Knock Down of Chlamydomonas reinhardtii Phytyl Ester Synthase α Triggers DGAT3 Overexpression and Triacylglycerol Accumulation Under Low-Light Conditions
Abstract
1. Introduction
2. Results
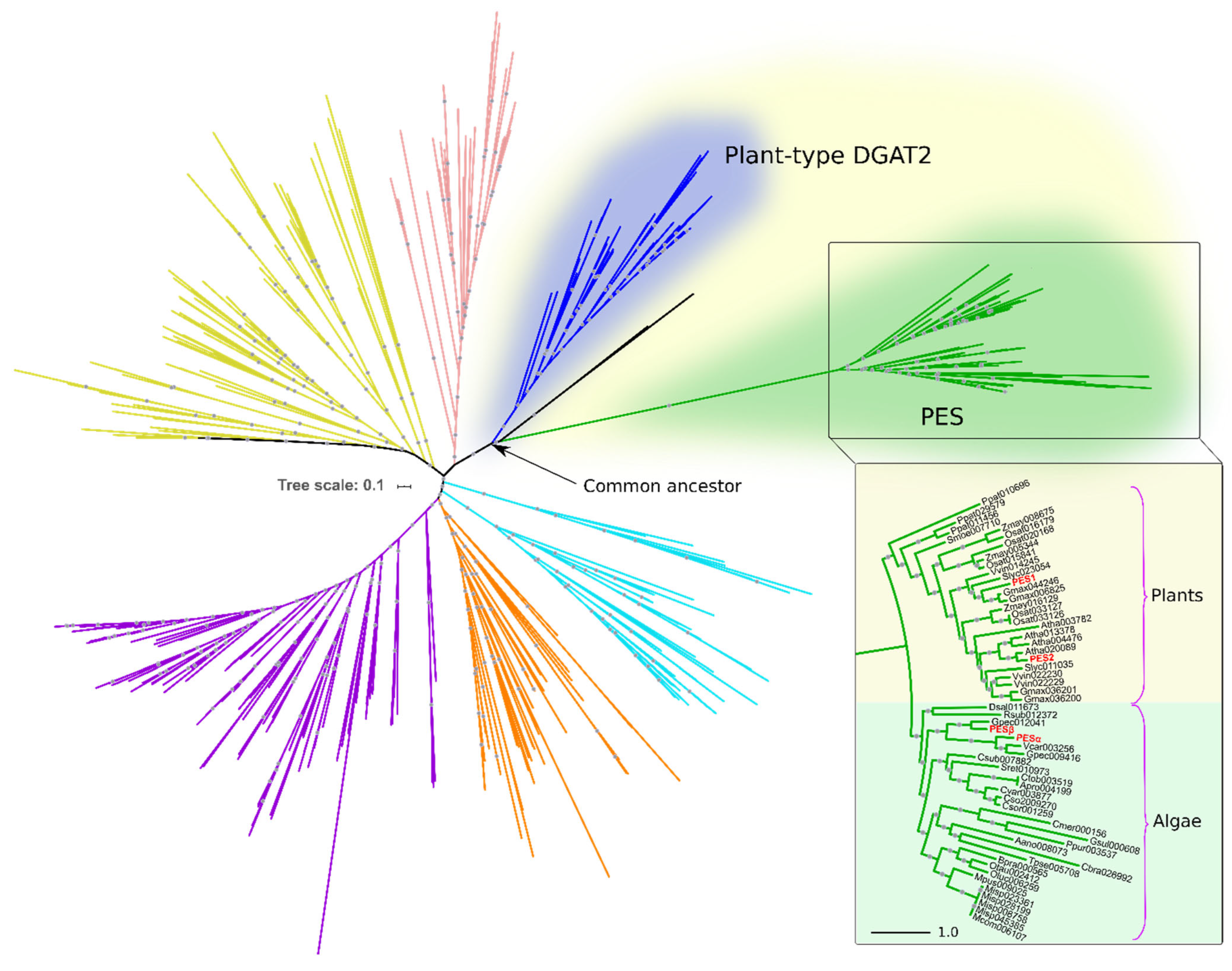
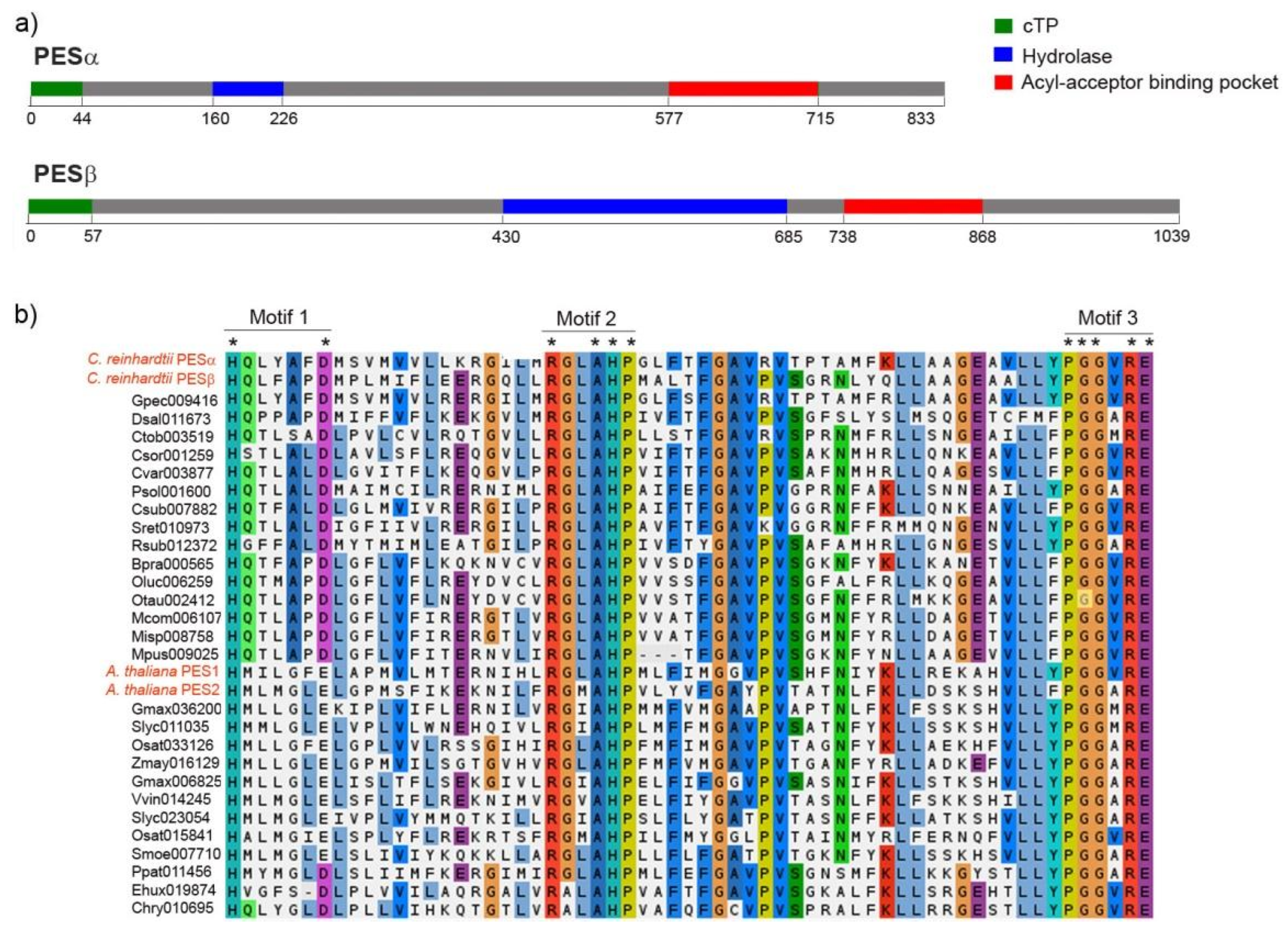
2.1. PESα and PESβ In Silico Analyses
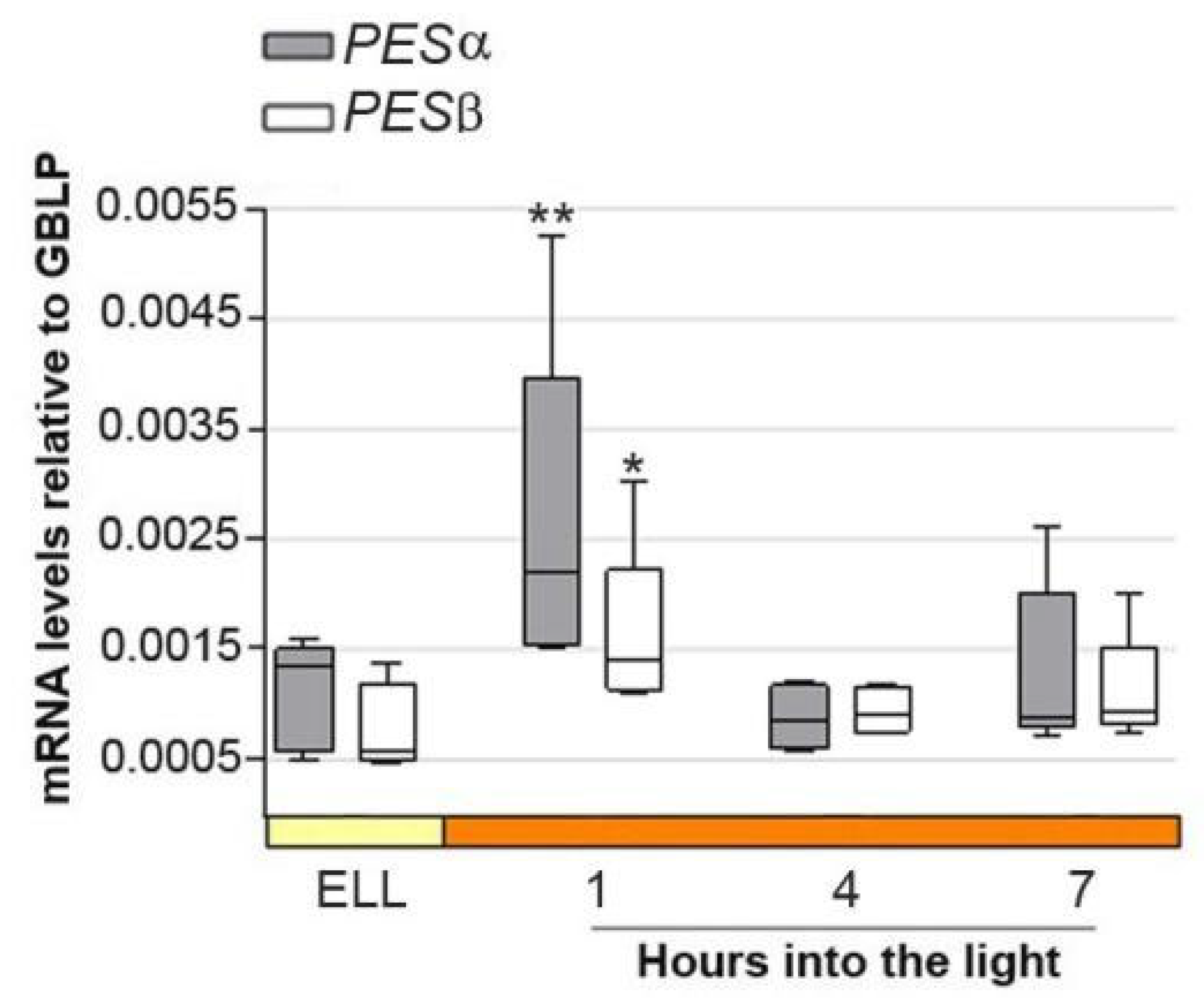
2.2. Analysis of PESα and PESβ Transcripts in the Parental Strain, cc-5325, After Shifting Cells from Low Light to High Light
2.3. Molecular Characterization of Pesα and Pesβ Mutants
2.4. Analysis of DGAT3, DGAT1, PDAT Expression and TAG Levels in cc-5325, Pesα and Pesβ Strains After Shifting Cells from Low Light to High Light
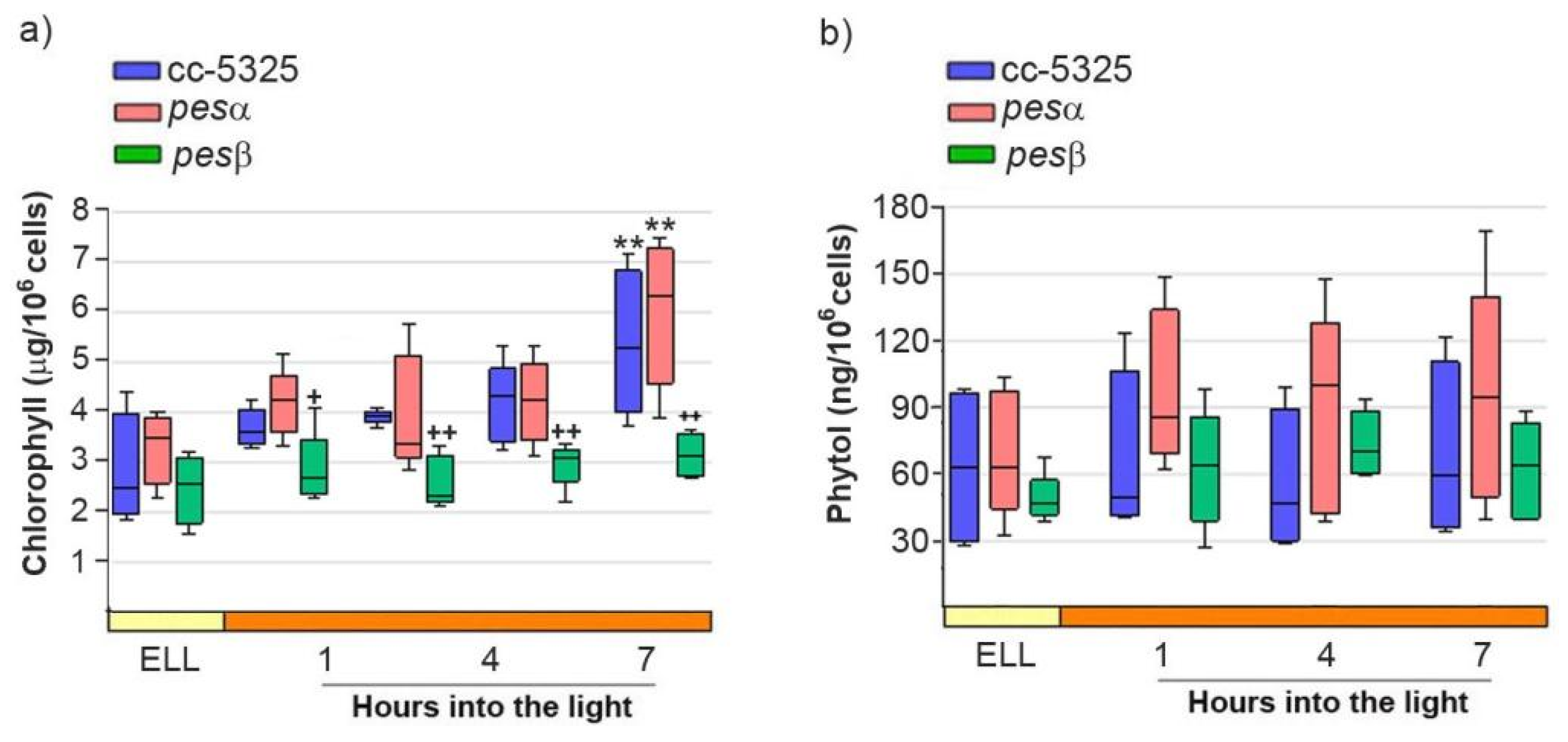
2.5. Analysis of FAPEs, Phytol and Chlorophyll Levels in cc-5325, Pesα and Pesβ Strains upon Shifting Cells from Low Light to High Light
3. Discussion
4. Materials and Methods
4.1. Chlamydomonas reinhardtii Strains and Culture Conditions
4.2. Determination of Growth Curve, Growth Rate and Cell Area
4.3. In Silico Analyses
4.4. DNA Isolation and PCR Mapping
4.5. RNA Isolation, cDNA Synthesis and Quantitative PCR Analyses
4.6. Lipid Extraction and Thin-Layer Chromatography
4.7. Detection of FAPEs by GC-MS
4.8. Quantification of Phytol by HPLC
4.9. Chlorophyll Quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mulgund, A. Increasing Lipid Accumulation in Microalgae through Environmental Manipulation, Metabolic and Genetic Engineering: A Review in the Energy NEXUS Framework. Energy Nexus 2022, 5, 100054. [Google Scholar] [CrossRef]
- Kong, F.; Blot, C.; Liu, K.; Kim, M.; Li-Beisson, Y. Advances in Algal Lipid Metabolism and Their Use to Improve Oil Content. Curr. Opin. Biotechnol. 2024, 87, 103130. [Google Scholar] [CrossRef]
- Du, Z.-Y.; Benning, C. Triacylglycerol Accumulation in Photosynthetic Cells in Plants and Algae. In Lipids in Plant and Algae Development; Nakamura, Y., Li-Beisson, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 179–205. ISBN 978-3-319-25979-6. [Google Scholar]
- Merchant, S.S.; Kropat, J.; Liu, B.; Shaw, J.; Warakanont, J. TAG, You’re It! Chlamydomonas as a Reference Organism for Understanding Algal Triacylglycerol Accumulation. Curr. Opin. Biotechnol. 2012, 23, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Rodolfi, L.; Chini Zittelli, G.; Bassi, N.; Padovani, G.; Biondi, N.; Bonini, G.; Tredici, M.R. Microalgae for Oil: Strain Selection, Induction of Lipid Synthesis and Outdoor Mass Cultivation in a Low-Cost Photobioreactor. Biotechnol. Bioeng. 2009, 102, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Benning, C. Lipid Metabolism in Microalgae Distinguishes Itself. Curr. Opin. Biotechnol. 2013, 24, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Goold, H.D.; Cuiné, S.; Légeret, B.; Liang, Y.; Brugière, S.; Auroy, P.; Javot, H.; Tardif, M.; Jones, B.; Beisson, F.; et al. Saturating Light Induces Sustained Accumulation of Oil in Plastidal Lipid Droplets in Chlamydomonas reinhardtii. Plant Physiol. 2016, 171, 2406–2417. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huff, J.; Crunkleton, D.W.; Johannes, T.W. Light Intensity and Spectral Quality Modulation for Improved Growth Kinetics and Biochemical Composition of Chlamydomonas reinhardtii. J. Biotechnol. 2023, 375, 28–39. [Google Scholar] [CrossRef]
- Roessler, P.G. Environmental Control of Glycerolipid Metabolism in Microalgae: Commercial Implications and Future Research Directions. J. Phycol. 1990, 26, 393–399. [Google Scholar] [CrossRef]
- Chen, G.; Harwood, J.L.; Lemieux, M.J.; Stone, S.J.; Weselake, R.J. Acyl-CoA:Diacylglycerol Acyltransferase: Properties, Physiological Roles, Metabolic Engineering and Intentional Control. Prog. Lipid Res. 2022, 88, 101181. [Google Scholar] [CrossRef]
- Martin, B.A.; Wilson, R.F. Subcellular Localization of Triacylglycerol Synthesis in Spinach Leaves. Lipids 1984, 19, 117–121. [Google Scholar] [CrossRef]
- Goodson, C.; Roth, R.; Wang, Z.T.; Goodenough, U. Structural Correlates of Cytoplasmic and Chloroplast Lipid Body Synthesis in Chlamydomonas reinhardtii and Stimulation of Lipid Body Production with Acetate Boost. Eukaryot. Cell 2011, 10, 1592–1606. [Google Scholar] [CrossRef]
- Davidi, L.; Shimoni, E.; Khozin-Goldberg, I.; Zamir, A.; Pick, U. Origin of β-Carotene-Rich Plastoglobuli in Dunaliella bardawil. Plant Physiol. 2014, 164, 2139–2156. [Google Scholar] [CrossRef]
- Kaup, M.T.; Froese, C.D.; Thompson, J.E. A Role for Diacylglycerol Acyltransferase during Leaf Senescence. Plant Physiol. 2002, 129, 1616–1626. [Google Scholar] [CrossRef]
- Bagnato, C.; Prados, M.B.; Franchini, G.R.; Scaglia, N.; Miranda, S.E.; Beligni, M.V. Analysis of Triglyceride Synthesis Unveils a Green Algal Soluble Diacylglycerol Acyltransferase and Provides Clues to Potential Enzymatic Components of the Chloroplast Pathway. BMC Genom. 2017, 18, 223. [Google Scholar] [CrossRef]
- Carro, M.; Gonorazky, G.; Soto, D.; Mamone, L.; Bagnato, C.; Pagnussat, L.; Beligni, M. Expression of Chlamydomonas reinhardtii Chloroplast Diacylglycerol Acyltransferase-3 Is Induced by Light in Concert with Triacylglycerol Accumulation. Plant J. 2022, 110, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Boyle, N.R.; Page, M.D.; Liu, B.; Blaby, I.K.; Casero, D.; Kropat, J.; Cokus, S.J.; Hong-Hermesdorf, A.; Shaw, J.; Karpowicz, S.J.; et al. Three Acyltransferases and Nitrogen-Responsive Regulator Are Implicated in Nitrogen Starvation-Induced Triacylglycerol Accumulation in Chlamydomonas. J. Biol. Chem. 2012, 287, 15811–15825. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.; Han, D.; Li, Y.; Sommerfeld, M.; Hu, Q. Phospholipid:Diacylglycerol Acyltransferase Is a Multifunctional Enzyme Involved in Membrane Lipid Turnover and Degradation While Synthesizing Triacylglycerol in the Unicellular Green Microalga Chlamydomonas reinhardtii. Plant Cell 2012, 24, 3708. [Google Scholar] [CrossRef] [PubMed]
- Chouhan, N.; Devadasu, E.; Yadav, R.M.; Subramanyam, R. Autophagy Induced Accumulation of Lipids in Pgrl1 and Pgr5 of Chlamydomonas reinhardtii Under High Light. Front. Plant Sci. 2022, 12, 3308. [Google Scholar] [CrossRef]
- Christ, B.; Hörtensteiner, S. Mechanism and Significance of Chlorophyll Breakdown. J. Plant Growth Regul. 2014, 33, 4–20. [Google Scholar] [CrossRef]
- Cook, R.; Lupette, J.; Benning, C. The Role of Chloroplast Membrane Lipid Metabolism in Plant Environmental Responses. Cells 2021, 10, 706. [Google Scholar] [CrossRef]
- Gutbrod, K.; Romer, J.; Dörmann, P. Phytol Metabolism in Plants. Prog. Lipid Res. 2019, 74, 1–17. [Google Scholar] [CrossRef]
- Lippold, F.; vom Dorp, K.; Abraham, M.; Hölzl, G.; Wewer, V.; Yilmaz, J.L.; Lager, I.; Montandon, C.; Besagni, C.; Kessler, F.; et al. Fatty Acid Phytyl Ester Synthesis in Chloroplasts of Arabidopsis. Plant Cell 2012, 24, 2001–2014. [Google Scholar] [CrossRef]
- Vidi, P.A.; Kanwischer, M.; Baginsky, S.; Austin, J.R.; Csucs, G.; Dörmann, P.; Kessler, F.; Bréhélin, C. Tocopherol Cyclase (VTE1) Localization and Vitamin E Accumulation in Chloroplast Plastoglobule Lipoprotein Particles. J. Biol. Chem. 2006, 281, 11225–11233. [Google Scholar] [CrossRef] [PubMed]
- Ytterberg, A.J.; Peltier, J.B.; Van Wijk, K.J. Protein Profiling of Plastoglobules in Chloroplasts and Chromoplasts. A Surprising Site for Differential Accumulation of Metabolic Enzymes. Plant Physiol. 2006, 140, 984–997. [Google Scholar] [CrossRef] [PubMed]
- Aslan, S.; Sun, C.; Leonova, S.; Dutta, P.; Dörmann, P.; Domergue, F.; Stymne, S.; Hofvander, P. Wax Esters of Different Compositions Produced via Engineering of Leaf Chloroplast Metabolism in Nicotiana benthamiana. Metab. Eng. 2014, 25, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Cranwell, A.; Jaworski, G.H.M.; Bickley, H.M. Hydrocarbons, Sterols, Esters and Fatty Acids in Six Freshwater Chlorophytes. Phytochemistry 1990, 29, 145–151. [Google Scholar] [CrossRef]
- Rager, M.N.; Metzger, P. Six Novel Tetraterpenoid Ethers, Lycopanerols B-G, and Some Other Constituents from the Green Microalga Botryococcus braunii. Phytochemistry 2000, 54, 427–437. [Google Scholar] [CrossRef]
- Wittkopp, T.M.; Schmollinger, S.; Saroussi, S.; Hu, W.; Zhang, W.; Fan, Q.; Gallaher, S.D.; Leonard, M.T.; Soubeyrand, E.; Basset, G.J.; et al. Bilin-Dependent Photoacclimation in Chlamydomonas reinhardtii. Plant Cell 2017, 29, 2711–2726. [Google Scholar] [CrossRef]
- Li, X.; Patena, W.; Fauser, F.; Jinkerson, R.E.; Saroussi, S.; Meyer, M.T.; Ivanova, N.; Robertson, J.M.; Yue, R.; Zhang, R.; et al. A Genome-Wide Algal Mutant Library and Functional Screen Identifies Genes Required for Eukaryotic Photosynthesis. Nat. Genet. 2019, 51, 627–635. [Google Scholar] [CrossRef]
- de Carpentier, F.; Lemaire, S.D.; Danon, A. When Unity Is Strength: The Strategies Used by Chlamydomonas to Survive Environmental Stresses. Cells 2019, 8, 1307. [Google Scholar] [CrossRef]
- Ischebeck, T.; Zbierzak, A.M.; Kanwischer, M.; Dörmann, P. A Salvage Pathway for Phytol Metabolism in Arabidopsis. J. Biol. Chem. 2006, 281, 2470–2477. [Google Scholar] [CrossRef]
- Shajil Das, A.; Shajil Das, A.; Chen, Z.; Peisker, H.; Gutbrod, K.; Hölzl, G.; Dörmann, P. Multifunctional Acyltransferases Involved in the Synthesis of Triacylglycerol, Fatty Acid Phytyl Esters and Plastoquinol Esters in Cyanobacteria. Planta 2025, 261, 123. [Google Scholar] [CrossRef]
- Maltsev, Y.; Maltseva, K.; Kulikovskiy, M.; Maltseva, S. Influence of Light Conditions on Microalgae Growth and Content of Lipids, Carotenoids, and Fatty Acid Composition. Biology 2021, 10, 1060. [Google Scholar] [CrossRef]
- Rottet, S.; Besagni, C.; Kessler, F. The Role of Plastoglobules in Thylakoid Lipid Remodeling during Plant Development. Biochim. Biophys. Acta (BBA)—Bioenerg. 2015, 1847, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Aizouq, M.; Peisker, H.; Gutbrod, K.; Melzer, M.; Hölzl, G.; Dörmann, P.; Niyogi, K.K. Triacylglycerol and Phytyl Ester Synthesis in Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. USA 2020, 117, 6216–6222. [Google Scholar] [CrossRef] [PubMed]
- Ollis, D.L.; Cheah, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Sybille, M.; Harel, M.; James Remington, S.; Silman, I.; Schrag, J.; et al. The α/β Hydrolase Fold. Protein Eng. Des. Sel. 1992, 5, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, A.P.; Rafferty, J.B.; Sedelnikova, S.E.; Slabas, A.R.; Schierer, T.P.; Kroon, J.T.M.; Simon, J.W.; Fawcett, T.; Nishida, I.; Murata, N.; et al. Analysis of the Structure, Substrate Specificity, and Mechanism of Squash Glycerol-3-Phosphate (1)-Acyltransferase. Structure 2001, 9, 347–353. [Google Scholar] [CrossRef]
- Grabsztunowicz, M.; Koskela, M.M.; Mulo, P. Post-Translational Modifications in Regulation of Chloroplast Function: Recent Advances. Front. Plant Sci. 2017, 8, 246787. [Google Scholar] [CrossRef]
- Wang, L.; Patena, W.; Van Baalen, K.A.; Xie, Y.; Singer, E.R.; Gavrilenko, S.; Warren-Williams, M.; Han, L.; Harrigan, H.R.; Hartz, L.D.; et al. A Chloroplast Protein Atlas Reveals Punctate Structures and Spatial Organization of Biosynthetic Pathways. Cell 2023, 186, 3499–3518.e14. [Google Scholar] [CrossRef]
- Beligni, M.V.; Yamaguchi, K.; Mayfield, S.P. The Translational Apparatus of Chlamydomonas reinhardtii Chloroplast. Photosynth. Res. 2004, 82, 315–325. [Google Scholar] [CrossRef]
- Strenkert, D.; Schmollinger, S.; Gallaher, S.D.; Salomé, P.A.; Purvine, S.O.; Nicora, C.D.; Mettler-Altmann, T.; Soubeyrand, E.; Weber, A.P.M.; Lipton, M.S.; et al. Multiomics Resolution of Molecular Events during a Day in the Life of Chlamydomonas. Proc. Natl. Acad. Sci. USA 2019, 116, 2374–2383. [Google Scholar] [CrossRef]
- Idoine, A.D.; Boulouis, A.; Rupprecht, J.; Bock, R. The Diurnal Logic of the Expression of the Chloroplast Genome in Chlamydomonas reinhardtii. PLoS ONE 2014, 9, e108760. [Google Scholar] [CrossRef] [PubMed]
- Coragliotti, A.T.; Beligni, M.V.; Franklin, S.E.; Mayfield, S.P. Molecular Factors Affecting the Accumulation of Recombinant Proteins in the Chlamydomonas reinhardtii Chloroplast. Mol. Biotechnol. 2011, 48, 60–75. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.C.; Sullivan, J.A.; Wang, J.-H.; Jerome, C.A.; MacLean, D. Coordination of Plastid and Nuclear Gene Expression. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2003, 358, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Salvador, M.L.; Klein, U.; Bogorad, L. Light-Regulated and Endogenous Fluctuations of Chloroplast Transcript Levels in Chlamydomonas. Regulation by Transcription and RNA Degradation. Plant J. 1993, 3, 213–219. [Google Scholar] [CrossRef]
- Spicher, L.; Almeida, J.; Gutbrod, K.; Pipitone, R.; Dörmann, P.; Glauser, G.; Rossi, M.; Kessler, F. Essential Role for Phytol Kinase and Tocopherol in Tolerance to Combined Light and Temperature Stress in Tomato. J. Exp. Bot. 2017, 68, 5845–5856. [Google Scholar] [CrossRef]
- Kamranfar, I.; Xue, G.P.; Tohge, T.; Sedaghatmehr, M.; Fernie, A.R.; Balazadeh, S.; Mueller-Roeber, B. Transcription Factor RD26 Is a Key Regulator of Metabolic Reprogramming during Dark-Induced Senescence. New Phytol. 2018, 218, 1543–1557. [Google Scholar] [CrossRef]
- Gonzalez-Ballester, D.; Pootakham, W.; Mus, F.; Yang, W.; Catalanotti, C.; Magneschi, L.; De Montaigu, A.; Higuera, J.J.; Prior, M.; Galván, A.; et al. Reverse Genetics in Chlamydomonas: A Platform for Isolating Insertional Mutants. Plant Methods 2011, 7, 24. [Google Scholar] [CrossRef]
- Cheng, X.; Liu, G.; Ke, W.; Zhao, L.; Lv, B.; Ma, X.; Xu, N.; Xia, X.; Deng, X.; Zheng, C.; et al. Building a Multipurpose Insertional Mutant Library for Forward and Reverse Genetics in Chlamydomonas. Plant Methods 2017, 13, 36. [Google Scholar] [CrossRef]
- El-Brolosy, M.A.; Stainier, D.Y.R. Genetic Compensation: A Phenomenon in Search of Mechanisms. PLoS Genet. 2017, 13, e1006780. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Park, R.; Miller, S.M.; Li, Y. Genetic Compensation of Triacylglycerol Biosynthesis in the Green Microalga Chlamydomonas reinhardtii. Plant J. 2022, 111, 1069–1080. [Google Scholar] [CrossRef]
- Peisker, C.; Düggelin, T.; Rentsch, D.; Matile, P. Phytol and the Breakdown of Chlorophyll in Senescent Leaves. J. Plant Physiol. 1989, 135, 428–432. [Google Scholar] [CrossRef]
- Gaude, N.; Bréhélin, C.; Tischendorf, G.; Kessler, F.; Dörmann, P. Nitrogen Deficiency in Arabidopsis Affects Galactolipid Composition and Gene Expression and Results in Accumulation of Fatty Acid Phytyl Esters. Plant J. 2007, 49, 729–739. [Google Scholar] [CrossRef]
- Baroli, I.; Gutman, B.L.; Ledford, H.K.; Shin, J.W.; Chin, B.L.; Havaux, M.; Niyogi, K.K. Photo-Oxidative Stress in a Xanthophyll-Deficient Mutant of Chlamydomonas. J. Biol. Chem. 2004, 279, 6337–6344. [Google Scholar] [CrossRef]
- Li, Z.; Keasling, J.D.; Niyogi, K.K. Overlapping Photoprotective Function of Vitamin E and Carotenoids in Chlamydomonas. Plant Physiol. 2012, 158, 313–323. [Google Scholar] [CrossRef]
- Gorman, D.S.; Levine, R.P. Cytochrome f and Plastocyanin: Their Sequence in the Photosynthetic Electron Transport Chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 1965, 54, 1665–1669. [Google Scholar] [CrossRef] [PubMed]
- Fogg, G.E.; Thake, B. Algal Cultures and Phytoplankton Ecology; University of Wisconsin Press: Madison, WI, USA, 1987; ISBN 0-299-10560-1. [Google Scholar]
- Eddy, S.R. Profile Hidden Markov Models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A New Software for Selection of Phylogenetic Informative Regions from Multiple Sequence Alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 1989; ISBN 978-0-87969-309-1. [Google Scholar]
- Ruijter, J.M.; Lorenz, P.; Tuomi, J.M.; Hecker, M.; van den Hoff, M.J.B. Fluorescent-Increase Kinetics of Different Fluorescent Reporters Used for qPCR Depend on Monitoring Chemistry, Targeted Sequence, Type of DNA Input and PCR Efficiency. Microchim. Acta 2014, 181, 1689–1696. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Krauß, S.; Hammann, S.; Vetter, W. Phytyl Fatty Acid Esters in the Pulp of Bell Pepper (Capsicum annuum). J. Agric. Food Chem. 2016, 64, 6306–6311. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Fukushima, H.; Tamaki, E. Metabolism of Chlorophyll in Higher Plants—I.: A Thin-Layer Chromatography for the Quantitative Determination of Phytol. Phytochemistry 1963, 3, 641–645. [Google Scholar] [CrossRef]
- Narai-Kanayama, A.; Yokosaka, S.; Seo, Y.; Mikami, K.; Yoshino, T.; Matsuda, H. Evidence of Increases of Phytol and Chlorophyllide by Enzymatic Dephytylation of Chlorophylls in Smoothie Made from Spinach Leaves. J. Food Sci. 2023, 88, 2385–2396. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and Carotenoids: Pigments of Photosynthetic Biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar] [CrossRef]
- Gao, C.; Wang, Y.; Shen, Y.; Yan, D.; He, X.; Dai, J.; Wu, Q. Oil Accumulation Mechanisms of the Oleaginous Microalga Chlorella Protothecoides Revealed through Its Genome, Transcriptomes, and Proteomes. BMC Genom. 2014, 15, 582. [Google Scholar] [CrossRef]
- Moreau, H.; Verhelst, B.; Couloux, A.; Derelle, E.; Rombauts, S.; Grimsley, N.; Van Bel, M.; Poulain, J.; Katinka, M.; Hohmann-Marriott, M.F.; et al. Gene Functionalities and Genome Structure in Bathycoccus Prasinos Reflect Cellular Specializations at the Base of the Green Lineage. Genome Biol. 2012, 13, R74. [Google Scholar] [CrossRef]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Maréchal-Drouard, L.; et al. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science 2007, 318, 245–251. [Google Scholar] [CrossRef]
- Hovde, B.T.; Deodato, C.R.; Hunsperger, H.M.; Ryken, S.A.; Yost, W.; Jha, R.K.; Patterson, J.; Monnat, R.J., Jr.; Barlow, S.B.; Starkenburg, S.R.; et al. Genome Sequence and Transcriptome Analyses of Chrysochromulina Tobin: Metabolic Tools for Enhanced Algal Fitness in the Prominent Order Prymnesiales (Haptophyceae). PLoS Genet. 2015, 11, e1005469. [Google Scholar] [CrossRef]
- Blanc, G.; Agarkova, I.; Grimwood, J.; Kuo, A.; Brueggeman, A.; Dunigan, D.D.; Gurnon, J.; Ladunga, I.; Lindquist, E.; Lucas, S.; et al. The Genome of the Polar Eukaryotic Microalga Coccomyxa subellipsoidea Reveals Traits of Cold Adaptation. Genome Biol. 2012, 13, R39. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Duncan, G.; Agarkova, I.; Borodovsky, M.; Gurnon, J.; Kuo, A.; Lindquist, E.; Lucas, S.; Pangilinan, J.; Polle, J.; et al. The Chlorella variabilis NC64A Genome Reveals Adaptation to Photosymbiosis, Coevolution with Viruses, and Cryptic Sex. Plant Cell 2010, 22, 2943. [Google Scholar] [CrossRef] [PubMed]
- Polle Juergen, E.W.; Barry, K.; Cushman, J.; Schmutz, J.; Tran, D.; Hathwaik Leyla, T.; Yim Won, C.; Jenkins, J.; McKie-Krisberg, Z.; Prochnik, S.; et al. Draft Nuclear Genome Sequence of the Halophilic and Beta-Carotene-Accumulating Green Alga Dunaliella salina Strain CCAP19/18. Genome Announc. 2017, 5, 10–1128. [Google Scholar] [CrossRef]
- Hanschen, E.R.; Marriage, T.N.; Ferris, P.J.; Hamaji, T.; Toyoda, A.; Fujiyama, A.; Neme, R.; Noguchi, H.; Minakuchi, Y.; Suzuki, M.; et al. The Gonium Pectorale Genome Demonstrates Co-Option of Cell Cycle Regulation during the Evolution of Multicellularity. Nat. Commun. 2016, 7, 11370. [Google Scholar] [CrossRef]
- Worden, A.Z.; Lee, J.-H.; Mock, T.; Rouzé, P.; Simmons, M.P.; Aerts, A.L.; Allen, A.E.; Cuvelier, M.L.; Derelle, E.; Everett, M.V.; et al. Green Evolution and Dynamic Adaptations Revealed by Genomes of the Marine Picoeukaryotes Micromonas. Science 2009, 324, 268–272. [Google Scholar] [CrossRef]
- Bogen, C.; Al-Dilaimi, A.; Albersmeier, A.; Wichmann, J.; Grundmann, M.; Rupp, O.; Lauersen, K.J.; Blifernez-Klassen, O.; Kalinowski, J.; Goesmann, A.; et al. Reconstruction of the Lipid Metabolism for the Microalga Monoraphidium neglectum from Its Genome Sequence Reveals Characteristics Suitable for Biofuel Production. BMC Genom. 2013, 14, 926. [Google Scholar] [CrossRef]
- Palenik, B.; Grimwood, J.; Aerts, A.; Rouzé, P.; Salamov, A.; Putnam, N.; Dupont, C.; Jorgensen, R.; Derelle, E.; Rombauts, S.; et al. The Tiny Eukaryote Ostreococcus Provides Genomic Insights into the Paradox of Plankton Speciation. Proc. Natl. Acad. Sci. USA 2007, 104, 7705–7710. [Google Scholar] [CrossRef]
- Gonzalez-Esquer, C.R.; Twary Scott, N.; Hovde Blake, T.; Starkenburg Shawn, R. Nuclear, Chloroplast, and Mitochondrial Genome Sequences of the Prospective Microalgal Biofuel Strain Picochlorum soloecismus. Genome Announc. 2018, 6, 10–1128. [Google Scholar] [CrossRef]
- Suzuki, S.; Yamaguchi, H.; Nakajima, N.; Kawachi, M. Raphidocelis Subcapitata (=Pseudokirchneriella subcapitata) Provides an Insight into Genome Evolution and Environmental Adaptations in the Sphaeropleales. Sci. Rep. 2018, 8, 8058. [Google Scholar] [CrossRef]
- Prochnik, S.E.; Umen, J.; Nedelcu, A.M.; Hallmann, A.; Miller, S.M.; Nishii, I.; Ferris, P.; Kuo, A.; Mitros, T.; Fritz-Laylin, L.K.; et al. Genomic Analysis of Organismal Complexity in the Multicellular Green Alga Volvox carteri. Science 2010, 329, 223–226. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome Initiative. Analysis of the Genome Sequence of the Flowering Plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Dorrell, R.G.; Kuo, A.; Füssy, Z.; Richardson, E.H.; Salamov, A.; Zarevski, N.; Freyria, N.J.; Ibarbalz, F.M.; Jenkins, J.; Pierella Karlusich, J.J.; et al. Convergent Evolution and Horizontal Gene Transfer in Arctic Ocean Microalgae. Life Sci. Alliance 2023, 6, e202201833. [Google Scholar] [CrossRef] [PubMed]
- Read, B.A.; Kegel, J.; Klute, M.J.; Kuo, A.; Lefebvre, S.C.; Maumus, F.; Mayer, C.; Miller, J.; Monier, A.; Salamov, A.; et al. Pan Genome of the Phytoplankton Emiliania Underpins Its Global Distribution. Nature 2013, 499, 209–213. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome Sequence of the Palaeopolyploid Soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- International Rice Genome Sequencing Project. The Map-Based Sequence of the Rice Genome. Nature 2005, 436, 793. [Google Scholar] [CrossRef]
- Lang, D.; Ullrich, K.K.; Murat, F.; Fuchs, J.; Jenkins, J.; Haas, F.B.; Piednoel, M.; Gundlach, H.; Van Bel, M.; Meyberg, R.; et al. The Physcomitrella Patens Chromosome-Scale Assembly Reveals Moss Genome Structure and Evolution. Plant J. 2018, 93, 515–533. [Google Scholar] [CrossRef]
- Hosmani, P.S.; Flores-Gonzalez, M.; van de Geest, H.; Maumus, F.; Bakker, L.V.; Schijlen, E.; van Haarst, J.; Cordewener, J.; Sanchez-Perez, G.; Peters, S.; et al. An Improved de Novo Assembly and Annotation of the Tomato Reference Genome Using Single-Molecule Sequencing, Hi-C Proximity Ligation and Optical Maps. bioRxiv 2019. [Google Scholar] [CrossRef]
- Banks, J.A.; Nishiyama, T.; Hasebe, M.; Bowman, J.L.; Gribskov, M.; de Pamphilis, C.; Albert, V.A.; Aono, N.; Aoyama, T.; Ambrose, B.A.; et al. The Selaginella Genome Identifies Genetic Changes Associated with the Evolution of Vascular Plants. Science 2011, 332, 960–963. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The Grapevine Genome Sequence Suggests Ancestral Hexaploidization in Major Angiosperm Phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef]
- Gore, M.A.; Chia, J.-M.; Elshire, R.J.; Sun, Q.; Ersoz, E.S.; Hurwitz, B.L.; Peiffer, J.A.; McMullen, M.D.; Grills, G.S.; Ross-Ibarra, J.; et al. A First-Generation Haplotype Map of Maize. Science 2009, 326, 1115–1117. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zegarra Borlando, F.E.; Oresti, G.M.; Pavia, N.; Beligni, M.V.; Gonorazky, G. Knock Down of Chlamydomonas reinhardtii Phytyl Ester Synthase α Triggers DGAT3 Overexpression and Triacylglycerol Accumulation Under Low-Light Conditions. Plants 2025, 14, 3044. https://doi.org/10.3390/plants14193044
Zegarra Borlando FE, Oresti GM, Pavia N, Beligni MV, Gonorazky G. Knock Down of Chlamydomonas reinhardtii Phytyl Ester Synthase α Triggers DGAT3 Overexpression and Triacylglycerol Accumulation Under Low-Light Conditions. Plants. 2025; 14(19):3044. https://doi.org/10.3390/plants14193044
Chicago/Turabian StyleZegarra Borlando, Félix Eduardo, Gerardo Martín Oresti, Natalia Pavia, María Verónica Beligni, and Gabriela Gonorazky. 2025. "Knock Down of Chlamydomonas reinhardtii Phytyl Ester Synthase α Triggers DGAT3 Overexpression and Triacylglycerol Accumulation Under Low-Light Conditions" Plants 14, no. 19: 3044. https://doi.org/10.3390/plants14193044
APA StyleZegarra Borlando, F. E., Oresti, G. M., Pavia, N., Beligni, M. V., & Gonorazky, G. (2025). Knock Down of Chlamydomonas reinhardtii Phytyl Ester Synthase α Triggers DGAT3 Overexpression and Triacylglycerol Accumulation Under Low-Light Conditions. Plants, 14(19), 3044. https://doi.org/10.3390/plants14193044





