Genome-Wide Analysis Reveals Chitinases as Putative Defense-Related Proteins Against Fungi in the Genomes of Coffea arabica and Its Progenitors
Abstract
1. Introduction
2. Results
2.1. Identification of Putative Chitinase
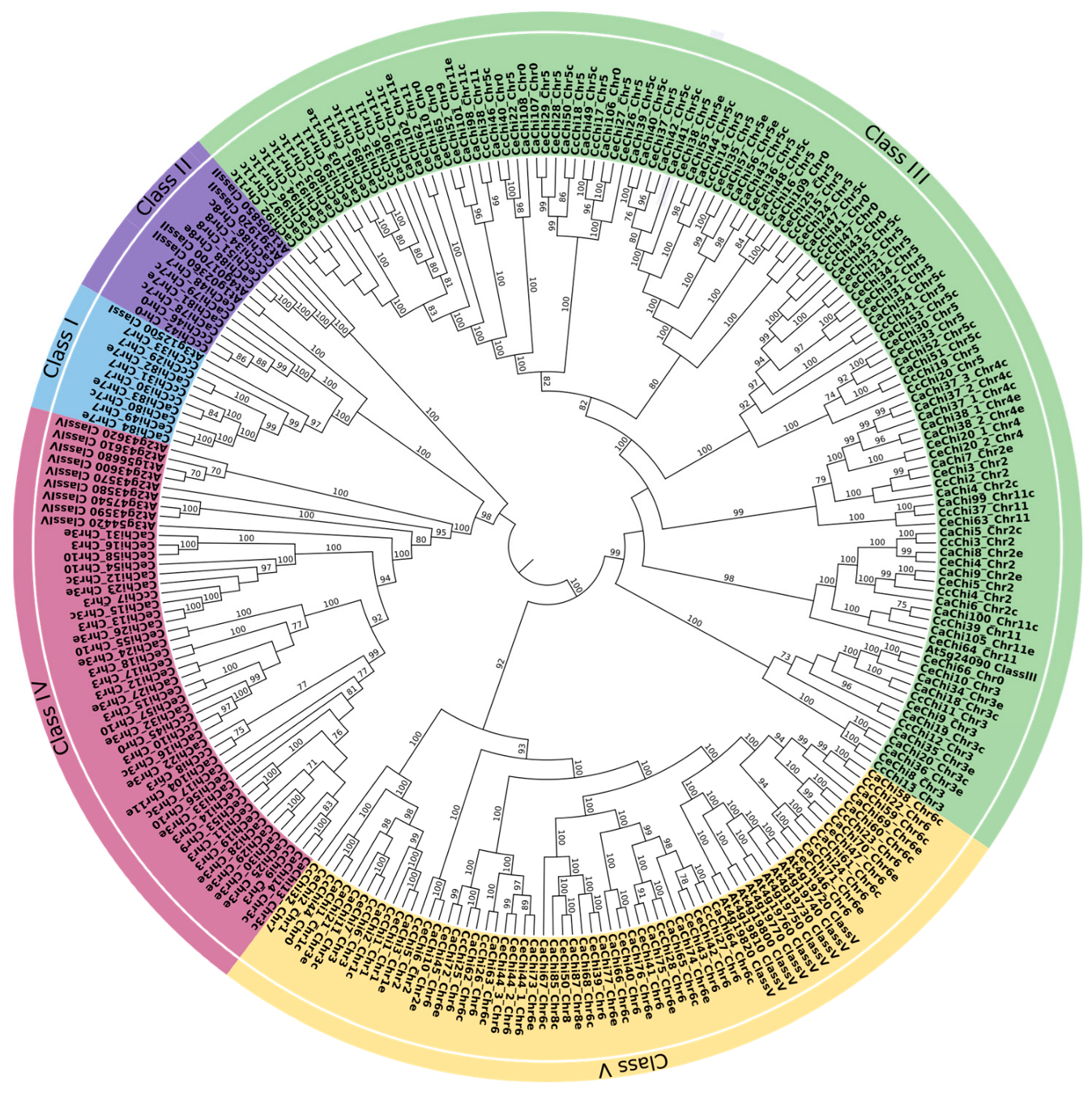
2.2. Phylogenetic Analysis
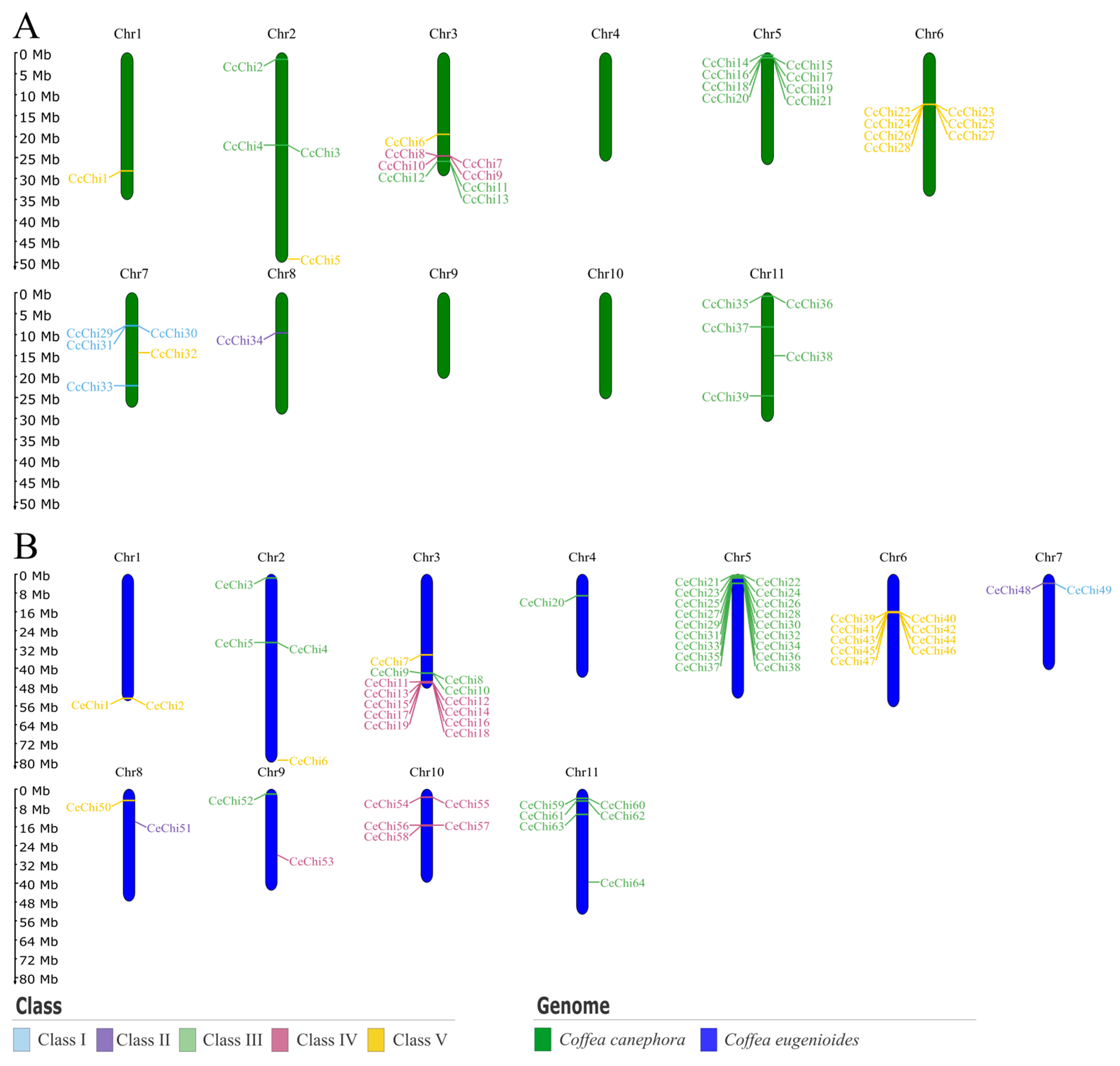
2.3. Chromosomal Location and Distribution
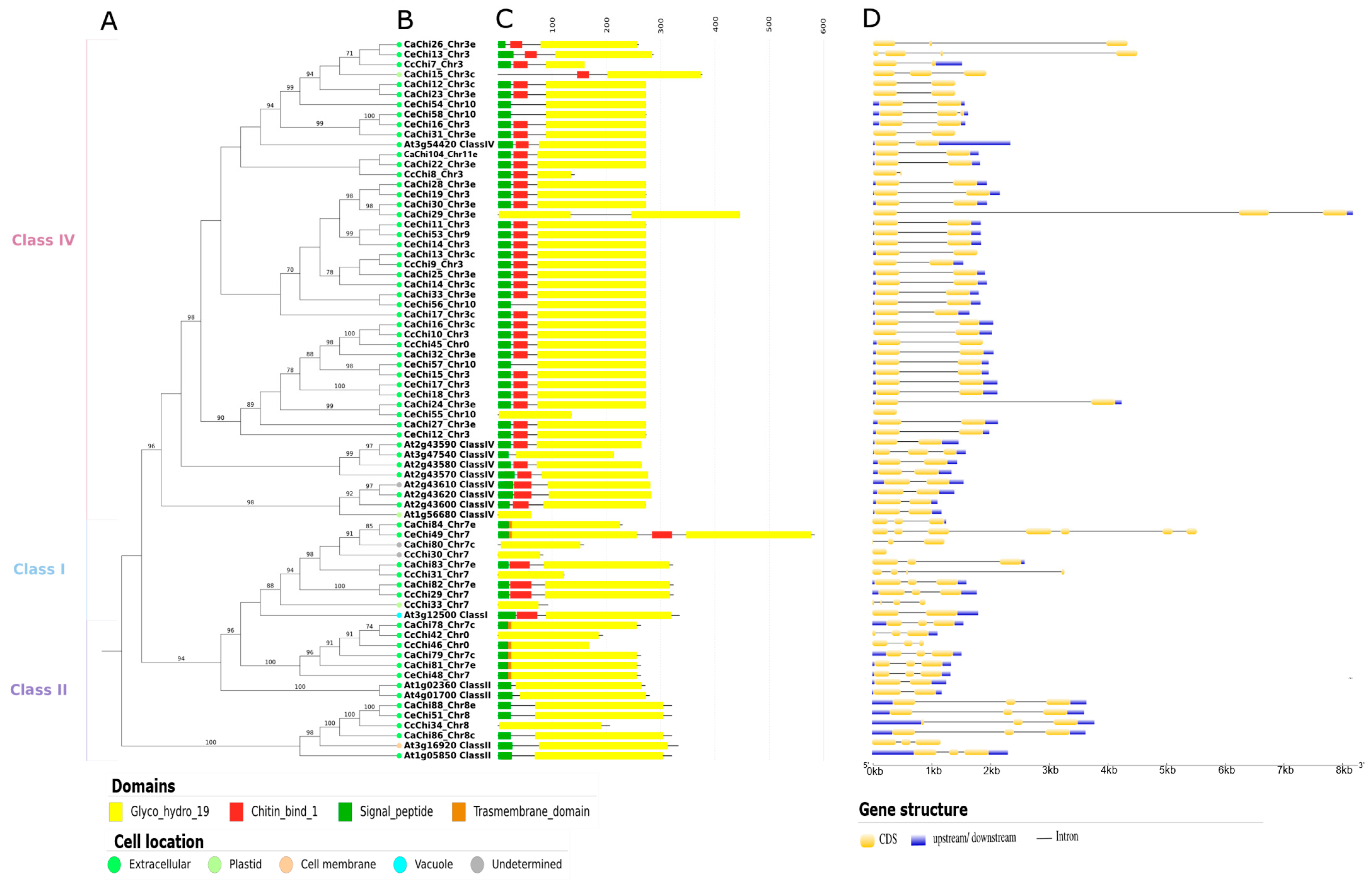
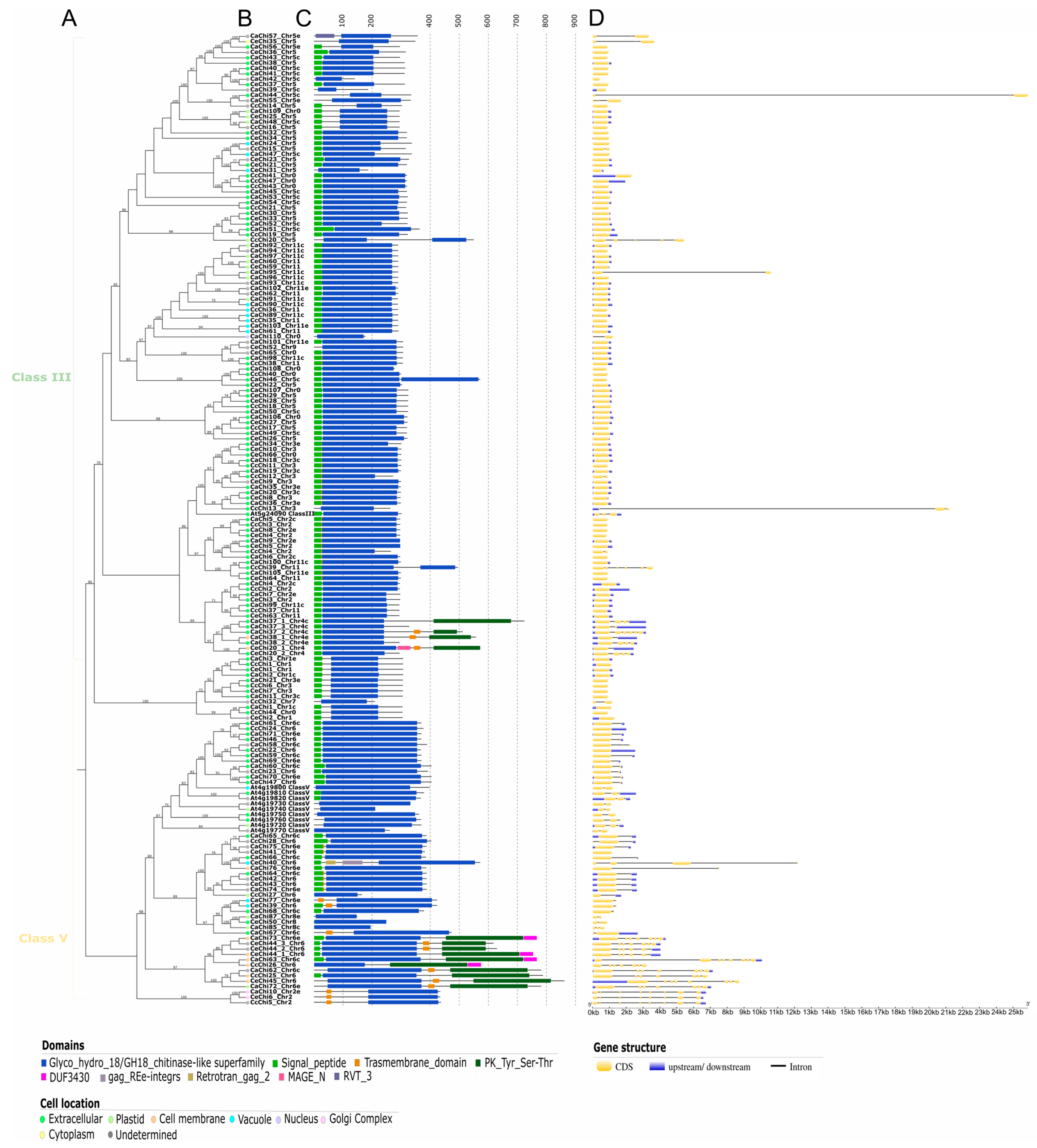
2.4. Domain Architecture Analysis and Subcellular Localization
2.5. Gene Structure Analysis
2.6. Orthologs Analysis
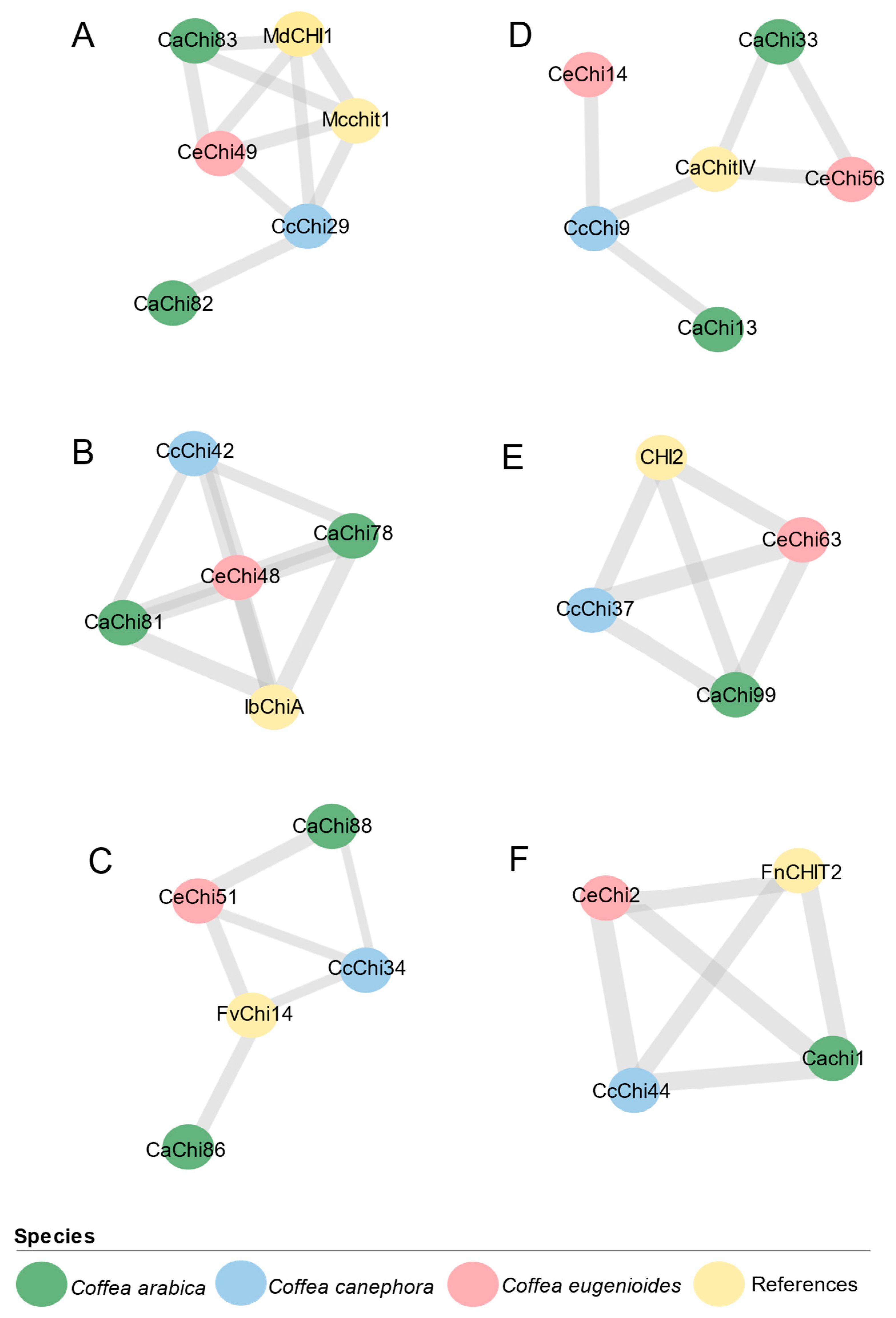
2.7. Putative Fungal Defense Response Chitinase in Coffea spp.
3. Discussion
4. Materials and Methods
4.1. Coffea spp. Genomes
4.2. In Silico Identification of Putative Chitinase
4.3. Phylogenetic Analyses
4.4. Chromosomal Mapping of Chitinase Genes
4.5. Domain Architecture Analysis and Subcellular Location
4.6. Gene Structure Annotation and Analysis
4.7. Orthogroups Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Coffee Organization. Coffee Market Report—March 2025; ICO: London, UK, 2025. [Google Scholar]
- Lashermes, P.; Combes, M.C.; Robert, J.; Trouslot, P.; D’Hont, A.; Anthony, F.; Charrier, A. Molecular characterisation and origin of the Coffea arabica L. genome. Mol. Genet. Genom. 1999, 261, 259–266. [Google Scholar] [CrossRef]
- Scalabrin, S.; Toniutti, L.; Di Gaspero, G.; Scaglione, D.; Magris, G.; Vidotto, M.; Pinosio, S.; Cattonaro, F.; Magni, F.; Jurman, I.; et al. A single polyploidization event at the origin of the tetraploid genome of Coffea arabica is responsible for the extremely low genetic variation in wild and cultivated germplasm. Sci. Rep. 2020, 10, 4642. [Google Scholar] [CrossRef]
- Lashermes, P.; Combes, M.C.; Topart, P.; Graziosi, G.; Bertrand, B.; Anthony, F. Molecular breeding in coffee (Coffea arabica L.). In Coffee Biotechnology and Quality; Springer: Dordrecht, The Netherlands, 2000; pp. 101–112. [Google Scholar] [CrossRef]
- Fazuoli, L.C.; Maluf, M.P.; Filho, O.G.; Filho, H.M.; Silvarolla, M.B. Breeding and biotechnology of coffee. In Coffee Biotechnology and Quality; Springer: Dordrecht, The Netherlands, 2000; pp. 27–45. [Google Scholar]
- Hall, R.D.; Trevisan, F.; de Vos, R.C.H. Coffee berry and green bean chemistry—Opportunities for improving cup quality and crop circularity. Food Res. Int. 2022, 151, 110825. [Google Scholar] [CrossRef]
- Seninde, D.R.; Chambers, E. Coffee flavor: A review. Beverages 2020, 6, 44. [Google Scholar] [CrossRef]
- International Coffee Organization. ICO December 2023: Coffee Report and Outlook; ICO: London, UK, 2023. [Google Scholar]
- Harvey, C.A.; Rakotobe, L.Z.; Rao, N.S.; Dave, R.; Razafimahatratra, H.; Rabarijohn, R.H.; Rajaofara, H.; MacKinnon, J.L. Extreme vulnerability of smallholder farmers to agricultural risks and climate change in Madagascar. Philos. Trans. R. Soc. B 2014, 369, 20130089. [Google Scholar] [CrossRef] [PubMed]
- Pham, Y.; Reardon-Smith, K.; Mushtaq, S.; Cockfield, G. The impact of climate change and variability on coffee production: A systematic review. Clim. Change 2019, 156, 609–630. [Google Scholar] [CrossRef]
- Lu, L.; Tibpromma, S.; Karunarathna, S.C.; Jayawardena, R.S.; Lumyong, S.; Xu, J.; Hyde, K.D. Comprehensive review of fungi on coffee. Pathogens 2022, 11, 411. [Google Scholar] [CrossRef]
- Talhinhas, P.; Batista, D.; Diniz, I.; Vieira, A.; Silva, D.; Loureiro, A.; Tavares, S.; Pereira, A.; Azinheira, H.; Guerra-Guimarães, L.; et al. The coffee leaf rust pathogen Hemileia vastatrix: One and a half centuries around the tropics. Mol. Plant Pathol. 2017, 18, 1039–1051. [Google Scholar] [CrossRef]
- Botelho, D.M.S.; de Resende, M.L.V.; Andrade, V.T.; Pereira, A.A.; Patricio, F.R.A.; Junior, P.M.R.; Ogoshi, C.; de Rezende, J.C. Cercosporiosis resistance in coffee germplasm collection. Euphytica 2017, 213, 117. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, C.; Si, J.; Han, Z.; Chen, D. Action mechanisms of effectors in plant–pathogen interaction. Int. J. Mol. Sci. 2022, 23, 6758. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Huffaker, A. Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 2011, 14, 351–357. [Google Scholar] [CrossRef]
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Igarashi, D.; Mase, K.; Lu, Y.; Tsuda, Y.; Chakravarthy, S.; Wei, H.; Foley, J.W.; Collmer, A.; Glazebrook, J.; et al. A plant effector-triggered immunity signaling sector is inhibited by pattern-triggered immunity. EMBO J. 2017, 36, 2758–2769. [Google Scholar] [CrossRef]
- Andersen, E.J.; Ali, S.; Byamukama, E.; Yen, Y.; Nepal, M.P. Disease resistance mechanisms in plants. Genes 2018, 9, 339. [Google Scholar] [CrossRef]
- Filyushin, M.A.; Anisimova, O.K.; Kochieva, E.Z.; Shchennikova, A.V. Genome-wide identification and expression of chitinase class I genes in garlic (Allium sativum L.) cultivars resistant and susceptible to Fusarium proliferatum. Plants 2021, 10, 720. [Google Scholar] [CrossRef]
- Kesari, P.; Patil, D.N.; Kumar, P.; Tomar, S.; Sharma, A.K.; Kumar, P. Structural and functional evolution of chitinase-like proteins from plants. Proteomics 2015, 15, 1693–1705. [Google Scholar] [CrossRef]
- Oliveira, S.T.; Azevedo, M.I.G.; Cunha, R.M.S.; Silva, C.F.B.; Muniz, C.R.; Monteiro-Júnior, J.E.; Carneiro, R.F.; Nagano, C.S.; Girão, M.S.; Freitas, C.D.T.; et al. Structural and functional features of a class VI chitinase from cashew (Anacardium occidentale L.) with antifungal properties. Phytochemistry 2020, 180, 112527. [Google Scholar] [CrossRef]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Mir, Z.A.; Ali, S.; Shivaraj, S.M.; Bhat, J.A.; Singh, A.; Yadav, P.; Rawat, S.; Paplao, P.K.; Grover, A. Genome-wide identification and characterization of chitinase gene family in Brassica juncea and Camelina sativa in response to Alternaria brassicae. Genomics 2020, 112, 749–763. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, J.-M.; Fritig, B.; Linthorst, H.J.M.; Meins, F.; Mikkelsen, J.D.; Ryals, J. A revised nomenclature of chitinase genes. Plant Mol. Biol. Report. 1996, 14, 102–104. [Google Scholar] [CrossRef]
- Bordoloi, K.S.; Krishnatreya, D.B.; Baruah, P.M.; Borah, A.K.; Mondal, T.K.; Agarwala, N. Genome-wide identification and expression profiling of chitinase genes in tea (Camellia sinensis (L.) O. Kuntze) under biotic stress conditions. Physiol. Mol. Biol. Plants 2021, 27, 369–385. [Google Scholar] [CrossRef]
- Cao, J.; Tan, X. Comprehensive analysis of the chitinase family genes in tomato (Solanum lycopersicum). Plants 2019, 8, 52. [Google Scholar] [CrossRef]
- Shinshi, H.; Neuhaus, J.-M.; Ryals, J.; Meins, F. Structure of a tobacco endochitinase gene: Evidence that different chitinase genes can arise by transposition of sequences encoding a cysteine-rich domain. Plant Mol. Biol. 1990, 14, 357–368. [Google Scholar] [CrossRef]
- Berglund, L.; Brunstedt, J.; Nielsen, K.K.; Chen, Z.; Mikkelsen, J.D.; Marcker, K.A. A proline-rich chitinase from Beta vulgaris. Plant Mol. Biol. 1995, 27, 211–216. [Google Scholar] [CrossRef]
- Arakane, Y.; Taira, T.; Ohnuma, T.; Fukamizo, T. Chitin-related enzymes in agro-biosciences. Curr. Drug Targets 2012, 13, 442–470. [Google Scholar] [CrossRef]
- Ohnuma, T.; Umemoto, N.; Kondo, K.; Numata, T.; Fukamizo, T. Complete subsite mapping of a “loopful” GH19 chitinase from rye seeds based on its crystal structure. FEBS Lett. 2013, 587, 2691–2697. [Google Scholar] [CrossRef]
- Zheng, T.; Zhang, K.; Sadeghnezhad, E.; Jiu, S.; Zhu, X.; Dong, T.; Liu, Z.; Guan, L.; Jia, H.; Fang, J. Chitinase family genes in grape differentially expressed in a manner specific to fruit species in response to Botrytis cinerea. Mol. Biol. Rep. 2020, 47, 7349–7363. [Google Scholar] [CrossRef]
- Lv, P.; Zhang, C.; Xie, P.; Yang, X.; El-Sheikh, M.; Hefft, D.; Ahmad, P.; Zhao, T.; Bhat, J. Genome-wide identification and expression analyses of the chitinase gene family in response to white mold and drought stress in soybean (Glycine max). Life 2022, 12, 1340. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.C.; Purugganan, M.D. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 2005, 8, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Grover, A. Plant chitinases: Genetic diversity and physiological roles. Crit. Rev. Plant Sci. 2012, 31, 57–73. [Google Scholar] [CrossRef]
- Bishop, J.G.; Dean, A.M.; Mitchell-Olds, T. Rapid evolution in plant chitinases: Molecular targets of selection in plant–pathogen coevolution. Proc. Natl. Acad. Sci. USA 2000, 97, 5322–5327. [Google Scholar] [CrossRef]
- Wen, Z.; Bai, J.; Wang, L.; Yao, L.; Ahmad, B.; Hanif, M.; Chen, Q. Overexpression of a chitinase 2 gene from Chinese wild strawberry improves resistance to anthracnose disease in transgenic Arabidopsis thaliana. Plant Biotechnol. Rep. 2020, 14, 725–736. [Google Scholar] [CrossRef]
- Haxim, Y.; Kahar, G.; Zhang, X.; Si, Y.; Waheed, A.; Liu, X.; Wen, X.; Li, X.; Zhang, D. Genome-wide characterization of the chitinase gene family in wild apple (Malus sieversii) and domesticated apple (Malus domestica) reveals its role in resistance to Valsa mali. Front. Plant Sci. 2022, 13, 1007936. [Google Scholar] [CrossRef]
- He, T.; Fan, J.; Jiao, G.; Liu, Y.; Zhang, Q.; Luo, N.; Ahmad, B.; Chen, Q.; Wen, Z. Bioinformatics and expression analysis of the chitinase genes in strawberry (Fragaria vesca) and functional study of FvChi-14. Plants 2023, 12, 1543. [Google Scholar] [CrossRef]
- Kishimoto, K.; Nishizawa, Y.; Tabei, Y.; Nakajima, M.; Hibi, T.; Akutsu, K. Transgenic cucumber expressing an endogenous class III chitinase gene has reduced symptoms from Botrytis cinerea. J. Gen. Plant Pathol. 2004, 70, 314–320. [Google Scholar] [CrossRef]
- Guerra-Guimarães, L.; Tenente, R.; Pinheiro, C.; Chaves, I.; Silva, M.C.; Cardoso, F.M.H.; Planchon, S.; Barros, D.R.; Renaut, J.; Ricardo, C.P. Proteomic analysis of apoplastic fluid of Coffea arabica leaves highlights novel biomarkers for resistance against Hemileia vastatrix. Front. Plant Sci. 2015, 6, 478. [Google Scholar] [CrossRef]
- Guerra-Guimarães, L.; Silva, M.C.; Struck, C.; Loureiro, A.; Nicole, M.; Rodrigues, C.J.; Ricardo, C.P.P. Chitinases of Coffea arabica genotypes resistant to orange rust Hemileia vastatrix. Biol. Plant. 2009, 53, 702–706. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Passarinho, P.A.; de Vries, S.C. Arabidopsis chitinases: A genomic survey. Arab. Book 2002, 1, e0023. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.-H.; Li, X.-B.; Yang, X.-Y.; Luo, M.; Hou, L.; Guo, S.-H.; Luo, X.-Y.; Pei, Y. Cloning and characterization of a balsam pear class I chitinase gene (McCHIT1) and its ectopic expression enhances fungal resistance in transgenic plants. Biosci. Biotechnol. Biochem. 2007, 71, 1211–1219. [Google Scholar] [CrossRef]
- Li, P.; Pei, Y.; Sang, X.; Ling, Y.; Yang, Z.; He, G. Transgenic indica rice expressing a bitter melon (Momordica charantia) class I chitinase gene (McCHIT1) confers enhanced resistance to Magnaporthe grisea and Rhizoctonia solani. Eur. J. Plant Pathol. 2009, 125, 533–543. [Google Scholar] [CrossRef]
- Wang, F.; Yang, S.; Wang, Y.; Zhang, B.; Zhang, F.; Xue, H.; Jiang, Q.; Ma, Y. Overexpression of chitinase gene enhances resistance to Colletotrichum gloeosporioides and Alternaria alternata in apple (Malus × domestica). Sci. Hortic. 2021, 277, 109779. [Google Scholar] [CrossRef]
- Liu, M.; Gong, Y.; Sun, H.; Zhang, J.; Zhang, L.; Sun, J.; Han, Y.; Huang, J.; Wu, Q.; Zhang, C.; et al. Characterization of a novel chitinase from sweet potato and its fungicidal effect against Ceratocystis fimbriata. J. Agric. Food Chem. 2020, 68, 7591–7600. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Kim, N.H.; Hwang, B.K. The Capsicum annuum class IV chitinase ChitIV interacts with receptor-like cytoplasmic protein kinase PIK1 to accelerate PIK1-triggered cell death and defence responses. J. Exp. Bot. 2015, 66, 1987–1999. [Google Scholar] [CrossRef] [PubMed]
- Chouhan, R.; Ahmed, S.; Gandhi, S.G. Over-expression of PR proteins with chitinase activity in transgenic plants for alleviation of fungal pathogenesis. J. Plant Pathol. 2023, 105, 69–81. [Google Scholar] [CrossRef]
- Sharma, S.; Gautam, N.; Thakur, A.K.; Srivastava, D.K. Transgenic lettuce (Lactuca sativa L.) harboring chitinase gene expressed resistance against a devastating fungus, Sclerotinia sclerotiorum. Vegetos 2023, 36, 1265–1274. [Google Scholar] [CrossRef]
- Salojärvi, J.; Rambani, A.; Yu, Z.; Guyot, R.; Strickler, S.; Lepelley, M.; Wang, C.; Rajaraman, S.; Rastas, P.; Zheng, C.; et al. The genome and population genomics of allopolyploid Coffea arabica reveal the diversification history of modern coffee cultivars. Nat. Genet. 2024, 56, 721–731. [Google Scholar] [CrossRef]
- Zheng, T.; Zhang, K.; Zhu, X.; Jiu, S.; Dong, T.; Liu, Z.; Guan, L.; Jia, H.; Fang, J. Genome-wide identification and functional analysis of chitinase gene family in grape. Res. Sq. 2020, preprint. [Google Scholar] [CrossRef]
- Kwon, C.; Bednarek, P.; Schulze-Lefert, P. Secretory pathways in plant immune responses. Plant Physiol. 2008, 147, 1575–1583. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The Conserved Domain Database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Sahu, S.S.; Loaiza, C.D.; Kaundal, R. Plant-mSubP: A computational framework for the prediction of single- and multi-target protein subcellular localization using integrated machine-learning approaches. AoB Plants 2021, 12, plz068. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-Y.; Sang, H.; Chilvers, M.I.; Wu, C.-H.; Chang, H.-X. Characterization of soybean chitinase genes induced by rhizobacteria involved in the defense against Fusarium oxysporum. Front. Plant Sci. 2024, 15, 1341181. [Google Scholar] [CrossRef]
- Sun, J.; Lu, F.; Luo, Y.; Bie, L.; Xu, L.; Wang, Y. OrthoVenn3: An integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 2023, 51, W397–W403. [Google Scholar] [CrossRef]
- Khan, A.; Nasir, I.A.; Tabassum, B.; Aaliya, K.; Tariq, M.; Rao, A.Q. Expression studies of chitinase gene in transgenic potato against Alternaria solani. Plant Cell Tissue Organ Cult. 2017, 128, 563–576. [Google Scholar] [CrossRef]
- Chopra, R.; Saini, R. Transformation of blackgram (Vigna mungo (L.) Hepper) by barley chitinase and ribosome-inactivating protein genes towards improving resistance to Corynespora leaf spot fungal disease. Appl. Biochem. Biotechnol. 2014, 174, 2791–2800. [Google Scholar] [CrossRef]
- Jach, G.; Görnhardt, B.; Mundy, J.; Logemann, J.; Pinsdorf, E.; Leah, R.; Schell, J.; Maas, C. Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 1995, 8, 97–109. [Google Scholar] [CrossRef]
- Shin, S.; Mackintosh, C.A.; Lewis, J.; Heinen, S.J.; Radmer, L.; Dill-Macky, R.; Baldridge, G.D.; Zeyen, R.J.; Muehlbauer, G.J. Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum. J. Exp. Bot. 2008, 59, 2371–2378. [Google Scholar] [CrossRef]
- Eissa, H.F.; Hassanien, S.E.; Ramadan, A.M.; El-Shamy, M.M.; Saleh, O.M.; Shokry, A.M.; Abdelsattar, M.; Morsy, Y.B.; El-Maghraby, M.A.; Alameldin, H.F.; et al. Developing transgenic wheat to encounter rusts and powdery mildew by overexpressing barley Chi26 gene for fungal resistance. Plant Methods 2017, 13, 1–12. [Google Scholar] [CrossRef]
- Tariq, M.; Khan, A.; Tabassum, B.; Toufiq, N.; Bhatti, M.U.; Riaz, S.; Nasir, I.A.; Husnain, T. Antifungal activity of chitinase II against Colletotrichum falcatum Went. causing red rot disease in transgenic sugarcane. Turk. J. Biol. 2018, 42, 45–53. [Google Scholar] [CrossRef]
- Girhepuje, P.V.; Shinde, G.B. Transgenic tomato plants expressing a wheat endochitinase gene demonstrate enhanced resistance to Fusarium oxysporum f. sp. lycopersici. Plant Cell Tissue Organ Cult. 2011, 105, 243–251. [Google Scholar] [CrossRef]
- Tohidfar, M.; Mohammadi, M.; Ghareyazie, B. Agrobacterium-mediated transformation of cotton (Gossypium hirsutum) using a heterologous bean chitinase gene. Plant Cell Tissue Organ Cult. 2005, 83, 83–96. [Google Scholar] [CrossRef]
- Benhamou, N.; Broglie, K.; Chet, I.; Broglie, R. Cytology of infection of 35S-bean chitinase transgenic canola plants by Rhizoctonia solani: Cytochemical aspects of chitin breakdown in vivo. Plant J. 1993, 4, 295–305. [Google Scholar] [CrossRef]
- Broglie, K.; Chet, I.; Holliday, M.; Cressman, R.; Biddle, P.; Knowlton, S.; Mauvais, C.J.; Broglie, R. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 1991, 254, 1194–1197. [Google Scholar] [CrossRef]
- Vellicce, G.R.; Ricci, J.C.D.; Hernández, L.; Castagnaro, A.P. Enhanced resistance to Botrytis cinerea mediated by the transgenic expression of the chitinase gene Ch5B in strawberry. Transgenic Res. 2006, 15, 57–68. [Google Scholar] [CrossRef]
- Maximova, S.N.; Marelli, J.P.; Young, A.; Pishak, S.; Verica, J.A.; Guiltinan, M.J. Overexpression of a cacao class I chitinase gene in Theobroma cacao L. enhances resistance against the pathogen Colletotrichum gloeosporioides. Planta 2006, 224, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Zhao, Y.; Ran, X.; Guo, L.; Zhao, D.G. Overexpression of a new chitinase gene EuCHIT2 enhances resistance to Erysiphe cichoracearum DC in tobacco plants. Int. J. Mol. Sci. 2017, 18, 2361. [Google Scholar] [CrossRef]
- Xin, Y.; Wang, D.; Han, S.; Li, S.; Gong, N.; Fan, Y.; Ji, X. Characterization of the chitinase gene family in mulberry (Morus notabilis) and MnChi18 involved in resistance to Botrytis cinerea. Genes 2022, 13, 98. [Google Scholar] [CrossRef] [PubMed]
- Dowd, P.F.; Naumann, T.A.; Price, N.P.J.; Johnson, E.T. Identification of a maize (Zea mays) chitinase allele sequence suitable for a role in ear rot fungal resistance. Agri Gene 2018, 7, 15–22. [Google Scholar] [CrossRef]
- Marchant, R.; Davey, M.R.; Lucas, J.A.; Lamb, C.J.; Dixon, R.A.; Power, J.B. Expression of a chitinase transgene in rose (Rosa hybrida L.) reduces development of blackspot disease (Diplocarpon rosae Wolf). Mol. Breed. 1998, 4, 187–194. [Google Scholar] [CrossRef]
- Núñez de Cáceres González, F.F.; Davey, M.R.; Cancho Sanchez, E.; Wilson, Z.A. Conferred resistance to Botrytis cinerea in Lilium by overexpression of the RCH10 chitinase gene. Plant Cell Rep. 2015, 34, 1201–1209. [Google Scholar] [CrossRef]
- Kovács, G.; Sági, L.; Jacon, G.; Arinaitwe, G.; Busogoro, J.P.; Thiry, E.; Strosse, H.; Swennen, R.; Remy, S. Expression of a rice chitinase gene in transgenic banana (“Gros Michel”, AAA genome group) confers resistance to black leaf streak disease. Transgenic Res. 2013, 22, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Takatsu, Y.; Nishizawa, Y.; Hibi, T.; Akutsu, K. Transgenic chrysanthemum (Dendranthema grandiflorum (Ramat.) Kitamura) expressing a rice chitinase gene shows enhanced resistance to gray mold (Botrytis cinerea). Sci. Hortic. 1999, 82, 113–123. [Google Scholar] [CrossRef]
- Tabei, Y.; Kitade, S.; Nishizawa, Y.; Kikuchi, N.; Kayano, T.; Hibi, T.; Akutsu, K. Transgenic cucumber plants harboring a rice chitinase gene exhibit enhanced resistance to gray mold (Botrytis cinerea). Plant Cell Rep. 1998, 17, 159–164. [Google Scholar] [CrossRef]
- Kishimoto, K.; Nishizawa, Y.; Tabei, Y.; Hibi, T.; Nakajima, M.; Akutsu, K. Detailed analysis of rice chitinase gene expression in transgenic cucumber plants showing different levels of disease resistance to gray mold (Botrytis cinerea). Plant Sci. 2002, 162, 655–662. [Google Scholar] [CrossRef]
- Nishizawa, Y.; Notsuka, K.; Hibi, T.; Yamamoto, T. Transgenic grapevine plants expressing a rice chitinase with enhanced resistance to fungal pathogens. Plant Cell Rep. 2000, 19, 639–646. [Google Scholar] [CrossRef]
- Takahashi, W.; Fujimori, M.; Miura, Y.; Komatsu, T.; Nishizawa, Y.; Hibi, T.; Takamizo, T. Increased resistance to crown rust disease in transgenic Italian ryegrass (Lolium multiflorum Lam.) expressing the rice chitinase gene. Plant Cell Rep. 2005, 23, 811–818. [Google Scholar] [CrossRef]
- Datta, K.; Tu, J.; Oliva, N.; Ona, I.; Velazhahan, R.; Mew, W.; Muthukrishnan, S.; Datta, S.K. Enhanced resistance to sheath blight by constitutive expression of infection-related rice chitinase in transgenic elite indica rice cultivars. Plant Sci. 2001, 160, 405–414. [Google Scholar] [CrossRef]
- Iqbal, M.M.; Nazir, F.; Ali, S.; Asif, M.A.; Zafar, Y.; Iqbal, J.; Ali, G.M. Overexpression of rice chitinase gene in transgenic peanut (Arachis hypogaea L.) improves resistance against leaf spot. Mol. Biotechnol. 2012, 50, 129–136. [Google Scholar] [CrossRef]
- Nishizawa, Y.; Nishio, Z.; Nakazono, K.; Soma, M.; Nakajima, E.; Ugaki, M.; Hibi, T. Enhanced resistance to blast (Magnaporthe grisea) in transgenic japonica rice by constitutive expression of rice chitinase. Theor. Appl. Genet. 1999, 99, 383–390. [Google Scholar] [CrossRef]
- Asao, H.; Nishizawa, Y.; Arai, S.; Sato, T.; Hirai, M.; Yoshida, K.; Shinmyo, A.; Hibi, T. Enhanced resistance against a fungal pathogen Sphaerotheca humuli in transgenic strawberry expressing a rice chitinase gene. Plant Biotechnol. 1997, 14, 145–149. [Google Scholar] [CrossRef]
- Jabeen, N.; Chaudhary, Z.; Gulfraz, M.; Rashid, H.; Mirza, B. Expression of rice chitinase gene in genetically engineered tomato confers enhanced resistance to Fusarium wilt and early blight. Plant Pathol. J. 2015, 31, 252–258. [Google Scholar] [CrossRef]
- Huang, X.; Wang, J.; Du, Z.; Zhang, C.; Li, L.; Xu, Z. Enhanced resistance to stripe rust disease in transgenic wheat expressing the rice chitinase gene RC24. Transgenic Res. 2013, 22, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Ignacimuthu, S.; Ceasar, S.A. Development of transgenic finger millet (Eleusine coracana (L.) Gaertn.) resistant to leaf blast disease. J. Biosci. 2012, 37, 135–147. [Google Scholar] [CrossRef]
- Nirala, N.K.; Das, D.K.; Srivastava, P.S.; Sopory, S.K.; Upadhyaya, K.C. Expression of a rice chitinase gene enhances antifungal potential in transgenic grapevine (Vitis vinifera L.). Vitis 2010, 49, 181–187. [Google Scholar] [CrossRef]
- Das, D.K.; Rahman, A. Expression of a rice chitinase gene enhances antifungal response in transgenic litchi (Litchi chinensis Sonn.) cv. Bedana. Am. J. Plant Sci. 2018, 9, 2256–2275. [Google Scholar] [CrossRef]
- Prasad, K.; Bhatnagar-Mathur, P.; Waliyar, F.; Sharma, K.K. Overexpression of a chitinase gene in transgenic peanut confers enhanced resistance to major soil-borne and foliar fungal pathogens. J. Plant Biochem. Biotechnol. 2013, 22, 222–233. [Google Scholar] [CrossRef]
- Rajesh, T.; Maruthasalam, S.; Kalpana, K.; Poovannan, K.; Kumar, K.K.; Kokiladevi, E.; Sudhakar, D.; Samiyappan, R.; Balasubramanian, P. Stability of sheath blight resistance in transgenic ASD16 rice lines expressing a rice Chi11 gene encoding chitinase. Biol. Plant. 2016, 60, 749–756. [Google Scholar] [CrossRef]
- Richa, K.; Tiwari, I.M.; Devanna, B.N.; Botella, J.R.; Sharma, V.; Sharma, T.R. Novel chitinase gene LOC_Os11g47510 from indica rice Tetep provides enhanced resistance against sheath blight pathogen Rhizoctonia solani in rice. Front. Plant Sci. 2017, 8, 596. [Google Scholar] [CrossRef] [PubMed]
- Durechova, D.; Jopcik, M.; Rajninec, M.; Moravcikova, J.; Libantova, J. Expression of Drosera rotundifolia chitinase in transgenic tobacco plants enhanced their antifungal potential. Mol. Biotechnol. 2019, 61, 916–928. [Google Scholar] [CrossRef]
- Pasonen, H.L.; Seppänen, S.K.; Degefu, Y.; Rytkönen, A.; von Weissenberg, K.; Pappinen, A. Field performance of chitinase transgenic silver birches (Betula pendula): Resistance to fungal diseases. Theor. Appl. Genet. 2004, 109, 562–570. [Google Scholar] [CrossRef]
- Pappinen, A.; Degefu, Y.; Syrjälä, L.; Keinonen, K.; von Weissenberg, K. Transgenic silver birch (Betula pendula) expressing sugarbeet chitinase 4 shows enhanced resistance to Pyrenopeziza betulicola. Plant Cell Rep. 2002, 20, 1046–1051. [Google Scholar] [CrossRef]
- Vierheilig, H. Colonization of transgenic Nicotiana sylvestris plants expressing different forms of Nicotiana tabacum chitinase by the root pathogen Rhizoctonia solani and by the mycorrhizal symbiont Glomus mosseae. Mol. Plant-Microbe Interact. 1993, 6, 261. [Google Scholar] [CrossRef]
- Rohini, V.K.; Rao, K.S. Transformation of peanut (Arachis hypogaea L.) with tobacco chitinase gene: Variable response of transformants to leaf spot disease. Plant Sci. 2001, 160, 889–898. [Google Scholar] [CrossRef]
- Pak, J.H.; Chung, E.S.; Shin, S.H.; Jeon, E.H.; Kim, M.J.; Lee, H.Y.; Jeung, J.U.; Hyung, N.I.; Lee, J.H.; Chung, Y.S. Enhanced fungal resistance in Arabidopsis expressing wild rice PR-3 (OgChitIVa) encoding chitinase class IV. Plant Biotechnol. Rep. 2009, 3, 147–155. [Google Scholar] [CrossRef]
- Chalavi, V.; Tabaeizadeh, Z.; Thibodeau, P. Enhanced resistance to Verticillium dahliae in transgenic strawberry plants expressing a Lycopersicon chilense chitinase gene. J. Am. Soc. Hortic. Sci. 2003, 128, 747–753. [Google Scholar] [CrossRef]
- Tabaeizadeh, Z.; Agharbaoui, Z.; Harrak, H.; Poysa, V. Transgenic tomato plants expressing a Lycopersicon chilense chitinase gene demonstrate improved resistance to Verticillium dahliae race 2. Plant Cell Rep. 1999, 19, 197–202. [Google Scholar] [CrossRef] [PubMed]


| Coffee Species | Proteins | Proteins Size | Genes | GH18 Genes | GH19 Genes |
|---|---|---|---|---|---|
| Coffea arabica | 113 | 141–782 | 110 | 82 | 28 |
| Coffea canephora | 47 | 84–788 | 47 | 35 | 12 |
| Coffea eugenioides | 69 | 187–862 | 66 | 48 | 18 |
| Cluster | Reference | Protein | Class | Size | Chr | Cell Localization | Domains 1 | Exons | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Mcchit1 [45,46] MdCHI1 [47] | CcChi29 | I | 324 | 7 | Extracellular | SP | CBD | GH19 | 3 | |
| 1 | Mcchit1 [45,46] MdCHI1 [47] | CeChi49 | I | 584 | 7 | Extracellular | SP | TMD | CBD | 2 GH19 | 7 |
| 1 | Mcchit1 [45,46] MdCHI1 [47] | CaChi83 | I | 323 | 7e | Extracellular | SP | CBD | GH19 | 3 | |
| 1 | Mcchit1 [45,46] MdCHI1 [47] | CaChi82 | I | 324 | 7e | Extracellular | SP | CBD | GH19 | 3 | |
| 2 | IbChiA [48] | CaChi81 | II | 264 | 7e | Extracellular | SP | TMD | GH19 | 3 | |
| 2 | IbChiA [48] | CaChi78 | II | 264 | 7c | Extracellular | SP | TMD | GH19 | 3 | |
| 2 | IbChiA [48] | CcChi42 | II | 194 | 0 | Extracellular | GH19 | 3 | |||
| 2 | IbChiA [48] | CeChi48 | II | 264 | 7 | Extracellular | SP | TMD | GH19 | 3 | |
| 3 | FvChi-14 [39] | CcChi34 | II | 207 | 8 | Extracellular | GH19 | 3 | |||
| 3 | FvChi-14 [39] | CeChi51 | II | 321 | 8 | Extracellular | SP | GH19 | 3 | ||
| 3 | FvChi-14 [39] | CaChi86 | II | 321 | 8c | Extracellular | SP | GH19 | 3 | ||
| 3 | FvChi-14 [39] | CaChi88 | II | 321 | 8e | Extracellular | SP | GH19 | 3 | ||
| 4 | CaChitIV [49] | CcChi9 | IV | 273 | 3 | Extracellular | SP | CBD | GH19 | 2 | |
| 4 | CaChitIV [49] | CeChi56 | IV | 273 | 10 | Extracellular | SP | GH19 | 2 | ||
| 4 | CaChitIV [49] | CaChi33 | IV | 273 | 3e | Extracellular | SP | CBD | GH19 | 2 | |
| 4 | CaChitIV [49] | CeChi14 | IV | 273 | 3 | Extracellular | SP | CBD | GH19 | 2 | |
| 4 | CaChitIV [49] | CaChi13 | IV | 273 | 3c | Extracellular | SP | CBD | GH19 | 2 | |
| 5 | CHI2 [40] | CcChi37 | III | 294 | 11 | Extracellular | SP | GH18 | 1 | ||
| 5 | CHI2 [40] | CeChi63 | III | 294 | 11 | Extracellular | SP | GH18 | 1 | ||
| 5 | CHI2 [40] | CaChi99 | III | 294 | 11c | Extracellular | SP | GH18 | 1 | ||
| 6 | FnCHIT2 [37] | CcChi44 | V | 305 | 0 | Extracellular | SP | GH18 | 1 | ||
| 6 | FnCHIT2 [37] | CeChi2 | V | 305 | 1 | Undetermined | SP | GH18 | 1 | ||
| 6 | FnCHIT2 [37] | CaChi1 | V | 305 | 1c | Extracellular | SP | GH18 | 1 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, F.R.; de Resende, M.L.V.; Xavier, K.V.; Brawner, J.T.; de Lima Santos, M. Genome-Wide Analysis Reveals Chitinases as Putative Defense-Related Proteins Against Fungi in the Genomes of Coffea arabica and Its Progenitors. Plants 2025, 14, 3130. https://doi.org/10.3390/plants14203130
Silva FR, de Resende MLV, Xavier KV, Brawner JT, de Lima Santos M. Genome-Wide Analysis Reveals Chitinases as Putative Defense-Related Proteins Against Fungi in the Genomes of Coffea arabica and Its Progenitors. Plants. 2025; 14(20):3130. https://doi.org/10.3390/plants14203130
Chicago/Turabian StyleSilva, Fernanda Rodrigues, Mario Lucio V. de Resende, Katia V. Xavier, Jeremy T. Brawner, and Mariana de Lima Santos. 2025. "Genome-Wide Analysis Reveals Chitinases as Putative Defense-Related Proteins Against Fungi in the Genomes of Coffea arabica and Its Progenitors" Plants 14, no. 20: 3130. https://doi.org/10.3390/plants14203130
APA StyleSilva, F. R., de Resende, M. L. V., Xavier, K. V., Brawner, J. T., & de Lima Santos, M. (2025). Genome-Wide Analysis Reveals Chitinases as Putative Defense-Related Proteins Against Fungi in the Genomes of Coffea arabica and Its Progenitors. Plants, 14(20), 3130. https://doi.org/10.3390/plants14203130









