2013, 8, e61705. [Google Scholar] [CrossRef]
- Aoi, Y.; Hira, H.; Hayakawa, Y.; Liu, H.; Fukui, K.; Dai, X.; Tanaka, K.; Hayashi, K.i.; Zhao, Y.; Kasahara, H. UDP-glucosyltransferase UGT84B1 regulates the levels of indole-3-acetic acid and phenylacetic acid in Arabidopsis. Biochem. Biophys. Res. Commun. 2020, 532, 244–250. [Google Scholar] [CrossRef]
- Wang, J.; Ma, X.M.; Kojima, M.; Sakakibara, H.; Hou, B.K. Glucosyltransferase UGT76C1 finely modulates cytokinin responses via cytokinin N-glucosylation in Arabidopsis thaliana. Plant Physiol. Biochem. 2013, 65, 9–16. [Google Scholar] [CrossRef]
- O’Neill, J.P.; Colon, K.T.; Jenik, P.D. The onset of embryo maturation in Arabidopsis is determined by its developmental stage and does not depend on endosperm cellularization. Plant J. 2019, 99, 286–301. [Google Scholar] [CrossRef]
- Collins, C.; Dewitte, W.; Murray, J.A.H. D-type cyclins control cell division and developmental rate during Arabidopsis seed development. J. Exp. Bot. 2012, 63, 3571–3586. [Google Scholar] [CrossRef]
- Arc, E.; Sechet, J.; Corbineau, F.o.; Rajjou, L.c.; Marion-Poll, A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013, 4, 63. [Google Scholar] [CrossRef]
- Leprince, O.; Pellizzaro, A.; Berriri, S.; Buitink, J. Late seed maturation: Drying without dying. J. Exp. Bot. 2016, 68, 827–841. [Google Scholar] [CrossRef]
- Verma, S.; Attuluri, V.P.S.; Robert, H.S. Transcriptional control of Arabidopsis seed development. Planta 2022, 255, 90. [Google Scholar] [CrossRef]
- Djami-Tchatchou, A.T.; Straker, C.J. The isolation of high quality RNA from the fruit of avocado (Persea americana Mill.). S. Afr. J. Bot. 2012, 78, 44–46. [Google Scholar] [CrossRef]
- Nath, O.; Fletcher, S.J.; Hayward, A.; Shaw, L.M.; Masouleh, A.K.; Furtado, A.; Henry, R.J.; Mitter, N. A haplotype resolved chromosomal level avocado genome allows analysis of novel avocado genes. Hortic. Res. 2022, 9, uhac157. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quintana-Escobar, A.O.; Couoh-Cauich, M.D.; Vargas-Morales, B.V.; Mata-Rosas, M.; Ruíz-May, E.; Loyola-Vargas, V.M. Identification of Auxin, Cytokinin, Transcription Factors, and Other Zygotic Embryogenesis-Related Genes in Persea americana: A Transcriptomic-Based Study. Plants 2025, 14, 3288. https://doi.org/10.3390/plants14213288
Quintana-Escobar AO, Couoh-Cauich MD, Vargas-Morales BV, Mata-Rosas M, Ruíz-May E, Loyola-Vargas VM. Identification of Auxin, Cytokinin, Transcription Factors, and Other Zygotic Embryogenesis-Related Genes in Persea americana: A Transcriptomic-Based Study. Plants. 2025; 14(21):3288. https://doi.org/10.3390/plants14213288
Chicago/Turabian StyleQuintana-Escobar, Ana O., Marcos David Couoh-Cauich, Brigitte Valeria Vargas-Morales, Martín Mata-Rosas, Eliel Ruíz-May, and Víctor M. Loyola-Vargas. 2025. "Identification of Auxin, Cytokinin, Transcription Factors, and Other Zygotic Embryogenesis-Related Genes in Persea americana: A Transcriptomic-Based Study" Plants 14, no. 21: 3288. https://doi.org/10.3390/plants14213288
APA StyleQuintana-Escobar, A. O., Couoh-Cauich, M. D., Vargas-Morales, B. V., Mata-Rosas, M., Ruíz-May, E., & Loyola-Vargas, V. M. (2025). Identification of Auxin, Cytokinin, Transcription Factors, and Other Zygotic Embryogenesis-Related Genes in Persea americana: A Transcriptomic-Based Study. Plants, 14(21), 3288. https://doi.org/10.3390/plants14213288







