Abstract
Ribosome hibernation is a conserved translational stress response in bacteria, regulated by the hibernation-promoting factor (HPF). Plastid-specific ribosomal protein 1 (PSRP1) is the chloroplast ortholog of bacterial HPF. Although bacterial HPFs have been extensively characterized, both structurally and mechanistically, the physiological roles and mechanisms of PSRP1 in plant chloroplasts remain unclear. Here, we aimed to clarify the role of PSRP1 in chloroplast ribosome hibernation by examining its function under dark-stress conditions in the moss Physcomitrium patens. The PSRP1 knockout mutant exhibited moderate but statistically significant growth defects under both long- and short-day conditions compared to those of the wild-type plants. Moreover, the mutant displayed pronounced growth delay when co-cultured with wild-type plants, indicating a competitive disadvantage. Under dark conditions, wild-type plants exhibit increased PSRP1 protein accumulation, whereas the knockout mutant displayed reduction in chloroplast rRNA content. Notably, although PSRP1 is capable of inducing 100S dimers, we detected no chloroplast 100S dimers either in vivo or in vitro, suggesting a chloroplast-specific ribosome protection mechanism distinct from that of bacteria. These findings reveal PSRP1-mediated chloroplast ribosome protection and could provide new insights into plant stress tolerance.
1. Introduction
Chloroplasts are primary sources of fixed carbon and chemical energy in most ecosystems on Earth [1]. The endosymbiotic theory states that chloroplasts evolved when ancestral eukaryotic cells engulfed a cyanobacterium [2,3,4,5]. This primary event occurred approximately one billion years ago and led to the emergence of three evolutionary lineages: glaucophytes, rhodophytes (red algae), and chlorophytes (green algae). Land plants diverged from chlorophytes approximately 400–475 million years ago [6,7]. Throughout evolutionary history, numerous cyanobacteria-derived genes have migrated to the nuclear genome. However, some genes are retained within the chloroplast genome, where bacterial transcription and translation systems continue [4,8].
Protein synthesis, a critical process that accounts for nearly half of the cellular resources [9,10,11,12], is executed in chloroplasts by 70S ribosomes composed of 50S and 30S subunits, which retain characteristics inherited from the cyanobacterial ancestor. Under limited light and nutrient conditions, reducing protein synthesis in chloroplasts may be crucial for conserving cellular energy and nutrient reserves, ultimately promoting plant survival.
Under specific stress conditions, bacterial protein synthesis is regulated by various mechanisms, including new ribosome synthesis inhibition [13], excess ribosome degradation [14,15], and rapid modulation or suppression of translation by specific factors [16]. A striking change in the bacterial translation apparatus under stress, such as nutrient starvation, is 70S ribosome dimerization to form 100S complexes [17,18]. This mechanism, called ribosome hibernation [19], occurs in almost all bacteria and is classified into two major types [20,21]. In γ-proteobacteria, including Escherichia coli, ribosome hibernation occurs when ribosome modulation factor (RMF) binds to 70S ribosomes. This process interferes with the Shine–Dalgarno sequence and induces conformational changes in the 30S subunit, forming a 90S ribosomal dimer. Short-form hibernation-promoting factor (SHPF) then binds to the decoding center, where mRNA and tRNA interact, facilitating conversion of the 90S dimer intermediate to the 100S dimer [20,22]. In contrast, many other bacteria lack RMF and possess long-form HPF (LHPF), a SHPF homolog with an additional C-terminal domain (CTD) that promotes 100S dimer formation [17,23]. LHPF forms a homodimer via the CTD and triggers 100S dimer formation by tethering the two 30S subunits [24,25,26]. In addition, cyanobacteria, which are considered the evolutionary origin of chloroplasts, also harbor an ortholog of HPF known as LrtA. This gene was initially identified as a light-repressed transcript, since its mRNA accumulates in darkness but is rapidly degraded upon illumination [27]. A phylogenetic analysis revealed that LrtA is more closely related to LHPF than to SHPF [28]. Consistent with this relationship, LrtA retains the ability to promote the formation of 100S ribosome dimers, thereby supporting a conserved role in ribosome stabilization [21,29].
In 1990, a putative HPF encoded in the nuclear genome that binds to the 30S subunit of the chloroplast ribosome was identified [30]. Since bacterial HPFs were not reported until the 2000s [22,31], this protein was initially thought to be chloroplast-specific and was thus named plastid-specific ribosomal protein 1 (PSRP1). However, subsequent cryo-electron microscopy (cryo-EM) studies of spinach chloroplast ribosomes revealed that PSRP1 binds to the decoding center of chloroplast ribosomes, similar to SHPF and LHPF [32]. Furthermore, phylogenetic analysis has revealed that PSRP1 is an LHPF ortholog [20]. The high resolution cryo-EM microscopy structures of spinach chloroplast ribosome-bound PSRP1 has been determined [32,33,34,35,36]. Particularly, using PSRP1 with the cryo-EM spinach chloroplast ribosomes by Boerema et al. (2018) revealed that bS1c is reduced and tightly bound via an N-terminal extension, precluding HPF-like chloroplast ribosome dimerization by occupying the interface [36]. Despite the potential structural importance of PSRP1, its physiological functions under specific stress conditions and precise biological function have yet to be fully elucidated.
In the present study, we comprehensively analyzed PSRP1 function using the model organism Physcomitrium patens, while emphasizing its role under dark-stress conditions. As a basal land plant, P. patens occupies an evolutionary position bridging cyanobacteria and higher plants, providing a unique opportunity to link insights from cyanobacterial LrtA to the function of PSRP1 in plants.
2. Results
2.1. PSRP1 Is a Well-Conserved Chloroplast Protein Throughout Plant Evolution
Using the amino acid sequence of spinach PSRP1, we searched plant genomes and confirmed the presence of PSRP1, from green algae to seed plants (Figure S1A). Furthermore, our phylogenetic analysis of PSRP1 shows that its evolutionary lineage aligned with major lineages of plants, from green algae to seed plants (Figure S1B). This demonstrates that plant PSRP1 genes are closely related to those of cyanobacteria, such as Synechococcus and Synechocystis, the ancestors of chloroplasts. This relationship supports the hypothesis that PSRP1 was inherited from cyanobacterial ancestors through endosymbiosis and conserved throughout plant evolution. The extensive evolutionary conservation of this gene suggests that PSRP1 is crucial for plant survival in competitive environments. To gain further insights into its function, we analyzed PSRP1 using the bryophyte model organism P. patens.
NCBI database analysis revealed two PSRP1 genes in P. patens, which we designated PSRP1a (NCBI Gene ID: 112274408) and PSRP1b (NCBI Gene ID: 112295268). PSRP1a and PSRP1b amino acid sequences share 94% identity, excluding the N-terminal chloroplast transit peptide (Figure S1C). We confirmed PSRP1a and PSRP1b mRNA and the presence of PSRP1 protein in P. patens using reverse transcription PCR (RT-PCR) and Western blot (Figure 1A,B). To investigate intracellular PSRP1 localization, we constructed strains expressing PSRP1a-GFP or PSRP1b-YFP fusion proteins controlled by their native promoters. Fluorescence microscopy showed that both PSRP1a-GFP and PSRP1b-YFP were localized in chloroplasts (Figure 1C,D). These results suggest that the P. patens genome harbors two PSRP1 gene copies, which encode functional proteins localized in chloroplasts.
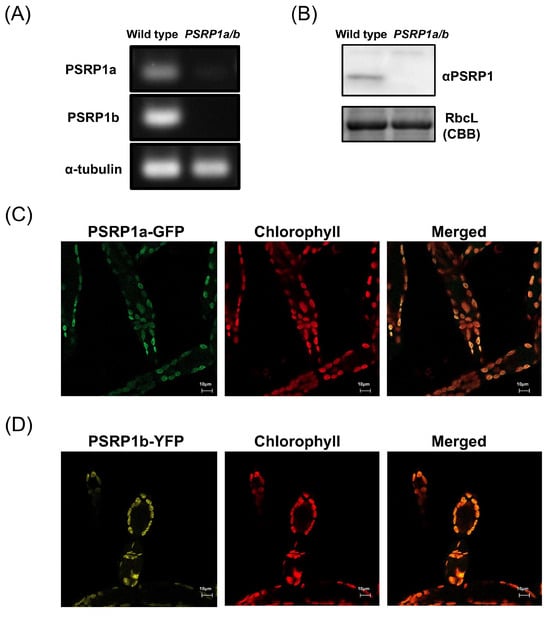
Figure 1.
Expression and chloroplast localization of PSRP1 in Physcomitrium patens. (A) PSRP1a and PSRP1b mRNA expression analysis by RT-PCR in wild-type and PSRP1a/b strains. The amplification products of PSRP1a, PSRP1b mRNA and internal control (α-tubulin mRNA) were shown. No detectable PSRP1a and PSRP1b mRNA was observed in PSRP1a/b strain. (B) Detection of PSRP1 protein by Western blot in wild-type and PSRP1a/b strains. Detection was performed using a specific antibody against PSRP1 protein. RbcL protein stained with Coomassie Brilliant Blue (CBB) was also shown as an internal control. No detectable PSRP1 protein was observed in the PSRP1a/b strain. (C,D) Intracellular localization of PSRP1a-GFP (C) and PSRP1b-YFP (D) fusion proteins in plant cells. GFP (green) or YFP (yellow) fluorescence co-localized with chlorophyll autofluorescence (red), confirming that PSRP1a or PSRP1b fusion proteins were localized to the chloroplasts. In the merged images, overlapping signals appear orage. Bars = 10 μm in each image.
2.2. Loss of PSRP1 Impairs Plant Growth
To investigate the physiological role of PSRP1 in plants, a double-knockout strain was generated by replacing PSRP1a and PSRP1b with nptII and aphIV, respectively. We confirmed the absence of the mRNAs and protein expression (Figure 1A,B). This double-knockout strain is referred to as PSRP1a/b. We compared the growth of the wild type and PSRP1a/b strains by cultivating protonemata on agar plates under long-day conditions (LD; 16 h light/8 h dark) and short-day conditions (SD; 8 h light/16 h dark) conditions for over 6 weeks. No differences in growth were observed during the early growth stages. However, after day 46 under LD conditions and day 36 under SD conditions, the PSRP1a/b strain showed statistically significant albeit modest decreases in growth relative to that of the wild type (Figure 2A,B, p < 0.05) under both conditions. To simulate natural environmental conditions, such as limited light, nutrients, and space, we co-cultured wild-type and PSRP1a/b protonemata under LD and SD conditions. After 3 weeks, real-time PCR of the extracted plant genomic DNA amplified strain-specific sequences: the PSRP1a gene for the wild type and the nptII gene for PSRP1a/b. The relative amplification levels of PSRP1a and nptII were normalized to the internal control (Actin) to estimate PSRP1a/b abundance relative to the wild type. After 3 weeks, the survival ratio of PSRP1a/b relative to that of the wild type decreased significantly under both conditions, indicating that PSRP1 plays a crucial role in plant growth, particularly in competitive environments.
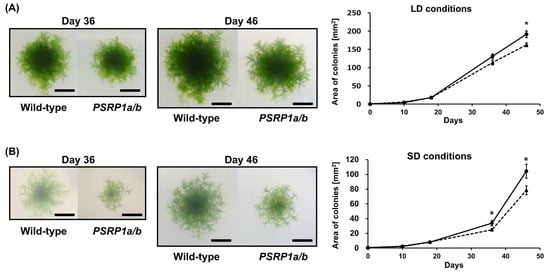
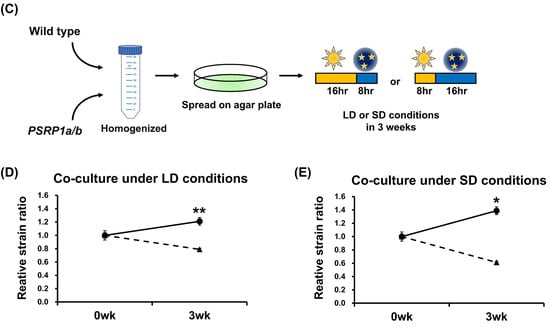
Figure 2.
Comparative growth of wild-type and PSRP1a/b mutant lines in separate and competitive co-cultures. (A) Separate culture under long-day (LD) conditions: Representative colonies of wild-type and PSRP1a/b strains grown on BCDAT agar plates for 46 d (16 h light/8 h dark). Images taken on Days 36 and 46 are shown. Scale bar = 5 mm. The graph on the right shows the average colony area ± SE (n = 10). Solid line, wild type; dotted line, PSRP1a/b. A significant difference was observed on Day 46 (p < 0.05, Student’s t-test), as indicated by an asterisk (*). (B) Separate culture under short-day (SD) conditions: Representative colonies of wild-type and PSRP1a/b strains grown on BCDAT agar plates for 46 d (8 h light/16 h dark). Images taken on Days 36 and 46 are shown. Scale bar = 5 mm. The graph on the right shows the average colony area ± SE (n = 9). Solid line, wild-type; dotted line, PSRP1a/b. Significant differences were observed on Days 36 and 46 (p < 0.05, Student’s t-test), as indicated by an asterisk (*). (C) Schematic of the co-culture assay: Wild-type and PSRP1a/b strains were homogenized, mixed, and spread on BCDAT agar plates. The plates were then incubated for 3 weeks under either LD (16 h light/8 h dark) or SD (8 h light/16 h dark) conditions. Genomic DNA was extracted and used for real-time PCR analysis with strain-specific primers (PSRP1a for wild-type, nptII for PSRP1a/b), and Actin as an internal control. (D) Co-culture under LD conditions: The graph shows the average relative strain ratio (wild type: PSRP1a/b) at 0 and 3 weeks (n = 3). Solid line, wild-type; dotted line, PSRP1a/b. Asterisks (**) indicate a significant difference at 3 weeks (p < 0.05, Student’s t-test). (E) Co-culture under SD conditions: The graph shows the average relative strain ratio (wild-type: PSRP1a/b) at 0 and 3 weeks (n = 3). Solid line, wild-type; dotted line, PSRP1a/b. Asterisks (*) indicate a significant difference at 3 weeks (p < 0.05, Student’s t-test).
2.3. PSRP1 Accumulates and Plays a Protective Role in Preventing Degradation of Chloroplast Ribosomes Under Dark Conditions
Bacterial HPFs associate with ribosomes under specific stress conditions or during stationary phases [18,21,37,38,39,40,41,42,43]. As shown in Figure S1, PSRP1 is an HPF ortholog that binds to chloroplast ribosomes under certain stress conditions, potentially modulating their activity. However, as shown in Figure 2, despite the absence of specific stress, the growth rates differed significantly. Thus, we hypothesized that darkness may act as a stressor that triggers the function of PSRP1. To investigate the effect of dark conditions on plant growth and the dynamics of PSRP1 protein, plants were exposed to prolonged darkness. To evaluate plant growth, wild-type and PSRP1a/b knockout lines were co-cultivated under dark conditions for 2 d within a one-week growth cycle, which was repeated for three consecutive weeks (Figure S2A). Despite the severity of these conditions, growth differences were still observed in the wild-type and PSRP1a/b lines under competitive conditions (Figure S2B).
Next, we examined the changes in PSRP1 protein levels under dark conditions. Plants were grown for a single week in darkness, lasting for 1, 2, or 3 d before sample collection (Figure 3A). The analysis revealed significant protein accumulation with increasing duration of dark exposure (Figure 3B).
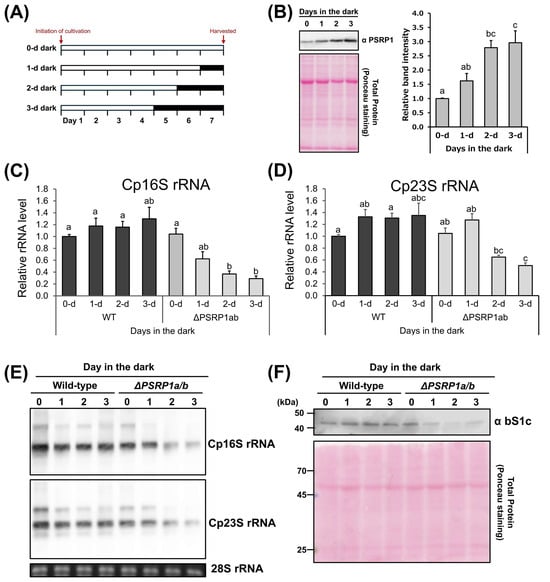
Figure 3.
Effects of prolonged darkness on PSRP1 levels and Cp rRNA levels in wild-type and PSRP1a/b in P. patens. (A) Schematic representation of the growth conditions in this assay. The growing period for each condition was 7 d, with the time in darkness varying from 0 to 3 d. The white and dark gray bars represent continuous light and continuous darkness, respectively. (B) The left panel shows representative Western blot images. Detection was performed using a specific antibody against PSRP1. Total protein stained with Ponceau S is also shown as an internal control. The graph on the right shows the relative amount of PSRP1 protein under each condition. The data are presented as means + standard error (n = 3). Significant differences were identified by Tukey’s HSD test, with different letters indicating significance (p < 0.05). (C,D) Quantification of chloroplast rRNA in the wild-type and PSRP1a/b strains. Total RNA was extracted from the plants grown under each condition. Real- time quantitative PCR (RT-qPCR) was performed using primers specific to chloroplast 16S (A) or 23S rRNA (B), with mitochondrial 18S rRNA (mt18S rRNA) serving as the internal control. Amplification ratios relative to mt18S rRNA were used to estimate the amount of rRNA per cell. The graph shows the average ratio and standard error of the mean (n = 4). Significant differences were identified using Tukey’s HSD test, with different letters indicating statistical significance (p < 0.05). (E) RNA gel blot analysis of chloroplast 16S and 23S rRNAs in P. patens. Total RNA was separated on agarose gels and hybridized with probes specific to Cp16S and Cp23S rRNAs. Ethidium bromide–stained cytosolic 28S rRNA is shown as a loading control. (F) Western blotting of chloroplast ribosomal protein S1 (bS1c) in wild-type and PSRP1a/b strains. Ponceau S staining of total protein is shown as a loading control.
Total RNA was extracted from plants grown under the conditions shown in Figure 3A, and RT-qPCR was performed to assess chloroplast rRNA levels. In wild-type plants, the chloroplast (Cp) 16S and 23S rRNA amounts did not differ significantly, regardless of the duration of darkness. In contrast, the PSRP1a/b strains exhibited significantly decreased Cp16S and Cp23S rRNA levels with increasing duration of darkness (Figure 3C,D). Consistent with these results, RNA gel blot analysis demonstrated degradation of chloroplast rRNAs (Cp16S and Cp23S), while immunoblot analysis further revealed that the levels of chloroplast ribosomal protein S1 (bS1c) were also decreased in the PSRP1a/b strains under prolonged darkness (Figure 3E,F). These results suggest that PSRP1 prevents chloroplast rRNA degradation in the dark.
2.4. Absence of 100S Dimers in Chloroplast Ribosomes Despite PSRP1 Dimerization and Induction of 100S Formation in Bacterial Ribosomes
In bacteria, HPF binding facilitates 100S dimer formation, which is resistant to RNase R-induced degradation [44,45]. Although our alignment results indicate that PSRP1 is closely related to LHPF (Figure S1A) and it is associated with chloroplast ribosomes in P. patens, no 100S dimers were detected under either 0-d or 2-d dark conditions (Figure S3). LHPF dimerization is critical for 100S ribosomal dimer formation [24,46]. Therefore, we examined whether PSRP1 dimerizes.
To determine the oligomeric states of PSRP1a and PSRP1b, we performed SEC-MALS, which is a reliable method for measuring absolute molar mass in solution. After removing the presumed signal peptides, mature PSRP1a and PSRP1b were found to comprise 219 and 221 residues, respectively. SEC-MALS revealed molecular masses of 48.2 kDa and 53.6 kDa, roughly twice their predicted masses of 26.7 and 26.8 kDa (Figure 4A,B). As a control, Bacillus subtilis LHPF (BsHPF), which forms dimers, was analyzed and showed a dimeric mass of 44.3 kDa, approximately twice the predicted value of 24.2 kDa (Figure 4C). Additionally, BSA had a mass of 61.2 kDa, which closely matches the predicted value (66.4 kDa) (Figure 4D). This analysis demonstrates the reliability of SEC-MALS and confirms that PSRP1 forms a stable dimer in solution. The SEC-MALS results, including predicted and observed masses and oligomeric states, are summarized in Table 1.
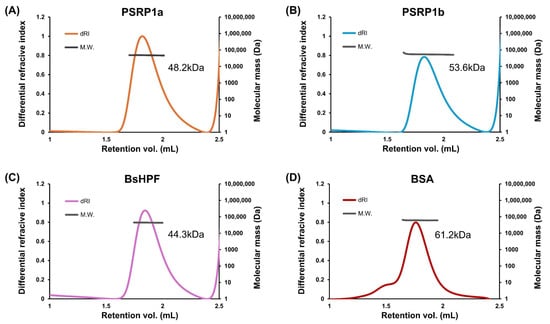
Figure 4.
Analysis of oligomeric states of PSRP1a, PSRP1b, and BsHPF. (A–D) PSRP1a (A), PSRP1b (B), BsHPF, a dimeric control protein (C), and BSA, a monomeric control protein (D), had molecular weights of 48.2, 53.6, 44.3, and 61.2 kDa, respectively. The differential refractive index (dRI) trace indicates the protein elution profile, and the molecular weight (MW) at each elution point was determined using multi-angle light scattering (MALS).

Table 1.
Comparison of calculated and experimentally determined molecular weights of PSRP1a, PSRP1b, BsHPF, and BSA by SEC-MALS.
Next, we tested whether PSRP1 dimers induce 100S ribosome dimerization in vitro. Chloroplast ribosomes purified from spinach were mixed with purified PSRP1a protein from P. patens at ratios of 1:1 and 1:5 in vitro. Although both endogenously bound PSRP1 from spinach and purified P. patens PSRP1a bound to chloroplast ribosomes, no chloroplast 100S dimer formation was observed (Figure 5A–C). This is consistent with the spinach chloroplast ribosome structure, in which bS1c occupies the anticipated dimer interface, thereby blocking 100S formation [36]. In contrast, incubation of PSRP1a with ribosomes purified from a Lactococcus lactis hpf knockout strain (Δlhpf) in vitro resulted in 100S dimer formation (Figure 5D–F). Thus, although PSRP1 retains intrinsic 100S-promoting capacity on bacterial ribosomes, chloroplastic bS1c likely prevents dimerization, which agrees with the prior structural model.
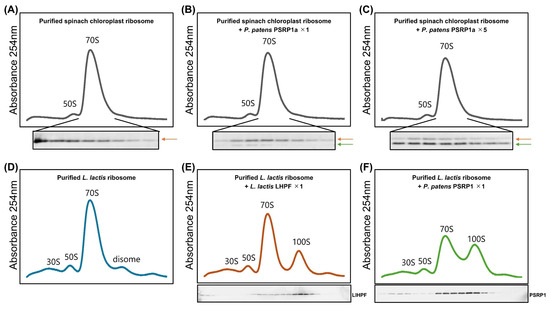
Figure 5.
In vitro analysis of 100S dimer formation in spinach chloroplast and Lactococcus lactis ribosomes with the addition of PSRP1a or LlHPF. (A–C) Sucrose density profile of purified spinach chloroplast ribosomes, with PSRP1a added at 1:1 (B) and 1:5 (C) ratios to ribosomes. The Western blot show PSRP1; the orange arrow indicates endogenous spinach PSRP1 while the green arrow indicates added purified P. patens PSRP1a. (D–F) Sucrose density profile of purified L. lactis ribosomes alone (D), with L. lactis LHPF (LlHPF) added (E), or with PSRP1a added (F), each at a 1:1 ratio. The Western blot show LlHPF (E) and PSRP1a (F).
3. Discussion
We investigated the physiological role of chloroplast PSRP1 by focusing on its response to dark stress. Our results show that PSRP1-deficient plants (PSRP1a/b) exhibited growth defects after more than one month under both LD and SD conditions when grown individually. In contrast, when co-cultivated with wild-type plants, the PSRP1a/b strain became more evident over a much shorter period. Furthermore, PSRP1 levels increased under dark conditions, and contributed to maintaining chloroplast ribosome content. This mechanism may enable reuse of inactive ribosomes when the stress conditions improve. Consequently, PSRP1 contributes to faster regrowth upon return of light, which likely accounts for the growth differences between wild-type and PSRP1a/b plants. Thus, evolutionary conservation of PSRP1 may be crucial for plant survival under harsh environmental conditions. A previous study on maize, however, observed plant growth over 7–9 d under a 16-h light/8-h dark cycle and reported that PSRP1 absence did not affect growth. No significant changes were found in PSRP1 protein abundance under light and dark conditions [47]. These discrepancies could be explained not only by differences in plant species (C4 maize versus C3 bryophyte P. patens), but also by the severity and duration of dark stress applied. While Swift et al. (2020) [47] monitored plants for only 7–9 d under standard LD cycles, our study involved not only prolonged dark treatments lasting several days but also repeated cycles of darkness extending over several weeks.
SHPF and RMF induce 100S dimer formation in E. coli and other γ-proteobacteria [22,48,49]. Most other bacteria possess LHPF, which retains an additional CTD. LHPF forms a homodimer via its CTD and induces 100S ribosome dimer formation by tethering the two 30S subunits [24,25,26,50,51]. These 100S dimers are more resistant to degradation than 70S ribosomes [44,45,52]. In contrast, our results show that chloroplast 100S dimer formation was not observed in vitro or in vivo (Figure 5A–C and Figure S3), although PSRP1 dimerized (Figure 4) and induced 100S dimer formation in bacterial ribosomes (Figure 5D–F). Boerema et al. found that the chloroplast ribosomal protein bS1c was tightly bound to the ribosome [36]. This cryo-EM analysis further revealed that bS1c is reduced and tightly bound via an N-terminal extension, with its binding region overlapped with the 30S–30S interface of the 100S dimer, thereby potentially inhibiting dimer formation (Figure S4). P. patens harbors the bS1c gene, and its sequence is well conserved with those of spinach and A. thaliana (Figure S5A). Structural comparison of P. patens and spinach bS1c based on AlphaFold3 predictions confirmed that the overall fold is highly conserved (Figure S5B,C). Therefore, inhibiting 100S dimer formation by bS1c is likely applicable to P. patens, consistent with our findings. These observations suggest that chloroplasts have evolved a ribosome protection mechanism distinct from that of bacteria.
At the same time, our study has some limitations. First, although we observed a clear reduction in chloroplast rRNA levels in PSRP1a/b strains, we cannot fully exclude alternative explanations such as reduced transcription or defects in rRNA processing. Second, our analyses were conducted primarily under prolonged darkness, and it remains to be determined whether PSRP1 has similar functions under other physiological stress conditions. Finally, this study focused on P. patens, and further investigations in other plant species will be important to determine whether the role of PSRP1 in maintaining chloroplast ribosome integrity is conserved across the plant kingdom.
An important direction for future research will be to elucidate the molecular basis underlying the absence of 100S dimer formation—potentially involving factors such as bS1c—and to clarify how PSRP1 protects ribosomes despite the lack of 100S dimers, as well as to determine whether these mechanisms operate under other environmental stresses and across diverse plant lineages.
In conclusion, our analysis revealed that PSRP1 stabilizes chloroplast ribosomes under dark conditions in the absence of 100S ribosome dimer formation, in contrast to HPF-mediated ribosome hibernation in bacteria. Our findings advance the understanding of chloroplast ribosome regulation and highlight the evolutionary divergence of plastid translation from its bacterial ancestors.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Physcomitrium patens (Hedw.) Bruch & Schimp. was used as the model organism in this study. Protonemata were cultivated on BCDAT agar medium [53] at 25 °C.
4.2. Construction of a P. patens PSRP1a and PSRP1b Double Knockout Strain
To create the PSRP1a and PSRP1b double knockout strain, the 5′ and 3′ flanking regions of each gene were amplified by PCR using the primers listed in Table S2. For PSRP1a, the 5′ region was amplified using PSRP1a-5′HR-KpnI Fwd and PSRP1a-5′HR-XhoI Rev, and the 3′ region with PSRP1a-3′HR-SacII Fwd and PSRP1a-3′HR-SacI Rev; these fragments were cloned into pTN3 [53] to construct pHAC7. For PSRP1b, 5′ region was amplified with PSRP1b-5′HR KpnI Fwd and PSRP1b-5′HR-XhoI Rev, and 3′ region with PSRP1b-3′HR SmaI Fwd and PSRP1b-3′HR-SacI Rev; the resulting fragments were inserted into pTN186 (a gift from Mitsuyasu Hasebe, Chiba, Japan, Addgene plasmid #34890) to yield pHAC8. The resulting constructs were linearized and introduced into P. patens protoplasts via PEG-mediated transformation [53]. Transformants were selected on BCDAT agar plates containing antibiotics and the insertion of a single copy of each gene was confirmed by PCR. The double knockout strain is referred to as PSRP1a/b.
4.3. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted using an RNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Reverse transcription was conducted using ReverTra Ace® qPCR RT Master Mix (Toyobo, Osaka, Japan) to generate cDNA from the isolated RNA. Gene-specific primers targeting PSRP1a, PSRP1b, and internal control Actin were used for subsequent PCR amplification. The PCR products were separated by agarose gel electrophoresis and visualized under UV light to verify the presence and expected size of the amplicons. The primer sequences are listed in Table S2.
4.4. Fluorescence Microscopy
The PSRP1a-sGFP knock-in plasmid (pGFPmutNPTII-PSRP1a) was constructed by amplifying the 5′ and 3′ homologous regions (5′HR and 3′HR) from the P. patens genome using primer pairs PSRP1a-5′HR Fwd/Rev and PSRP1a-3′HR Fwd/Rev. The resulting PCR fragments were digested with SalI/ClaI (5′HR) and XbaI/SacI (3′HR). These fragments were then ligated into the corresponding sites of the pGFPmutNPTII vector [54]. pCTRN-aphIV was constructed by replacing the antibiotic resistance gene in pCTRN-NPTII 2 (NCBI Accession No. AB697058) with the aphIV gene. Specifically, aphIV was amplified from pTN186 using aphIV Fwd/Rev primers, while pCTRN-NPTII 2 was amplified with pCTRN-NPTII 2 Fwd/Rev primers. The resulting fragments were assembled using a NEBuilder® HiFi DNA Assembly Kit. Using the P. patens genome as the template, 5′HR and 3′HR were amplified with PSRP1b-5′HR Fwd/Rev and PSRP1b-3′HR Fwd/Rev, respectively. In parallel, fragments No.1 and No.2 of pCTRN-aphIV were amplified using pCTRN-aphIV No.1 Fwd/Rev and pCTRN-aphIV No.2 Fwd/Rev. These fragments were subsequently assembled with NEBuilder® HiFi DNA Assembly Kit to generate the final plasmid, pTaK16. All primer sequences are listed in Table S2. The plasmids were linearized and introduced into plant cells via polyethylene glycol (PEG)-mediated protoplast transformation [53]. Transformants were selected on BCDAT agar plates containing antibiotics, and the insertion of a single copy of each gene was confirmed by PCR. The intracellular localization of PSRP1a-GFP or PSRP1b-Citrine in P. patens was observed using an LSM 800 microscope (Carl Zeiss, Jena, Germany). Additionally, chlorophyll autofluorescence was monitored to assess its localization within the chloroplasts.
4.5. Western Blotting
The protonemata were collected, drained, and immediately frozen in liquid nitrogen. Tissues were homogenized at 1750 rpm for 10 s using a Multi-beads shocker® (Yasui Kikai, Osaka, Japan). Total protein was extracted using extraction buffer (200 mM Tris-HCl, pH 8.0, 200 mM KCl, 35 mM MgCl2, 25 mM EGTA, 1% Triton X, 100 mM β-mercaptoethanol (β-ME), 2 mM PMSF). Cell debris was removed by centrifugation at 17,800× g for 20 min at 4 °C. The supernatant was analyzed by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on a 15% polyacrylamide gel, followed by immunoblotting with polyclonal anti-PSRP1 antibody (1/1000, custom-made) and polyclonal anti-bS1c antibody (1/1000, PhytoAB, San Jose, CA, USA). Alkaline phosphatase-conjugated goat anti-rabbit IgG (1/10,000, Sigma-Aldrich, St. Louis, MO, USA) was used as the secondary antibody, and detection was performed with AttoPhos® substrate (Promega, Madison, WI, USA) using a Typhoon™ FLA 9500 imager (GE Healthcare, Chicago, IL, USA). Band intensity of PSRP1 was measured from scanned immunoblot images using ImageJ software (Fiji, version 2.9.0; Image J 1.54p, https://imagej.net/ij/), accessed on 29 July 2020.
4.6. Northern Blotting
Total RNA was electrophoresed on MOPS-formaldehyde denaturing 1.2% agarose gels and transferred onto a Hybond™ N+ membrane (GE Healthcare, Chicago, IL, USA) via capillary transfer for at least 3 h. RNA was detected using a DIG-labeled probe with DIG reagents and kits for nonradioactive nucleic acid labeling and detection (Roche, Basel, Switzerland), according to the manufacturer’s protocol. The probe sequences are listed in Table S2.
4.7. Growth Comparison and Competitive Survival Analysis
For growth comparisons, small pieces of protonemal cells were inoculated on BCDAT agar plates and cultivated at 25 °C under long-day (LD; 16 h light/8 h dark) and short-day (SD; 8 h light/16 h dark) conditions for over 1 month. For competitive survival analysis, the wild-type and PSRP1a/b strains were grown separately under continuous white light for 7 d. Wild-type and PSRP1a/b strains were combined at equal fresh weights, homogenized, and spread on BCDAT agar plates. The cultures were incubated under LD or SD conditions for three weeks. For growth comparisons under prolonged dark conditions, the cultures were first grown under continuous light for 5 d, followed by 2 d in the dark, homogenized again, and spread on fresh BCDAT agar plates. This process was repeated weekly for 3 weeks. Genomic DNA was extracted using cetyl-trimethyl-ammonium bromide [55]. Real-time PCR was performed using PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) and a QuantStudio™ 12 K Flex Real-Time PCR System (Thermo Fisher Scientific) to amplify strain-specific sequences (PSRP1a for wild-type and nptII for PSRP1a/b), with Actin serving as the internal control. Sequences of the gene-specific primers are listed in Table S2.
4.8. Real-Time Quantitative PCR (RT-qPCR)
Total RNA was extracted using the RNeasy® Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized from the isolated RNA using ReverTra Ace® qPCR RT Master Mix (Toyobo, Osaka, Japan). The cDNA was subsequently amplified using PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific) on a QuantStudio™ 12 K Flex Real-Time PCR System (Thermo Fisher Scientific), following the manufacturer’s guidelines. The abundance of chloroplast 16S and 23S rRNA were normalized to mitochondrial 18S rRNA. The sequences of gene-specific primers are listed in Table S2.
4.9. Protein Purification
PSRP1a and PSRP1b from P. patens were amplified from the cDNA, whereas BsHPF from B. subtilis, LlHPF from L. lactis, and EcHPF from E. coli were amplified from the genomic DNA. Each gene was cloned into pET15b vector using gene-specific primers and the appropriate restriction enzyme sites. Details of the primers used for each gene are summarized in Table S2. The recombinant plasmid, pET15b, was transformed into E. coli strains ER2566 and BL21(DE3) for protein expression. Cultures were grown in LB medium supplemented with 0.2% glucose and 100 µg/mL ampicillin at 37 °C until the optical density at 600 nm (OD600) reached 0.4–0.5, at which point protein expression was induced by adding 0.4 mM isopropyl β-D-1-thiogalactopyranoside. Following induction, the cultures were incubated for another 3 h at 37 °C before harvesting the cells via centrifugation. The cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5; 500 mM NaCl; 10% (w/v) sucrose; 5 mM β-ME), frozen in liquid nitrogen, and stored at −80 °C. Lysis was performed by freeze-thawing with lysozyme (1 mg/mL). Cell debris was removed by centrifugation at 40,000× g for 30 min at 4 °C. Proteins were purified using TALON® Metal Affinity Resin (Takara Bio, Shiga, Japan) and eluted in a buffer containing 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 5 mM β-ME, and 100 mM imidazole. Protein concentrations were measured at 280 nm using a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific).
4.10. Size-Exclusion Chromatography with Multi-Angle Light Scattering (SEC-MALS)
SEC-MALS measurements were performed using a DAWN® HELEOS® II (Wyatt Technology Corporation, Santa Barbara, CA, USA) downstream of an Alliance liquid chromatography system (Waters, Milford, MA, USA) connected to a Superdex™ 200 5/150 GL (Cytiva, Uppsala, Sweden) gel filtration column. The differential refractive index downstream of the MALS detector was used to determine protein concentration. The column was equilibrated with a running buffer containing 100 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, and 10% (v/v) glycerol. The flow rate was set to 0.15 mL/min, and 90 µL of each sample at 1.6 mg/mL was injected. Data analysis was performed using ASTRA version 6 software (Wyatt Technology Corporation, Santa Barbara, CA, USA).
4.11. Isolation of Spinach Chloroplast 70S Ribosomes
Chloroplast ribosomes were isolated as described by Bartsch et al. [56]. Briefly, 4 kg of spinach leaves were purchased from a local supermarket. The leaves were homogenized using Buffer A (10 mM Tris-HCl, pH 7.5, 50 mM KCl, 10 mM Mg(OAc)2, 6 mM β-ME) containing 0.7 M sorbitol. The homogenate was filtered through four layers of gauze. The suspension was cleared by centrifugation at 1200× g for 15 min. The pellet was resuspended in Buffer A with 0.4 M sorbitol and re-centrifuged at 1200× g for 15 min. This step was repeated thrice. The washed chloroplast pellet was resuspended in Buffer A supplemented with 2% (v/v) Triton X-100 and incubated at 4 °C for 30 min. The lysed suspension was clarified by centrifugation at 26,000× g for 30 min at 4 °C. This step was repeated. The supernatant was layered onto a 30% sucrose cushion in Buffer II (20 mM Tris-HCl, pH 7.5, 10 mM Mg(OAc)2, 1 M NH4OAc, 6 mM β-ME) and centrifuged at 206,000× g for 3 h at 4 °C. The green pellet was resuspended in Buffer II. After mixing for 1 h at 4 °C, the high-salt washed ribosomes were layered onto a 30% sucrose cushion in Buffer I (20 mM Tris-HCl, pH 7.5, 15 mM Mg(OAc)2, 100 mM NH4OAc, 6 mM β-ME) and centrifuged at 206,000× g for 4 h at 4 °C. The pellet was resuspended in Buffer I.
4.12. Isolation of Lactococcus lactis Ribosomes
The Lactococcus lactis strain NZ9000 ΔyfiA used in this experiment was kindly provided by Prof. Bert Poolman (University of Groningen) and originally constructed in a previous study [39]. L. lactis 70S ribosomes were isolated as described by Ueta et al. [21]. L. lactis Δlhpf strains were cultured statically at 30 °C in M17 medium (Difco Laboratories, Detroit, MA, USA) supplemented with 0.5% (w/v) glucose. After reaching an OD600 of 1.5, the cells were harvested by centrifugation at 5210× g for 15 min at 4 °C. The resulting cell pellets were ground with approximately equal volumes of quartz sand (Wako Pure Chemical Industries) and extracted using Buffer I. The homogenate was subjected to centrifugation at 9000× g for 15 min at 4 °C, and this step was repeated to ensure thorough separation. The resulting supernatant was layered onto a 30% sucrose cushion prepared in Buffer II. Ribosome washing and subsequent purification steps were performed as outlined under “Section 4.11. Isolation of Spinach Chloroplast Ribosomes.”
4.13. Analysis of 100S Dimers of Chloroplasts and L. lactis Ribosomes
Reactions were performed as described by Usachev et al. [57]. For the in vitro reaction, 64 pmol of chloroplast ribosomes was mixed with 64 or 320 pmol of purified PSRP1a protein. Similarly, 64 pmol L. lactis ribosomes were mixed with 64 pmol of purified LlHPF or PSRP1a protein. After 30 min of incubation in Buffer I (20 mM Tris-HCl pH 7.5, 15 mM Mg(OAc)2, 100 mM NH4OAc, 6 mM β-ME) at 37 °C, the complexes were layered onto a 5–20% sucrose gradient prepared in Buffer I and centrifuged at 11,900 rpm for 20 h at 4 °C using a Hitachi P40ST rotor. The gradient was fractionated using a Piston Gradient Fractionator (BioComp, San Antonio, TX, USA), and the absorbance at 254 nm was monitored using a Bio-mini UV Monitor (ATTO, Amherst, NY, USA). Each fraction was subjected to Western blot.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants14203155/s1, Figure S1: Phylogenetic relationship among chloroplast PSRP1, cyanobacterial LrtA, and bacterial HPF proteins; Figure S2: Growth comparison between wild-type and PSRP1a/b strains in competitive co-culture under prolonged dark conditions; Figure S3: Polysome profiling in P. patens grown under 0-d or 2-d dark conditions; Figure S4: Structural comparison of the chloroplast ribosome and S. aureus 100S dimer; Figure S5: Sequence alignment and structural comparison of chloroplast bS1c proteins; Table S1: List of plasmids used in the study; Table S2: List of oligonucleotides used in the study; Method S1: Supplementary Materials and methods. References [53,54,58,59,60] are cited in the Supplementary Materials.
Author Contributions
K.T. and Y.S. designed the research; K.T., Y.Y. and T.O. performed the research; T.O. performed the size-exclusion chromatography with multi-angle light scattering (SEC-MALS) experiments and analyzed the resulting data; K.T. and Y.Y. analyzed additional data; K.T. wrote the paper; Y.S. reviewed and edited the manuscript; and Y.S. supervised the research. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials, and further inquiries can be directed to the corresponding author.
Acknowledgments
We gratefully acknowledge Bert Poolman (University of Groningen, Groningen, The Netherlands) for providing the Lactococcus lactis strain; Mitsuyasu Hasebe (National Institute for Basic Biology, Okazaki, Japan) for providing plasmids pTN3, pTN186, pCTRN-NPTII 2, and pGFPmutNPTII; Keiko Sakakibara (Rikkyo University, Tokyo, Japan) for advice on moss culture and experimental methods; Ryo Hanai (Rikkyo University, Tokyo, Japan) for valuable discussions and helpful advice on this study; and Masami Ueta (Yoshida Biological Laboratory, Kyoto, Japan) for advice on the purification of bacterial ribosomes and the experimental procedures for in vitro 100S dimer formation. The size-exclusion chromatography with multi-angle light scattering (SEC-MALS) measurements at the Photon Factory were performed under proposal number 2019G034.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Eberhard, S.; Finazzi, G.; Wollman, F.A. The dynamics of photosynthesis. Annu. Rev. Genet. 2008, 42, 463–515. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Parkinson, C.L. Jumping genes and containment. Nat. Biotechnol. 2003, 21, 374–375. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Ayliffe, M.A.; Timmis, J.N. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 2003, 422, 72–76. [Google Scholar] [CrossRef]
- Timmis, J.N.; Ayliffe, M.A.; Huang, C.Y.; Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 2004, 5, 123–135. [Google Scholar] [CrossRef]
- Sagan, L. On the origin of mitosing cells. 1967. J. NIH Res. 1993, 5, 65–72. [Google Scholar]
- Gould, S.B.; Waller, R.F.; McFadden, G.I. Plastid evolution. Annu. Rev. Plant Biol. 2008, 59, 491–517. [Google Scholar] [CrossRef]
- Jensen, P.E.; Leister, D. Chloroplast evolution, structure and functions. F1000Prime Rep. 2014, 6, 40. [Google Scholar] [CrossRef]
- Bock, R.; Timmis, J.N. Reconstructing evolution: Gene transfer from plastids to the nucleus. Bioessays 2008, 30, 556–566. [Google Scholar] [CrossRef]
- Wilson, D.N.; Nierhaus, K.H. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 187–219. [Google Scholar] [CrossRef]
- Buttgereit, F.; Brand, M.D. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 1995, 312 Pt 1, 163–167. [Google Scholar] [CrossRef]
- Russell, J.B.; Cook, G.M. Energetics of bacterial growth: Balance of anabolic and catabolic reactions. Microbiol. Rev. 1995, 59, 48–62. [Google Scholar] [CrossRef]
- Szaflarski, W.; Nierhaus, K.H. Question 7: Optimized energy consumption for protein synthesis. Orig. Life Evol. Biosph. 2007, 37, 423–428. [Google Scholar] [CrossRef]
- Bremer, H.; Dennis, P.P. Modulation of Chemical Composition and Other Parameters of the Cell at Different Exponential Growth Rates. EcoSal Plus 2008, 3, 10-1128. [Google Scholar] [CrossRef]
- Zundel, M.A.; Basturea, G.N.; Deutscher, M.P. Initiation of ribosome degradation during starvation in Escherichia coli. RNA 2009, 15, 977–983. [Google Scholar] [CrossRef]
- Fessler, M.; Gummesson, B.; Charbon, G.; Svenningsen, S.L.; Sørensen, M.A. Short-term kinetics of rRNA degradation in Escherichia coli upon starvation for carbon, amino acid or phosphate. Mol. Microbiol. 2020, 113, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Starosta, A.L.; Lassak, J.; Jung, K.; Wilson, D.N. The bacterial translation stress response. FEMS Microbiol. Rev. 2014, 38, 1172–1201. [Google Scholar] [CrossRef] [PubMed]
- Matzov, D.; Bashan, A.; Yap, M.F.; Yonath, A. Stress response as implemented by hibernating ribosomes: A structural overview. FEBS J. 2019, 286, 3558–3565. [Google Scholar] [CrossRef] [PubMed]
- Prossliner, T.; Skovbo Winther, K.; Sørensen, M.A.; Gerdes, K. Ribosome Hibernation. Annu. Rev. Genet. 2018, 52, 321–348. [Google Scholar] [CrossRef]
- Yoshida, H.; Maki, Y.; Kato, H.; Fujisawa, H.; Izutsu, K.; Wada, C.; Wada, A. The ribosome modulation factor (RMF) binding site on the 100S ribosome of Escherichia coli. J. Biochem. 2002, 132, 983–989. [Google Scholar] [CrossRef]
- Ueta, M.; Ohniwa, R.L.; Yoshida, H.; Maki, Y.; Wada, C.; Wada, A. Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J. Biochem. 2008, 143, 425–433. [Google Scholar] [CrossRef]
- Ueta, M.; Wada, C.; Daifuku, T.; Sako, Y.; Bessho, Y.; Kitamura, A.; Ohniwa, R.L.; Morikawa, K.; Yoshida, H.; Kato, T.; et al. Conservation of two distinct types of 100S ribosome in bacteria. Genes. Cells 2013, 18, 554–574. [Google Scholar] [CrossRef]
- Ueta, M.; Yoshida, H.; Wada, C.; Baba, T.; Mori, H.; Wada, A. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes. Cells 2005, 10, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Ueta, M.; Wada, C.; Wada, A. Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells 2010, 15, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Beckert, B.; Abdelshahid, M.; Schäfer, H.; Steinchen, W.; Arenz, S.; Berninghausen, O.; Beckmann, R.; Bange, G.; Turgay, K.; Wilson, D.N. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J. 2017, 36, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Franken, L.E.; Oostergetel, G.T.; Pijning, T.; Puri, P.; Arkhipova, V.; Boekema, E.J.; Poolman, B.; Guskov, A. A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy. Nat. Commun. 2017, 8, 722. [Google Scholar] [CrossRef]
- Flygaard, R.K.; Boegholm, N.; Yusupov, M.; Jenner, L.B. Cryo-EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein-mediated dimerization mechanism. Nat. Commun. 2018, 9, 4179. [Google Scholar] [CrossRef]
- Tan, X.; Varughese, M.; Widger, W.R. A light-repressed transcript found in Synechococcus PCC 7002 is similar to a chloroplast-specific small subunit ribosomal protein and to a transcription modulator protein associated with sigma 54. J. Biol. Chem. 1994, 269, 20905–20912. [Google Scholar] [CrossRef]
- Galmozzi, C.V.; Florencio, F.J.; Muro-Pastor, M.I. The Cyanobacterial Ribosomal-Associated Protein LrtA Is Involved in Post-Stress Survival in Synechocystis sp. PCC 6803. PLoS ONE 2016, 11, e0159346. [Google Scholar] [CrossRef]
- Hood, R.D.; Higgins, S.A.; Flamholz, A.; Nichols, R.J.; Savage, D.F. The stringent response regulates adaptation to darkness in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. USA 2016, 113, E4867–E4876. [Google Scholar] [CrossRef]
- Johnson, C.H.; Kruft, V.; Subramanian, A.R. Identification of a plastid-specific ribosomal protein in the 30 S subunit of chloroplast ribosomes and isolation of the cDNA clone encoding its cytoplasmic precursor. J. Biol. Chem. 1990, 265, 12790–12795. [Google Scholar] [CrossRef]
- Maki, Y.; Yoshida, H.; Wada, A. Two proteins, YfiA and YhbH, associated with resting ribosomes in stationary phase Escherichia coli. Genes Cells 2000, 5, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.R.; Wilson, D.N.; Datta, P.P.; Barat, C.; Schluenzen, F.; Fucini, P.; Agrawal, R.K. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 19315–19320. [Google Scholar] [CrossRef]
- Bieri, P.; Leibundgut, M.; Saurer, M.; Boehringer, D.; Ban, N. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 2017, 36, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Graf, M.; Arenz, S.; Huter, P.; Dönhöfer, A.; Novácek, J.; Wilson, D.N. Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res. 2017, 45, 2887–2896. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.; Shi, J.; Bhushan, S. Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation. Nucleic Acids Res. 2017, 45, 8581–8595. [Google Scholar] [CrossRef]
- Perez Boerema, A.; Aibara, S.; Paul, B.; Tobiasson, V.; Kimanius, D.; Forsberg, B.O.; Wallden, K.; Lindahl, E.; Amunts, A. Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat. Plants 2018, 4, 212–217. [Google Scholar] [CrossRef]
- Akanuma, G.; Kazo, Y.; Tagami, K.; Hiraoka, H.; Yano, K.; Suzuki, S.; Hanai, R.; Nanamiya, H.; Kato-Yamada, Y.; Kawamura, F. Ribosome dimerization is essential for the efficient regrowth of Bacillus subtilis. Microbiology 2016, 162, 448–458. [Google Scholar] [CrossRef]
- Basu, A.; Yap, M.N. Ribosome hibernation factor promotes Staphylococcal survival and differentially represses translation. Nucleic Acids Res. 2016, 44, 4881–4893. [Google Scholar] [CrossRef]
- Puri, P.; Eckhardt, T.H.; Franken, L.E.; Fusetti, F.; Stuart, M.C.; Boekema, E.J.; Kuipers, O.P.; Kok, J.; Poolman, B. Lactococcus lactis YfiA is necessary and sufficient for ribosome dimerization. Mol. Microbiol. 2014, 91, 394–407. [Google Scholar] [CrossRef]
- Li, Y.; Sharma, M.R.; Koripella, R.K.; Yang, Y.; Kaushal, P.S.; Lin, Q.; Wade, J.T.; Gray, T.A.; Derbyshire, K.M.; Agrawal, R.K.; et al. Zinc depletion induces ribosome hibernation in mycobacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 8191–8196. [Google Scholar] [CrossRef]
- Yoshida, H.; Wada, A. The 100S ribosome: Ribosomal hibernation induced by stress. Wiley Interdiscip. Rev. RNA 2014, 5, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Kline, B.C.; McKay, S.L.; Tang, W.W.; Portnoy, D.A. The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J. Bacteriol. 2015, 197, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liang, C.; Zheng, M.; Liu, L.; An, Y.; Xu, H.; Xiao, S.; Nie, L. Ribosome Hibernation as a Stress Response of Bacteria. Protein Pept. Lett. 2020, 27, 1082–1091. [Google Scholar] [CrossRef] [PubMed]
- Prossliner, T.; Gerdes, K.; Sørensen, M.A.; Winther, K.S. Hibernation factors directly block ribonucleases from entering the ribosome in response to starvation. Nucleic Acids Res. 2021, 49, 2226–2239. [Google Scholar] [CrossRef]
- Lipońska, A.; Yap, M.F. Hibernation-Promoting Factor Sequesters Staphylococcus aureus Ribosomes to Antagonize RNase R-Mediated Nucleolytic Degradation. mBio 2021, 12, e0033421. [Google Scholar] [CrossRef]
- Feaga, H.A.; Kopylov, M.; Kim, J.K.; Jovanovic, M.; Dworkin, J. Ribosome Dimerization Protects the Small Subunit. J. Bacteriol. 2020, 202, 10-1128. [Google Scholar] [CrossRef]
- Swift, K.; Chotewutmontri, P.; Belcher, S.; Williams-Carrier, R.; Barkan, A. Functional Analysis of PSRP1, the Chloroplast Homolog of a Cyanobacterial Ribosome Hibernation Factor. Plants 2020, 9, 209. [Google Scholar] [CrossRef]
- Polikanov, Y.S.; Blaha, G.M.; Steitz, T.A. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 2012, 336, 915–918. [Google Scholar] [CrossRef]
- Beckert, B.; Turk, M.; Czech, A.; Berninghausen, O.; Beckmann, R.; Ignatova, Z.; Plitzko, J.M.; Wilson, D.N. Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nat. Microbiol. 2018, 3, 1115–1121. [Google Scholar] [CrossRef]
- Khusainov, I.; Vicens, Q.; Ayupov, R.; Usachev, K.; Myasnikov, A.; Simonetti, A.; Validov, S.; Kieffer, B.; Yusupova, G.; Yusupov, M.; et al. Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF. EMBO J. 2017, 36, 2073–2087. [Google Scholar] [CrossRef]
- Matzov, D.; Aibara, S.; Basu, A.; Zimmerman, E.; Bashan, A.; Yap, M.F.; Amunts, A.; Yonath, A.E. The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus. Nat. Commun. 2017, 8, 723. [Google Scholar] [CrossRef] [PubMed]
- Lipońska, A.; Lee, H.; Yap, M.F. Staphylococcal exoribonuclease YhaM destabilizes ribosomes by targeting the mRNA of a hibernation factor. Nucleic Acids Res. 2024, 52, 8998–9013. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Hiwatashi, Y.; Sakakibara, I.; Kato, M.; Hasebe, M. Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res. 2000, 7, 9–17. [Google Scholar] [CrossRef]
- Hiwatashi, Y.; Obara, M.; Sato, Y.; Fujita, T.; Murata, T.; Hasebe, M. Kinesins are indispensable for interdigitation of phragmoplast microtubules in the moss Physcomitrella patens. Plant Cell 2008, 20, 3094–3106. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- Bartsch, M.; Kimura, M.; Subramanian, A.R. Purification, primary structure, and homology relationships of a chloroplast ribosomal protein. Proc. Natl. Acad. Sci. USA 1982, 79, 6871–6875. [Google Scholar] [CrossRef]
- Usachev, K.S.; Fatkhullin, B.F.; Klochkova, E.A.; Miftakhov, A.K.; Golubev, A.A.; Bikmullin, A.G.; Nurullina, L.I.; Garaeva, N.S.; Islamov, D.R.; Gabdulkhakov, A.G.; et al. Dimerization of long hibernation promoting factor from Staphylococcus aureus: Structural analysis and biochemical characterization. J. Struct. Biol. 2020, 209, 107408. [Google Scholar] [CrossRef]
- Odahara, M.; Inouye, T.; Nishimura, Y.; Sekine, Y. RECA plays a dual role in the maintenance of chloroplast genome stability in Physcomitrella patens. Plant J. 2015, 84, 516–526. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).