TaCML49-B, a Calmodulin-like Protein, Interacts with TaIQD23 to Positively Regulate Salt Tolerance in Wheat
Abstract
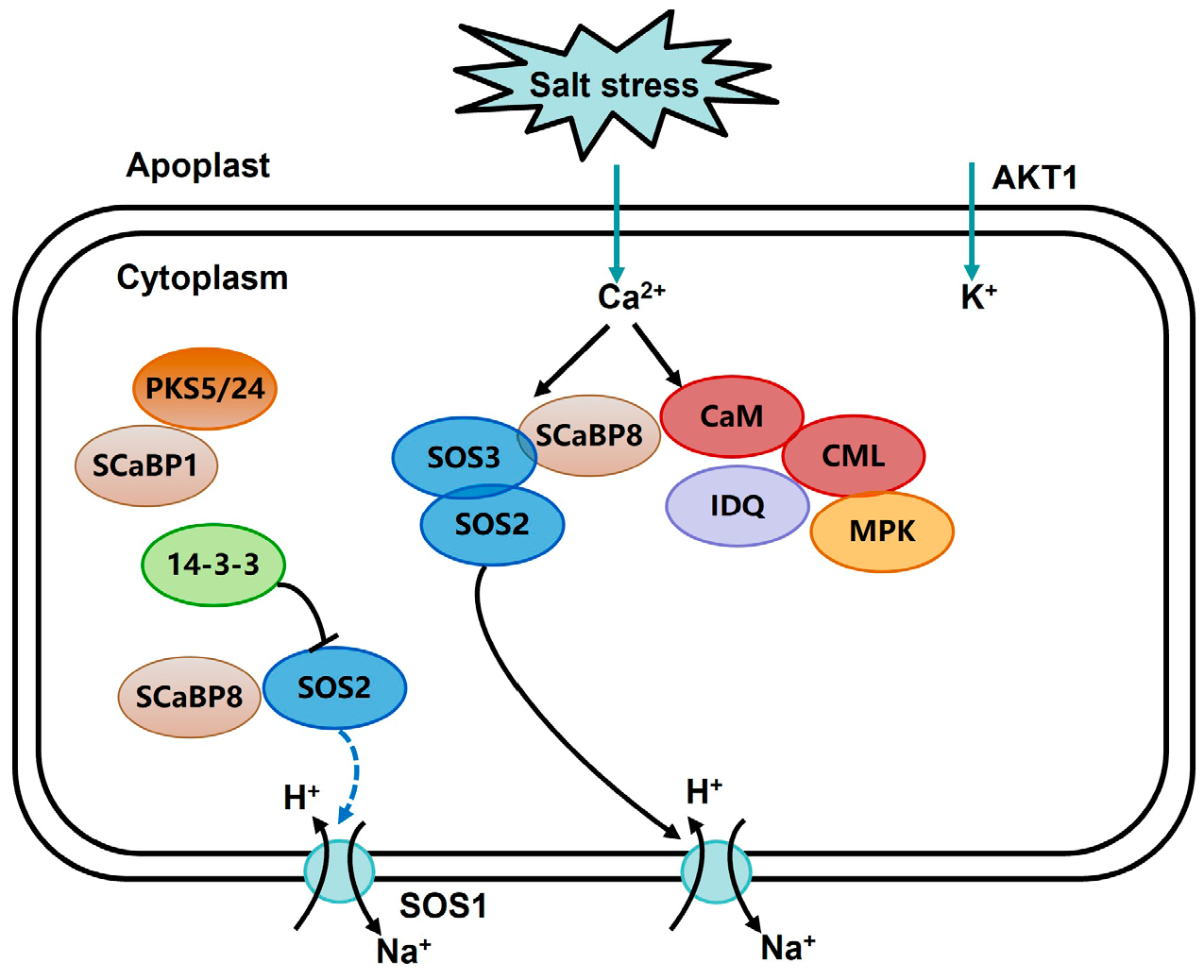
1. Introduction
2. Results
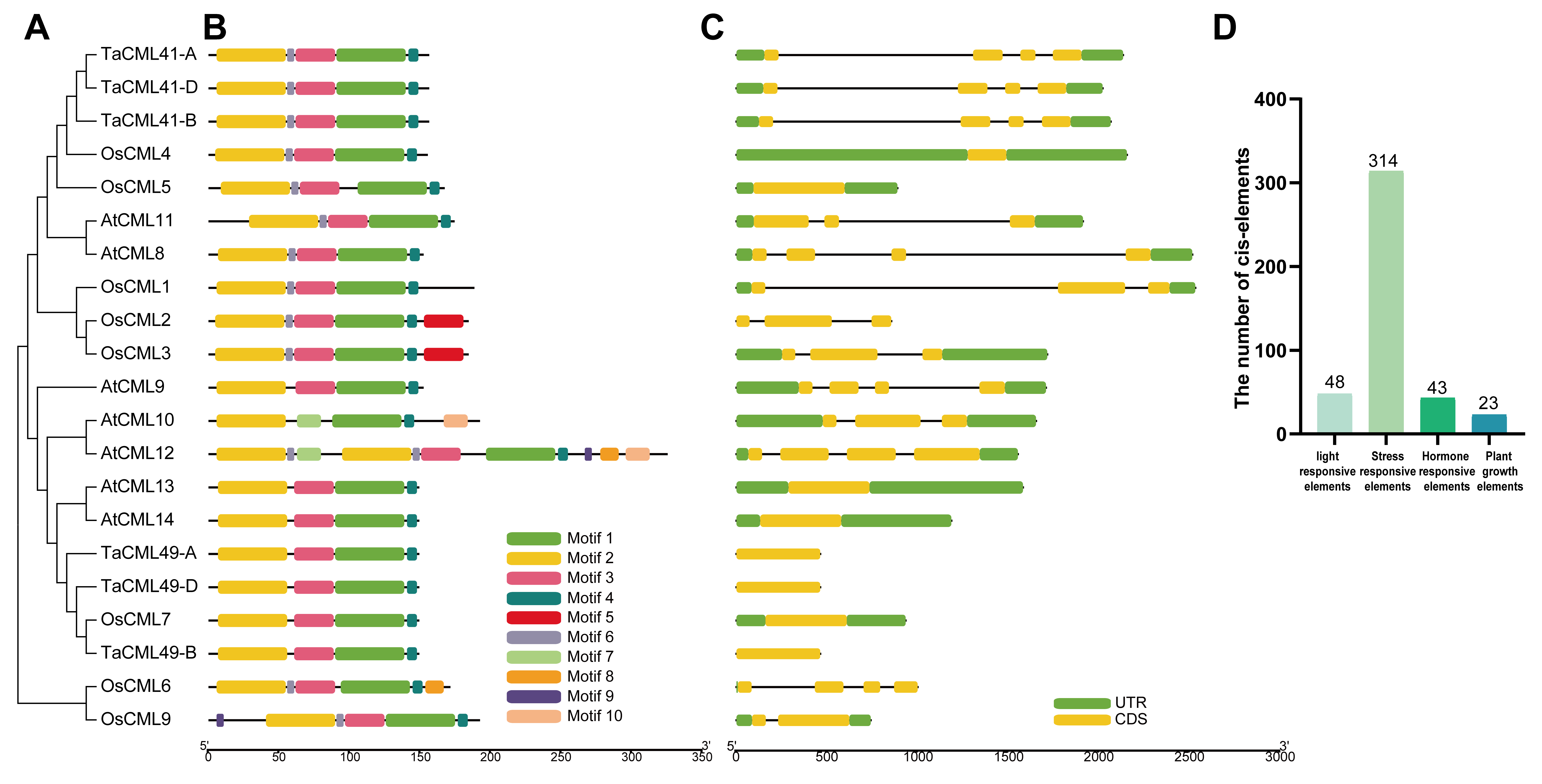
2.1. Subgroup II CaM/CMLs Are Involved in Responses to Multiple Abiotic Stresses
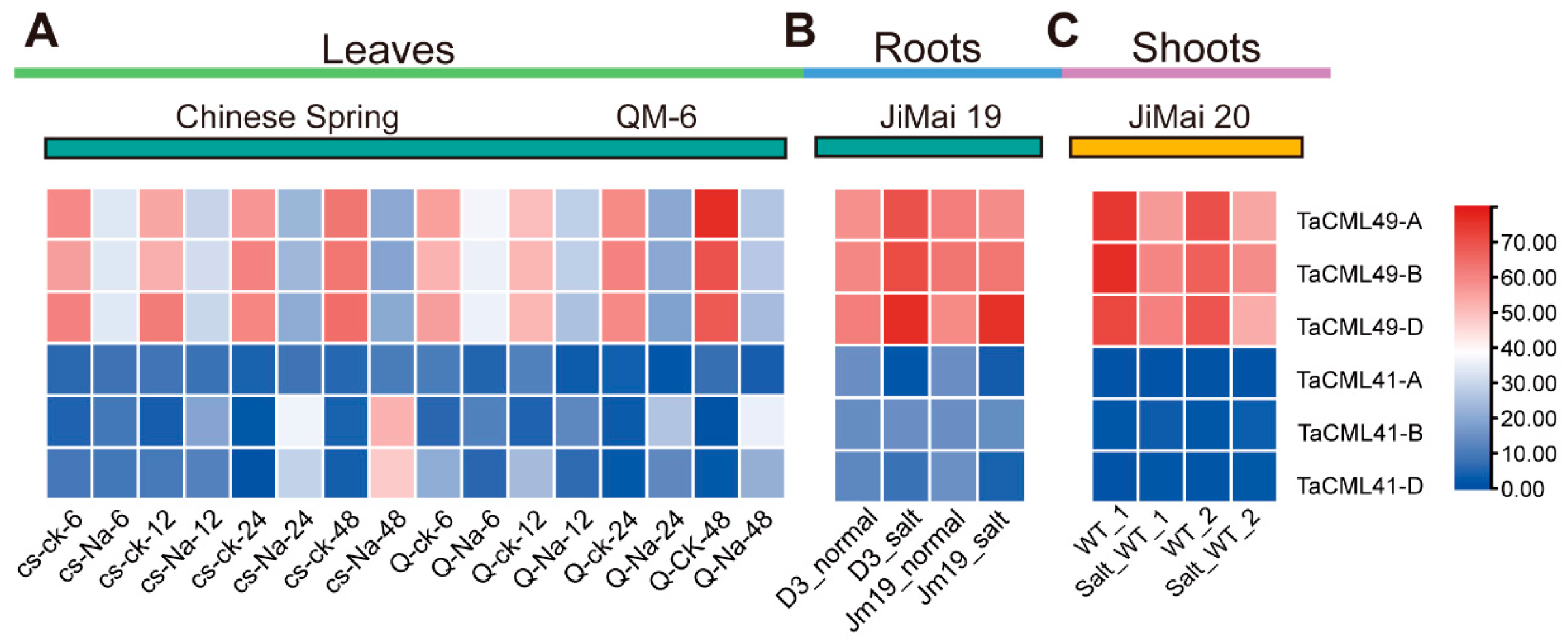
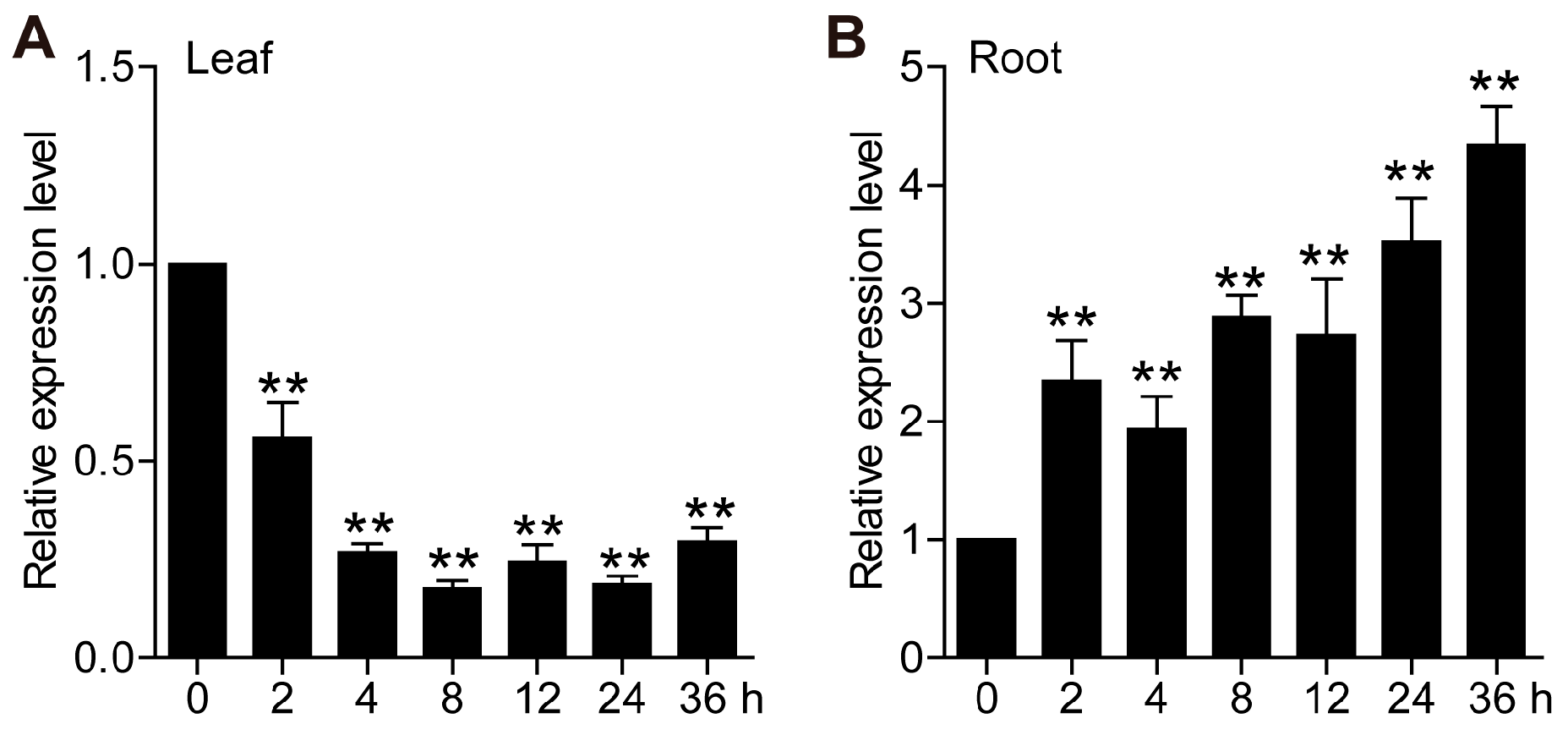
2.2. Expression Pattern Analysis of TaCML49-B Under Salt Stress
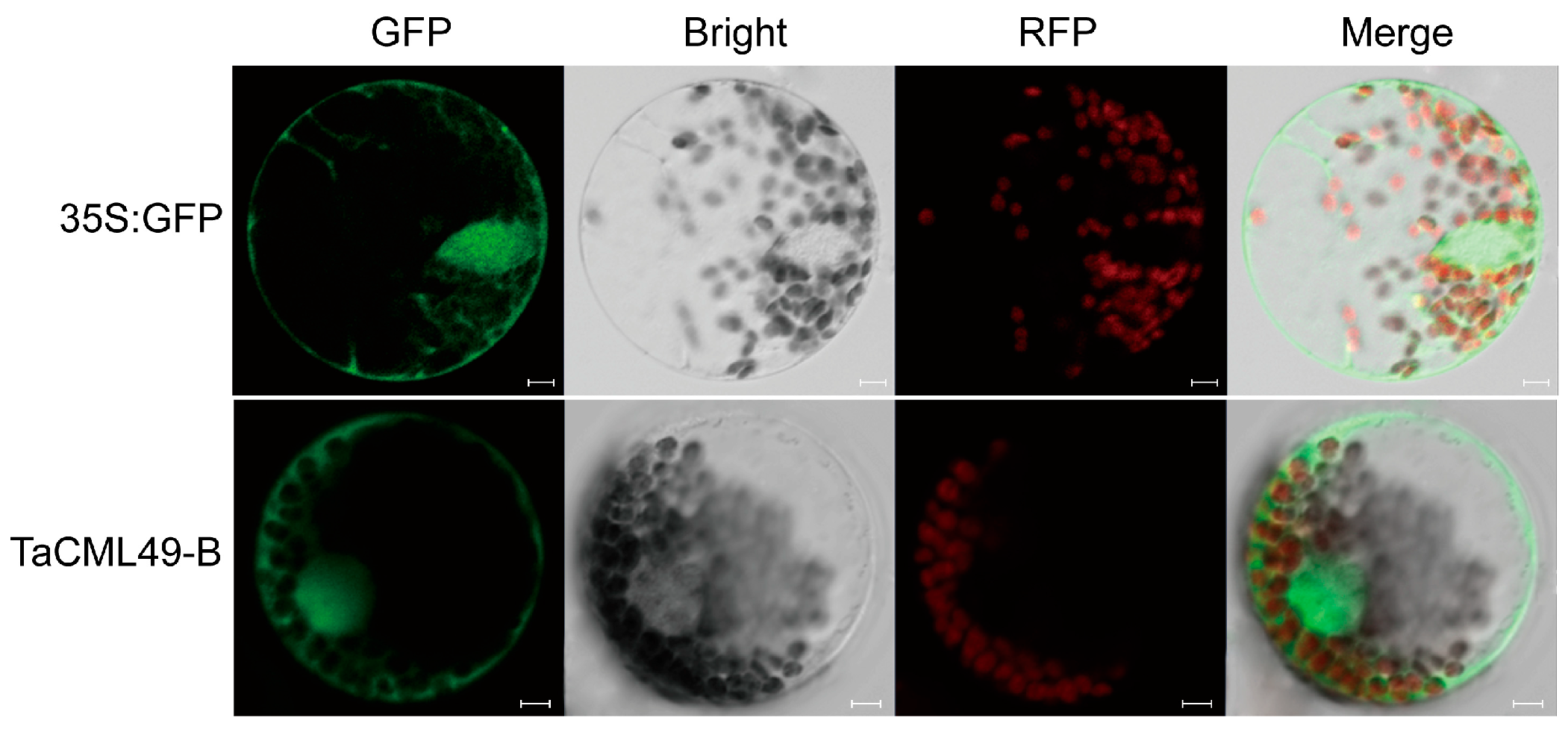
2.3. TaCML49-B Was Located to Nucleus, Cytoplasm and Membrane
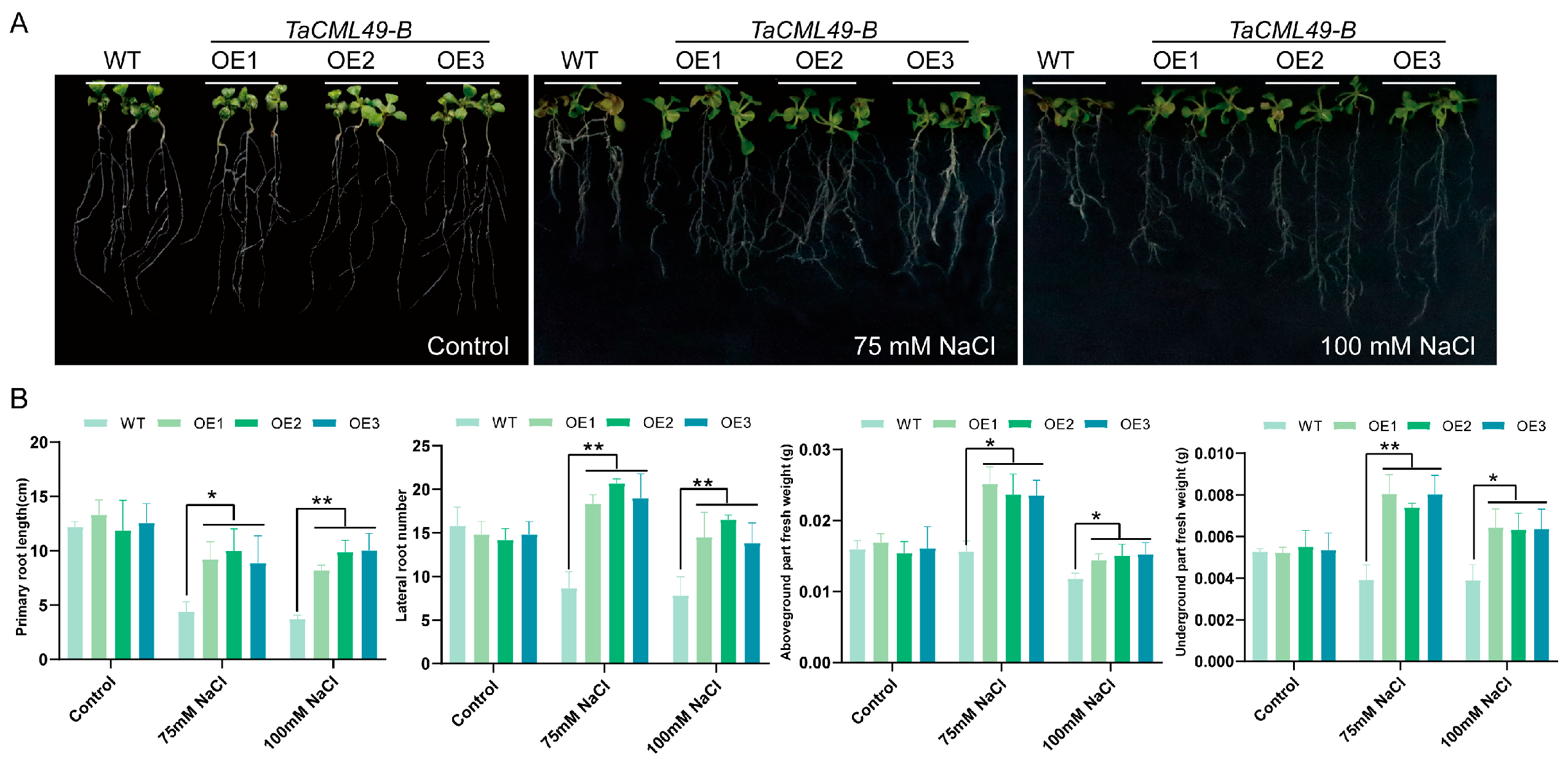
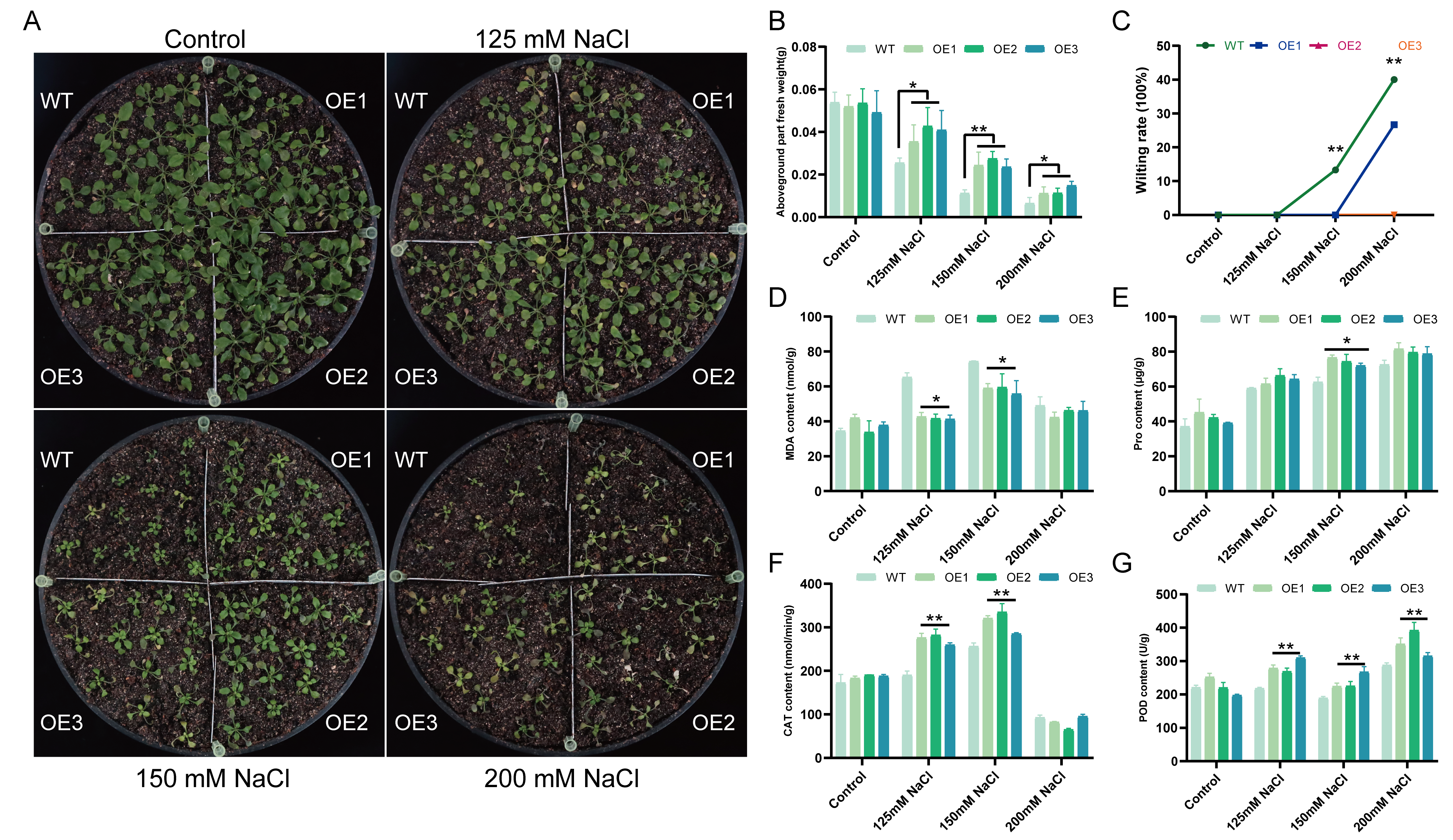
2.4. Overexpression of TaCML49-B Enhances Salt Tolerance in Arabidopsis
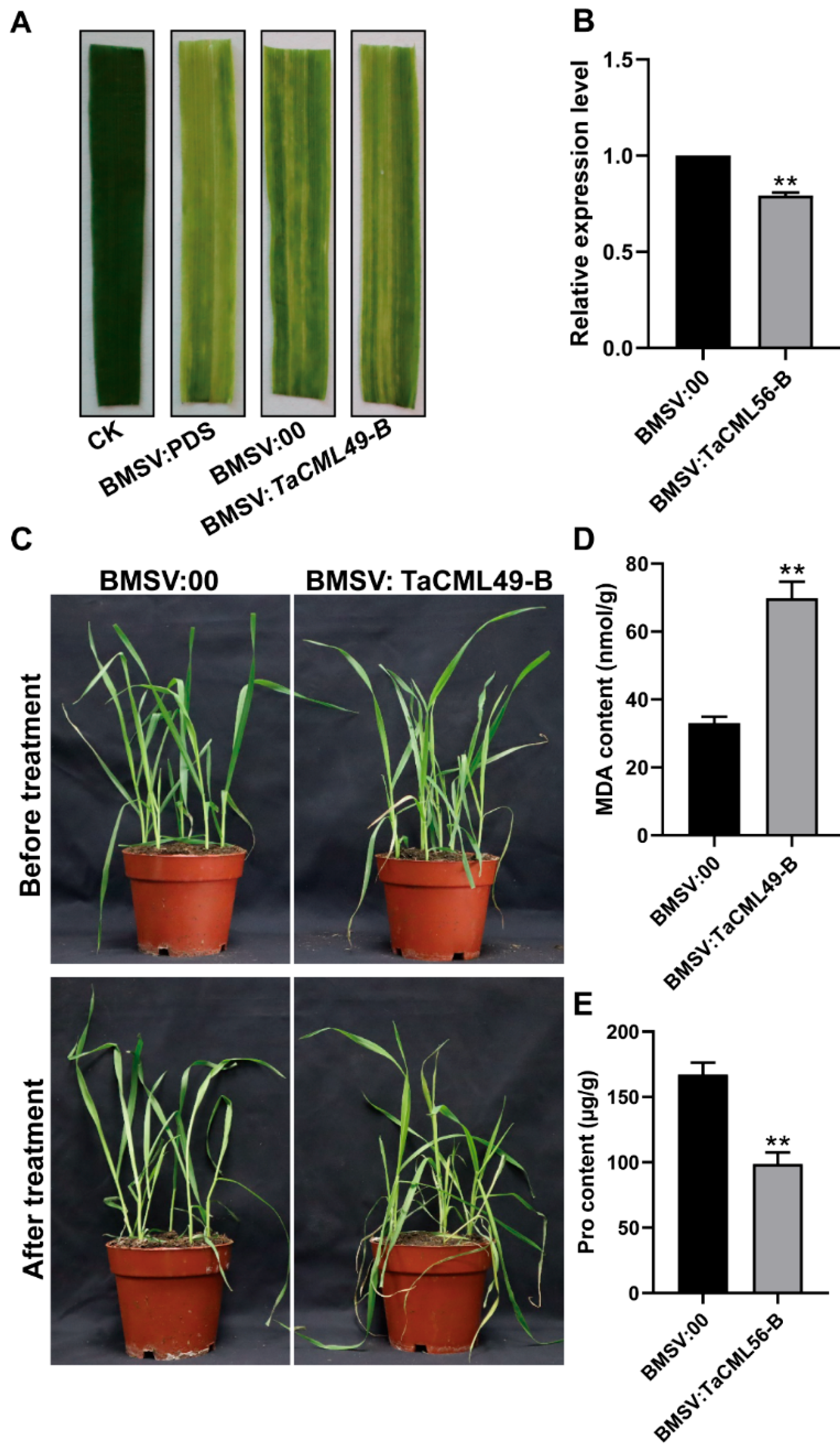
2.5. Silencing of TaCML49-B Alters Salt Response in Wheat
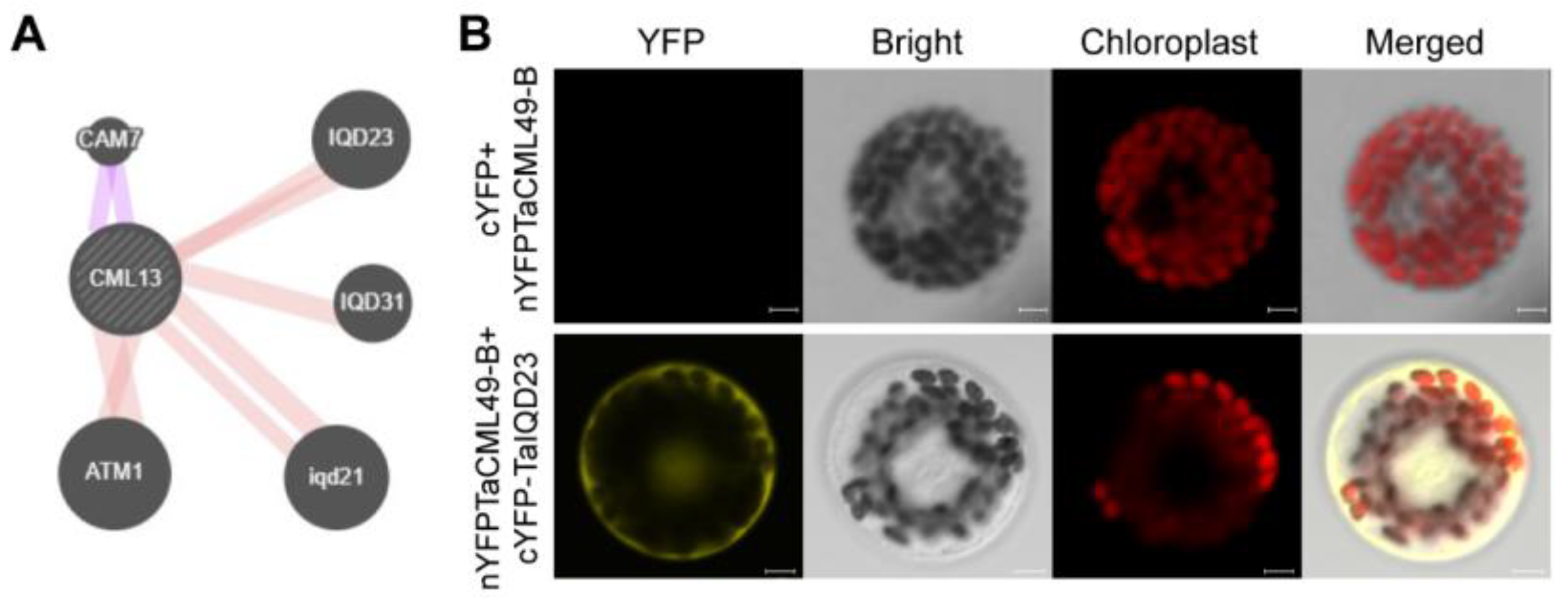
2.6. Protein Interaction Network of TaCML49-B
3. Discussion
4. Materials and Methods
4.1. Sequence Characteristics and Phylogenetic Analysis of 21 CaM/CMLs
4.2. Exon-Intron Structure, Motif Analysis and Cis-Elements Analysis of 21 CaM/CMLs
4.3. Expression Patterns Analysis of TaCML41 and TaCML49
4.4. Plant Material and Stress Treatments
4.5. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
4.6. Subcellular Localization of TaCML49-B
4.7. Transformation of Arabidopsis and Propagation of Positive Seedlings
4.8. Salt-Stress Assays of Transgenic Arabidopsis Plants
4.9. BSMV-VIGS-Induced TaCML49-B Silencing Assays
4.10. Prediction and Validation of TaCML49-B Interaction Networks
4.11. Bimolecular Fluorescence Complementation (BiFC) Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- McAinsh, M.R.; Pittman, J.K. Shaping the calcium signature. New Phytol. 2009, 181, 275–294. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Gao, Y.; Du, Y.; Du, J.; Han, Y. Genome-wide analysis of the CML gene family and its response to melatonin in common bean (Phaseolus vulgaris L.). Sci. Rep. 2023, 13, 1196. [Google Scholar] [CrossRef]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Bergey, D.R.; Kandel, R.; Tyree, B.K.; Dutt, M.; Dhekney, S.A. The Role of Calmodulin and Related Proteins in Plant Cell Function: An Ever-Thickening Plot. Springer Sci. Rev. 2014, 2, 145–159. [Google Scholar] [CrossRef]
- Zeng, H.; Xu, L.; Singh, A.; Wang, H.; Du, L.; Poovaiah, B.W. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 2015, 6, 600. [Google Scholar] [CrossRef] [PubMed]
- Perochon, A.; Aldon, D.; Galaud, J.P.; Ranty, B. Calmodulin and calmodulin-like proteins in plant calcium signaling—ScienceDirect. Biochimie 2011, 93, 2048–2053. [Google Scholar] [CrossRef]
- Adenike, O.; Kim, M.; Alexandra, D.; Midhat, U.; Snedden, W.A. Arabidopsis calmodulin-like proteins, cml15 and cml16 possess biochemical properties distinct from calmodulin and show non-overlapping tissue expression patterns. Front. Plant Sci. 2017, 8, 2175. [Google Scholar] [CrossRef]
- Xu, B.; Cheval, C.; Laohavisit, A.; Hocking, B.; Chiasson, D.; Olsson, T.S.G.; Shirasu, K.; Faulkner, C.; Gilliham, M. A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytol. 2017, 215, 77–84. [Google Scholar] [CrossRef]
- Nakahara, K.S.; Masuta, C.; Yamada, S.; Shimura, H.; Kashihara, Y.; Wada, T.S.; Meguro, A.; Goto, K.; Tadamura, K.; Sueda, K.; et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA 2012, 109, 10113–10118. [Google Scholar] [CrossRef]
- Munir, S.; Liu, H.; Xing, Y.; Hussain, S.; Ouyang, B.; Zhang, Y.; Li, H.; Ye, Z. Overexpression of calmodulin-like (ShCML44) stress-responsive gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses. Sci. Rep. 2016, 6, 31772. [Google Scholar] [CrossRef]
- Cho, K.M.; Nguyen, H.T.; Kim, S.Y.; Shin, J.S.; Cho, D.H.; Hong, S.B.; Shin, J.S.; Ok, S.H. CML10, a variant of calmodulin, modulates ascorbic acid synthesis. New Phytol. 2016, 209, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Vallone, R.; Verde, V.L.; D’Onofrio, M.; Giorgetti, A.; Astegno, A. Metal binding affinity and structural properties of calmodulin-like protein 14 from Arabidopsis thaliana. Protein Sci. 2016, 25, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Chung, P.J.; Park, S.H.; Redillas, M.C.F.R.; Kim, Y.S.; Suh, J.W.; Kim, J.K. Overexpression of OsERF48 causes regulation of OsCML16, a calmodulin-like protein gene that enhances root growth and drought tolerance. Plant Biotechnol. J. 2017, 15, 1295–1308. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Sun, X.; Duanmu, H.; Zhu, D.; Yu, Y.; Cao, L.; Liu, A.; Jia, B.; Xiao, J.; Zhu, Y. GsCML27, a Gene Encoding a Calcium-Binding Ef-Hand Protein from Glycine soja, Plays Differential Roles in Plant Responses to Bicarbonate, Salt and Osmotic Stresses. PLoS ONE 2015, 10, e0141888. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, W.; Liu, L.; Su, Y.; Li, Y.; Jia, W.; Jiao, B.; Wang, J.; Yang, F.; Dong, F.; et al. Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes in wheat (Triticum aestivum L.). Plant Signal. Behavior 2022, 17, 2013646. [Google Scholar] [CrossRef]
- Kalaipandian, S.; Xue, G.P.; Rae, A.L.; Glassop, D.; Bonnett, G.D.; Mcintyre, L.C. Overexpression of TaCML20, a calmodulin-like gene, enhances water soluble carbohydrate accumulation and yield in wheat. Physiol. Plant. 2019, 165, 790–799. [Google Scholar] [CrossRef]
- Lu, L.; Rong, W.; Zhou, R.; Huo, N.; Zhang, Z.J. TaCML36, a wheat calmodulin-like protein, positively participates in an immune response to Rhizoctonia cerealis. Crop J. 2019, 7, 608–618. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Dong, F.; Zou, J.; Gao, C.; Zhu, Z.; Liu, Y. Multiple roles of wheat calmodulin genes during stress treatment and TaCAM2-D as a positive regulator in response to drought and salt tolerance. Int. J. Biol. Macromol. 2022, 220, 985–997. [Google Scholar] [CrossRef]
- Guo, C.; Zhou, J.; Li, D. New Insights Into Functions of IQ67-Domain Proteins. Front. Plant Sci. 2020, 11, 614851. [Google Scholar] [CrossRef]
- Xu, F.; Wang, L.; Xu, J.; Chen, Q.; Ma, C.; Huang, L.; Li, G.; Luo, M. GhIQD10 interacts with GhCaM7 to control cotton fiber elongation via calcium signaling. Crop J. 2023, 11, 447–456. [Google Scholar] [CrossRef]
- Li, X.; Su, F.; Xiang, J.; Zhang, M.; Chen, X.; Hu, D.; Yu, D.; Wang, H. GmIQD63 functions as a novel GmCDPK38-interacting protein in soybean defense against the common cutworm. Planta 2025, 262, 103. [Google Scholar] [CrossRef] [PubMed]
- Kirlioğlu, T.; Okay, A.; Aras, E.S.; Büyük, I. Investigation and computational analysis of the IQD gene family in common bean (Phaseolus vulgaris L.): Expression profiling under salt stress. Turk. J. Bot. 2024, 48, 532–550. [Google Scholar] [CrossRef]
- Farooq, M.; Zahra, N.; Ullah, A.; Nadeem, F.; Rehman, A.; Kapoor, R.; Al-Hinani, M.S.; Siddique, K.H. Salt Stress in Wheat: Effects, Tolerance Mechanisms, and Management. J. Soil Sci. Plant Nutr. 2025, 24, 8151–8173. [Google Scholar] [CrossRef]
- Guo, X.; Wu, C.; Wang, D.; Wang, G.; Jin, K.; Zhao, Y.; Tian, J.; Deng, Z. Conditional QTL mapping for seed germination and seedling traits under salt stress and candidate gene prediction in wheat. Sci. Rep. 2022, 12, 21010. [Google Scholar] [CrossRef]
- Abdellaoui, R.; Elkelish, A.; El-Keblawy, A.; Mighri, H.; Boughalleb, F.; Bakhshandeh, E. Editorial: Halophytes: Salt stress tolerance mechanisms and potential use. Front. Plant Sci. 2023, 14, 1218184. [Google Scholar] [CrossRef]
- Khan, M.S.; Rizvi, A.; Saif, S.; Zaidi, A. Phosphate-Solubilizing Microorganisms in Sustainable Production of Wheat: Current Perspective. Probiotics Agroecosyst. 2017, 3, 51–81. [Google Scholar] [CrossRef]
- Zafar, S.; Hasnain, Z.; Anwar, S.; Perveen, S.; Iqbal, N.; Noman, A.; Ali, M. Influence of melatonin on antioxidant defense system and yield of wheat (Triticum aestivum L.) genotypes under saline condition. Pak. J. Bot. 2019, 51, 1987–1994. [Google Scholar] [CrossRef]
- Symonds, K.; Teresinski, H.; Hau, B.; Chiasson, D.; Benidickson, K.; Plaxton, W.; Snedden, W.A. Arabidopsis CML13 and CML14 Have Essential and Overlapping Roles inPlant Development. Plant Cell Physiol. 2024, 65, 228–242. [Google Scholar] [CrossRef]
- Yan, Z.; Deng, R.; Zhang, H.; Li, J.; Zhu, S. Transcriptome analysis of floret opening and closure both Indica and Japonica rice. 3 Biotech 2022, 12, 188. [Google Scholar] [CrossRef]
- Chinpongpanich, A.; Limruengroj, K.; Phean, O.P.S.; Limpaseni, T.; Buaboocha, T. Expression analysis of calmodulin and calmodulin-like genes from rice, Oryza sativa L. BMC Res. Notes 2012, 5, 625. [Google Scholar] [CrossRef]
- Berridge, M.J.; Lipp, P.; Bootman, M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000, 1, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Hetherington, A.M.; Brownlee, C. The generation of Ca(2+) signals in plants. Annu. Rev. Plant Biol. 2004, 55, 401–427. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Pandey, G.K. Expressional analysis and role of calcium regulated kinases in abiotic stress signaling. Curr. Genom. 2010, 11, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Magnan, F.; Ranty, B.; Charpenteau, M.; Sotta, B.; Galaud, J.P.; Aldon, D. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 2008, 56, 575–589. [Google Scholar] [CrossRef]
- Lucchin, A.; Fouassier, H.; Robe, E.; Mbengue, M.; Aguilar, M.; San Clemente, H.; Vert, G.; Galaud, J.P.; Aldon, D. The calcium sensor AtCML8 contributes to Arabidopsis plant cell growth by modulating the brassinosteroid signaling pathway. Plant J. 2025, 121, e17179. [Google Scholar] [CrossRef]
- Bürstenbinder, K.; Savchenko, T.; Müller, J.; Adamson, A.W.; Stamm, G.; Kwong, R.; Zipp, B.J.; Dinesh, D.C.; Abel, S. Arabidopsis calmodulin-binding protein IQ67-domain 1 localizes to microtubules and interacts with kinesin light chain-related protein-1. J. Biol. Chem. 2013, 288, 1871–1882. [Google Scholar] [CrossRef]
- Yang, X.; Kirungu, J.N.; Magwanga, R.O.; Xu, Y.; Pu, L.; Zhou, Z.; Hou, Y.; Cai, X.; Wang, K.; Liu, F. Knockdown of GhIQD31 and GhIQD32 increases drought and salt stress sensitivity in Gossypium hirsutum. Plant Physiol. Biochem. 2019, 144, 166–177. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; Mcwilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2. 0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Shi, W.-Y.; Du, Y.-T.; Ma, J.; Min, D.-H.; Jin, L.-G.; Chen, J.; Chen, M.; Zhou, Y.-B.; Ma, Y.-Z.; Xu, Z.-S.; et al. The WRKY Transcription Factor GmWRKY12 Confers Drought and Salt Tolerance in Soybean. Int. J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Gu, B.; Huang, G.; Tian, Y.; Quan, J.; Lindqvist-Kreuze, H.; Shan, W. Conserved RXLR Effector Genes of Phytophthora infestans Expressed at the Early Stage of Potato Infection Are Suppressive to Host Defense. Front. Plant Sci. 2017, 8, 2155. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.-F.; Zhao, W.-Y.; Fu, J.-D.; Liu, Y.-W.; Chen, M.; Zhou, Y.-B.; Ma, Y.-Z.; Xu, Z.-S.; Xi, Y.-J. Genome-Wide Analysis of CDPK Family in Foxtail Millet and Determination of SiCDPK24 Functions in Drought Stress. Front. Plant Sci. 2018, 9, 691. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, G.; Li, G.; Yuan, S.; Wang, C.; Xie, Y.; Guo, T.; Kang, G.; Wang, D. TaPHT1;9-4B and its transcriptional regulator TaMYB4-7D contribute to phosphate uptake and plant growth in bread wheat. New phytol. 2021, 231, 1968–1983. [Google Scholar] [CrossRef]
- Wang, P.; Cui, Y.; Ren, Q.; Yan, B.; Zhao, Y.; Yu, P.; Gao, G.; Shi, H.; Chang, S.; Chang, Y.Z. Mitochondrial ferritin attenuates cerebral ischaemia/reperfusion injury by inhibiting ferroptosis. Cell Death Dis. 2021, 12, 447. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ru, J.; Hao, J.; Hao, B.; Ji, X.; Yang, J.; Wang, H.; Quan, B.; Guo, P.; Zhao, J.; Shi, H.; et al. TaCML49-B, a Calmodulin-like Protein, Interacts with TaIQD23 to Positively Regulate Salt Tolerance in Wheat. Plants 2025, 14, 3163. https://doi.org/10.3390/plants14203163
Ru J, Hao J, Hao B, Ji X, Yang J, Wang H, Quan B, Guo P, Zhao J, Shi H, et al. TaCML49-B, a Calmodulin-like Protein, Interacts with TaIQD23 to Positively Regulate Salt Tolerance in Wheat. Plants. 2025; 14(20):3163. https://doi.org/10.3390/plants14203163
Chicago/Turabian StyleRu, Jingna, Jiamin Hao, Bingqing Hao, Xiaoqian Ji, Jiale Yang, Hongtao Wang, Baoquan Quan, Pengyan Guo, Jiping Zhao, Huawei Shi, and et al. 2025. "TaCML49-B, a Calmodulin-like Protein, Interacts with TaIQD23 to Positively Regulate Salt Tolerance in Wheat" Plants 14, no. 20: 3163. https://doi.org/10.3390/plants14203163
APA StyleRu, J., Hao, J., Hao, B., Ji, X., Yang, J., Wang, H., Quan, B., Guo, P., Zhao, J., Shi, H., & Xu, Z. (2025). TaCML49-B, a Calmodulin-like Protein, Interacts with TaIQD23 to Positively Regulate Salt Tolerance in Wheat. Plants, 14(20), 3163. https://doi.org/10.3390/plants14203163






