Satellite DNA in Populus and Molecular Karyotyping of Populus xiaohei and Its Derived Double Haploids
Abstract
1. Introduction
2. Results
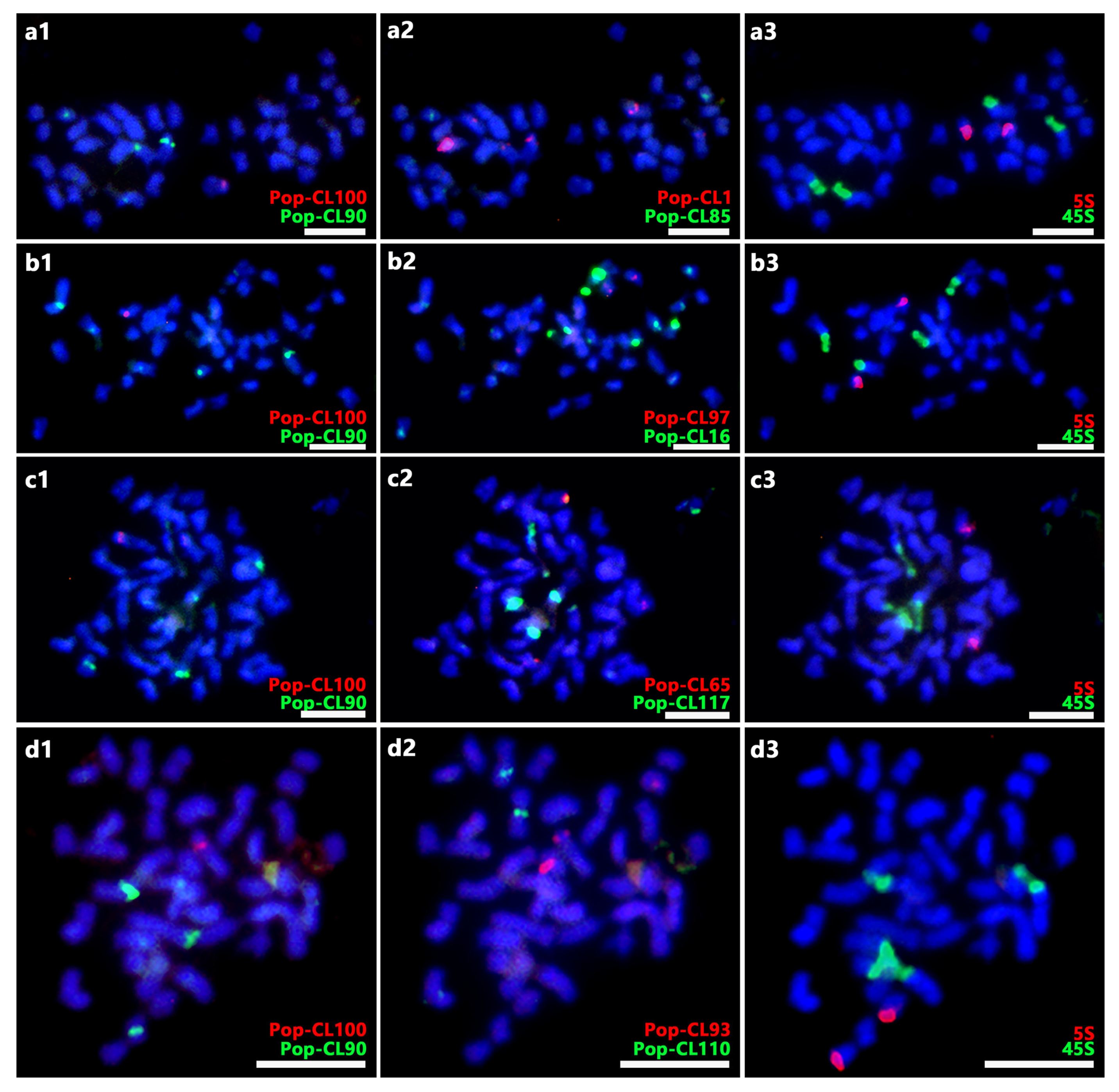
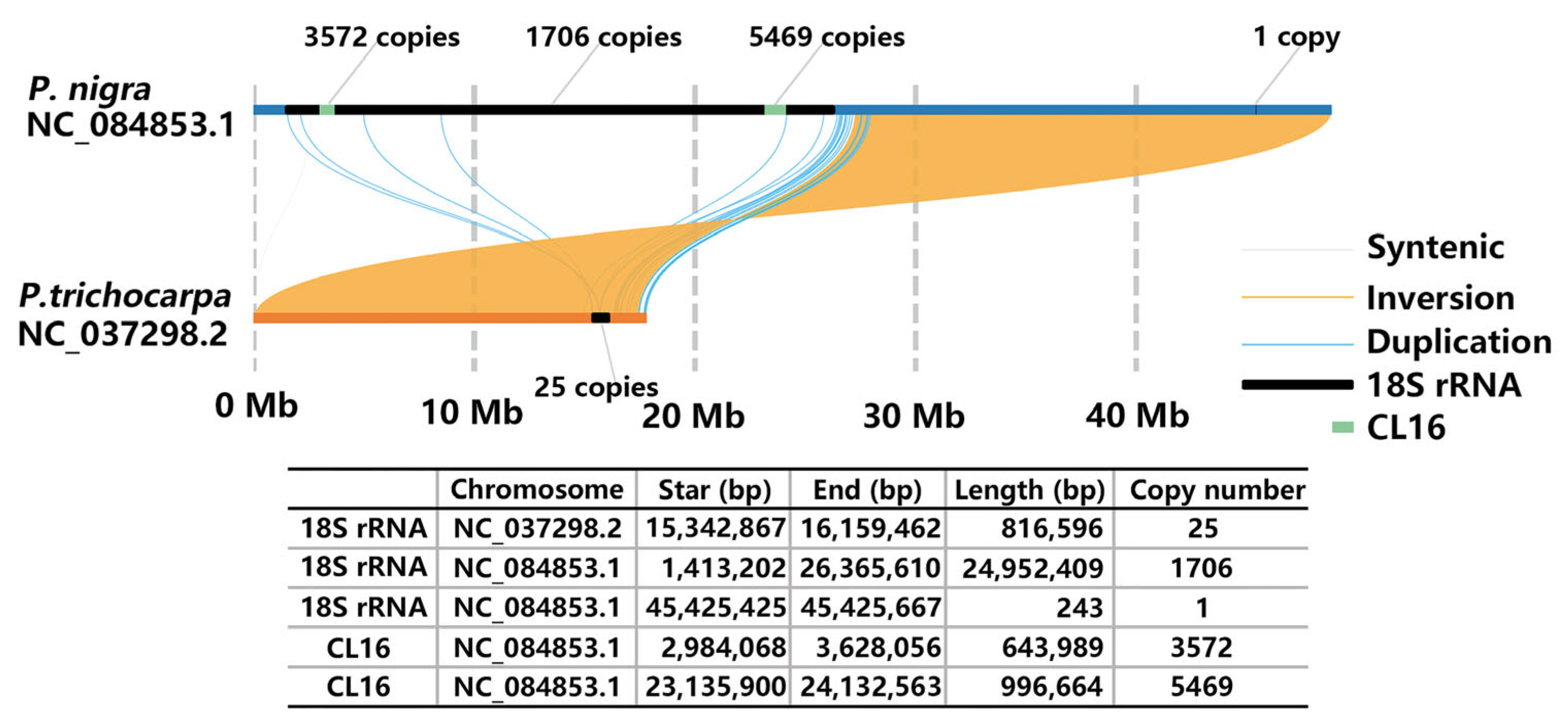
2.1. Identification and Distribution Analysis of SatDNA
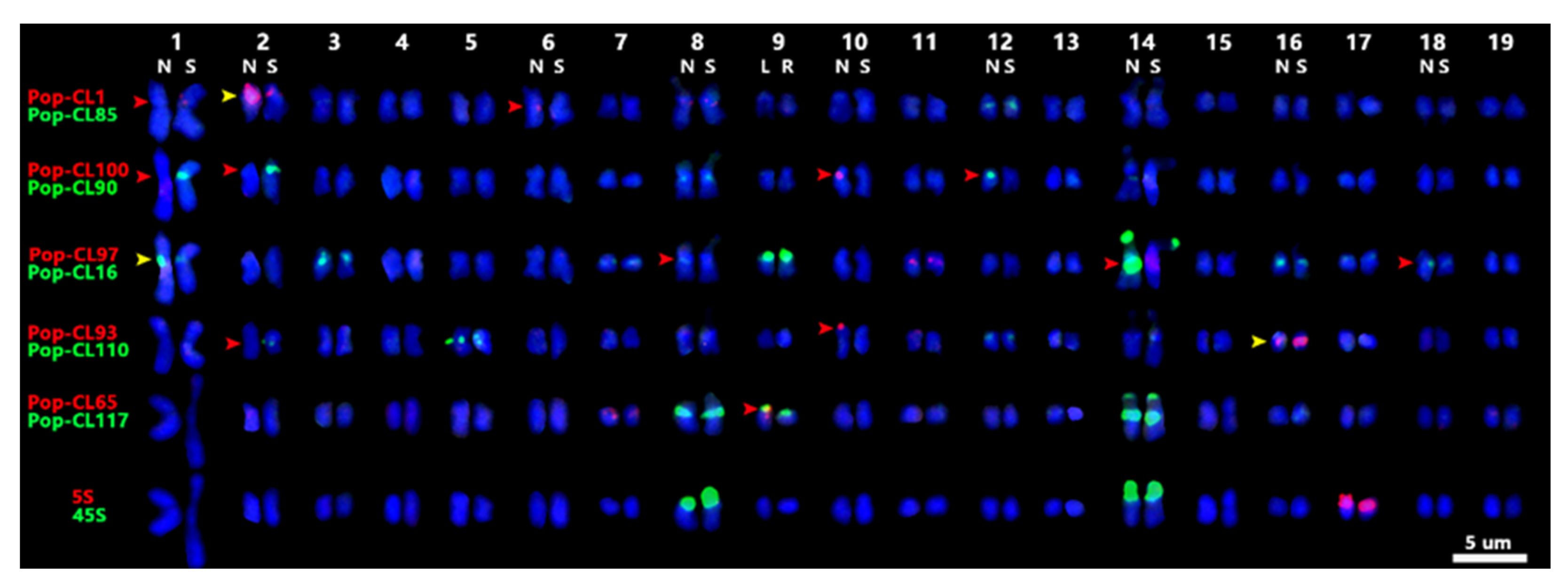
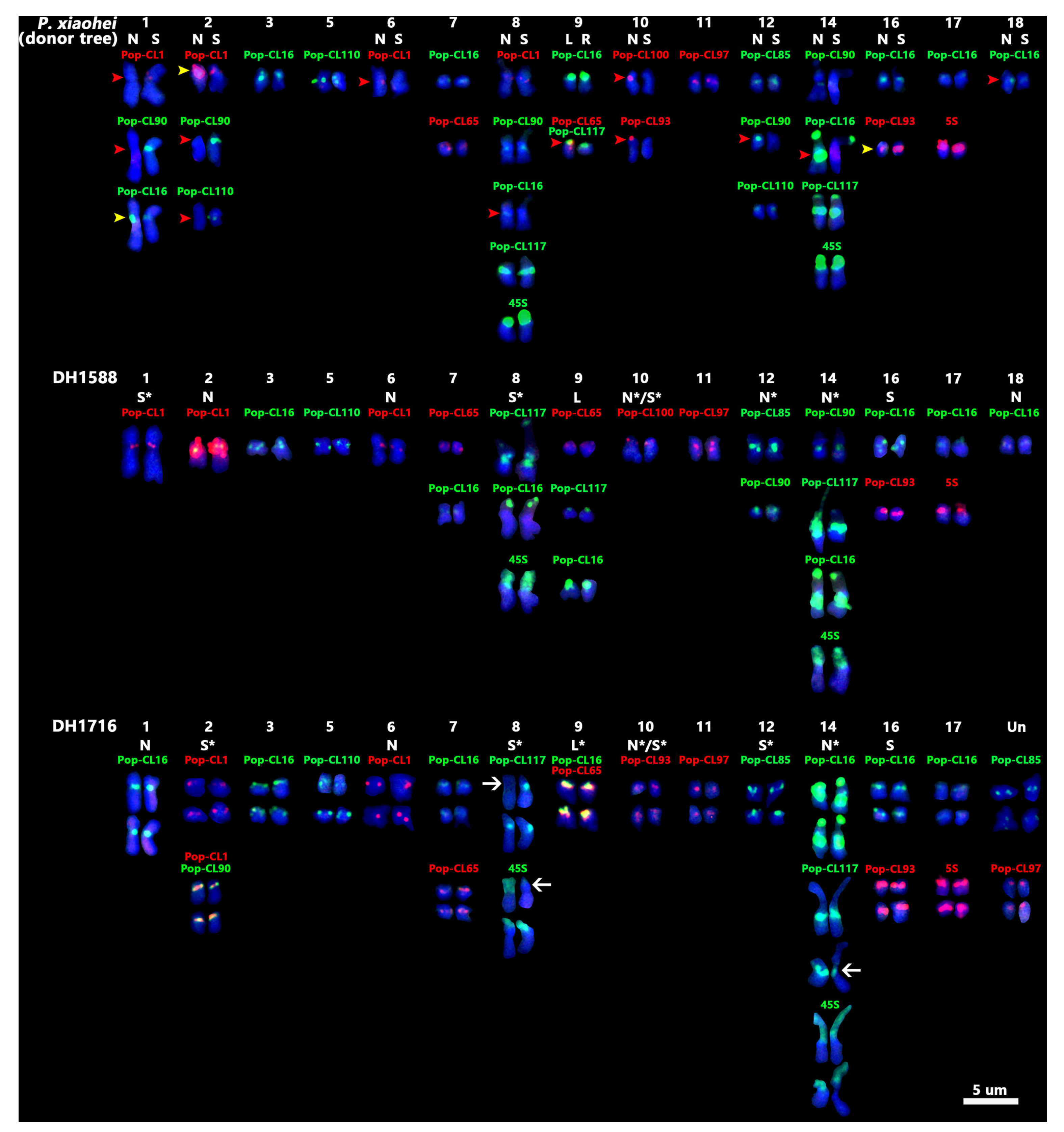
2.2. Karyotyping of P. xiaohei and Reference Genomes Limitations
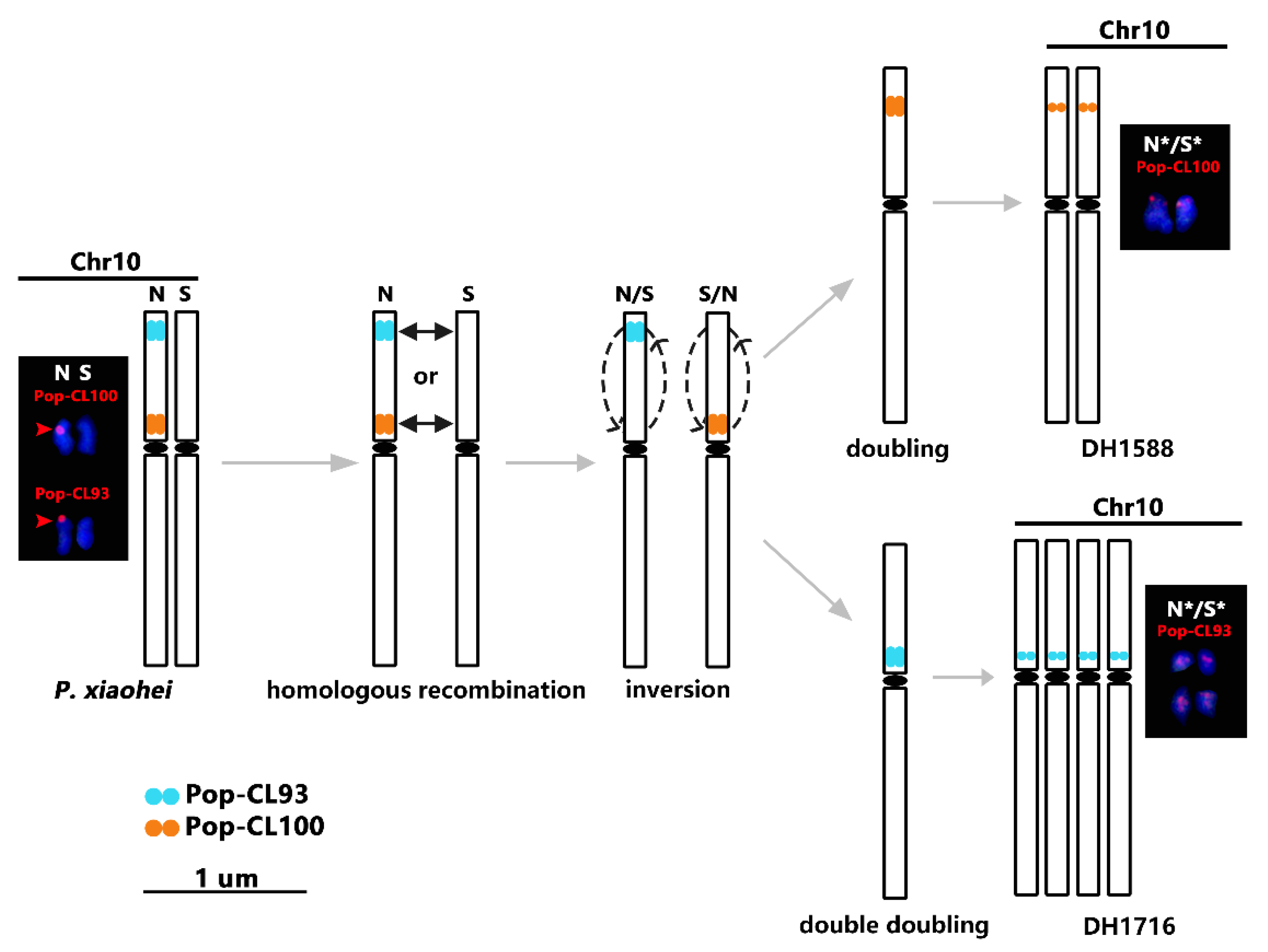
2.3. Chromosome Variation Analysis of DH Materials of P. xiaohei
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. satDNA Identification and Genome Distribution Analysis
4.3. Probe Design and Probe Labeling
4.4. Chromosome Preparation
4.5. Fluorescent In Situ Hybridization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, C.; Tung, S. New Taxa of Populus (II). Bull. Bot. Res. 1982, 2, 105–120. [Google Scholar]
- Du, Z.W.; Zheng, T.C.; Li, S.; Zang, L.N.; Qu, G.Z.; You, X.L. Rapid Propagation and Regeneration System of Populus simonii × Populus nigra. Bull. Bot. Res. 2015, 35, 904–907. [Google Scholar]
- Qu, Y.; Fernie, A.R.; Liu, J.; Yan, J. Doubled haploid technology and synthetic apomixis: Recent advances and applications in future crop breeding. Mol. Plant 2024, 17, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.L.; Britt, A.B.; Tripathi, L.; Sharma, S.; Upadhyaya, H.D.; Ortiz, R. Haploids: Constraints and opportunities in plant breeding. Biotechnol. Adv. 2015, 33, 812–829. [Google Scholar] [CrossRef]
- Liu, C.; Wang, S.; Liu, Y.; Wang, M.; Fan, E.; Liu, C.; Zhang, S.; Yang, C.; Wang, J.; Sederoff, H.W.; et al. Exceptionally high genetic variance of the doubled haploid (DH) population of poplar. J. For. Res. 2023, 34, 1941–1950. [Google Scholar] [CrossRef]
- Younis, A.; Ramzan, F.; Hwang, Y.J.; Lim, K.B. FISH and GISH: Molecular cytogenetic tools and their applications in ornamental plants. Plant Cell Rep. 2015, 34, 1477–1488. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Lower, S.S.; McGurk, M.P.; Clark, A.G.; Barbash, D.A. Satellite DNA evolution: Old ideas, new approaches. Curr. Opin. Genet. Dev. 2018, 49, 70–78. [Google Scholar] [CrossRef]
- Klemme, S.; Banaei-Moghaddam, A.M.; Macas, J.; Wicker, T.; Novák, P.; Houben, A. High-copy sequences reveal distinct evolution of the rye B chromosome. New Phytol. 2013, 199, 550–558. [Google Scholar] [CrossRef]
- Schmidt, T.; Heitkam, T.; Liedtke, S.; Schubert, V.; Menzel, G. Adding color to a century-old enigma: Multi-color chromosome identification unravels the autotriploid nature of saffron (Crocus sativus) as a hybrid of wild Crocus cartwrightianus cytotypes. New Phytol. 2019, 222, 1965–1980. [Google Scholar] [CrossRef]
- Wei, L.; Liu, B.; Zhang, C.; Yu, Y.; Yang, X.; Dou, Q.; Dong, Q. Identification and characterization of satellite DNAs in Poa L. Mol. Cytogenet. 2020, 13, 47. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; He, J.; Zhao, Y.; Zhang, S.; Rao, X.; Wang, H.; Wang, Z.; Song, A.; Jiang, J.; Chen, S.; et al. Divergence of 10 satellite repeats in Artemisia (Asteraceae: Anthemideae) based on sequential fluorescence in situ hybridization analysis: Evidence for species identification and evolution. Chromosome Res. 2024, 32, 5. [Google Scholar] [CrossRef] [PubMed]
- Heitkam, T.; Weber, B.; Walter, I.; Liedtke, S.; Ost, C.; Schmidt, T. Satellite DNA landscapes after allotetraploidization of quinoa (Chenopodium quinoa) reveal unique A and B subgenomes. Plant J. 2020, 103, 32–52. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef]
- Xin, H.; Zhang, T.; Wu, Y.; Zhang, W.; Zhang, P.; Xi, M.; Jiang, J. An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting. Plant J. 2020, 101, 253–264. [Google Scholar] [CrossRef]
- Hu, B.; Dong, F.; Wang, C.; Qi, L.; Song, W.; Chen, C. Multicolor Fluorescence in Situ Hybridization of Seven Populus Species-ribosomal DNA and Telomere Repeat Sequence. Acta Sci. Nat. Univ. Nankaiensis 2012, 45, 58–64. [Google Scholar]
- Liu, B.; Wang, S.; Tao, X.; Liu, C.; Qu, G.; Dou, Q. Molecular Karyotyping on Populus simonii × P. nigra and the Derived Doubled Haploid. Int. J. Mol. Sci. 2021, 22, 11424. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinf. 2009, 10, 421. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, G.; Wang, Z.; Ning, Y.; Ni, R.; Xi, M. Oligo-FISH of Populus simonii Pachytene Chromosomes Improves Karyotyping and Genome Assembly. Int. J. Mol. Sci. 2023, 24, 9950. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A. Satellite DNA in Plants: More than Just Rubbish. Cytogenet. Genome Res. 2015, 146, 153–170. [Google Scholar] [CrossRef]
- Neumann, P.; Oliveira, L.; Čížková, J.; Jang, T.-S.; Klemme, S.; Novák, P.; Stelmach, K.; Koblížková, A.; Doležel, J.; Macas, J. Impact of parasitic lifestyle and different types of centromere organization on chromosome and genome evolution in the plant genus Cuscuta. New Phytol. 2021, 229, 2365–2377. [Google Scholar] [CrossRef]
- Usai, G.; Mascagni, F.; Natali, L.; Giordani, T.; Cavallini, A. Comparative genome-wide analysis of repetitive DNA in the genus Populus L. Tree Genet. Genomes 2017, 13, 96. [Google Scholar] [CrossRef]
- Zhang, B.; Zhu, W.; Diao, S.; Wu, X.; Lu, J.; Ding, C.; Su, X. The poplar pangenome provides insights into the evolutionary history of the genus. Commun. Biol. 2019, 2, 215. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.; Barrela, R.M.; Bergès, H.; Marques, C.; Loureiro, J.; Morais-Cecílio, L.; Paiva, J.A. Advancing Eucalyptus Genomics: Cytogenomics Reveals Conservation of Eucalyptus Genomes. Front. Plant Sci. 2016, 7, 510. [Google Scholar] [CrossRef]
- Acosta, M.C.; Premoli, A.C. Understanding the extensive hybridization in South American Nothofagus through karyotype analysis. Bot. J. Linn. Soc. 2018, 188, 74–86. [Google Scholar] [CrossRef]
- Liu, W.; Liu, C.; Chen, S.; Wang, M.; Wang, X.; Yu, Y.; Sederoff, R.R.; Wei, H.; You, X.; Qu, G.; et al. A nearly gapless, highly contiguous reference genome for a doubled haploid line of Populus ussuriensis, enabling advanced genomic studies. For. Res. 2024, 4, e019. [Google Scholar] [CrossRef]
- Gao, W.; Wang, S.; Jiang, T.; Hu, H.; Gao, R.; Zhou, M.; Wang, G. Chromosome-scale and haplotype-resolved genome assembly of Populus trichocarpa. Hortic. Res. 2025, 12, uhaf012. [Google Scholar] [CrossRef]
- Gao, S.; Yu, H.; Wu, S.; Wang, S.; Geng, J.; Luo, Y.; Hu, S. Advances of sequencing and assembling technologies for complex genomes. Hereditas 2018, 40, 944–963. [Google Scholar]
- Bairu, M.W.; Aremu, A.O.; Van Staden, J. Somaclonal variation in plants: Causes and detection methods. Plant Growth Regul. 2011, 63, 147–173. [Google Scholar] [CrossRef]
- Akagbuo, N.; Jaja, E. A review of somaclonal variation in plantain (Musa spp.): Mechanism and applications. J. Appl. Biosci. 2013, 67, 5252–5260. [Google Scholar] [CrossRef]
- Aswathi, T.P.; Kumari, I.P.; Rafeekher, M.; Reshmi, C.R.; Anuradha, T. Somaclonal Variants in Ornamental Foliage: A Review. J. Adv. Biol. Biotechnol. 2025, 28, 460–472. [Google Scholar] [CrossRef]
- Lee, M.; Phillips, R.L. The Chromosomal Basis of Somaclonal Variation. Annu. Rev. Plant Biol. 1988, 39, 413–437. [Google Scholar] [CrossRef]
- Gahan, P. Molecular Aspects of Gene Expression in Plants. Biochem. Soc. Trans. 1976, 5, 1793. [Google Scholar] [CrossRef]
- Tang, H.; Krishnakumar, V.; Zeng, X.; Xu, Z.; Taranto, A.; Lomas, J.S.; Zhang, Y.; Huang, Y.; Wang, Y.; Yim, W.C.; et al. JCVI: A versatile toolkit for comparative genomics analysis. iMeta 2024, 3, e211. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Waminal, N.E.; Pellerin, R.J.; Kim, N.S.; Jayakodi, M.; Park, J.Y.; Yang, T.J.; Kim, H.H. Rapid and Efficient FISH using Pre-Labeled Oligomer Probes. Sci. Rep. 2018, 8, 8224. [Google Scholar] [CrossRef] [PubMed]
| Satellite DNA | Probe Name | Fluorochrome Label | Sequence (5′ to 3′) |
|---|---|---|---|
| CL1 | Pop-CL1 | TAMRA | TTCGACAGCCCAAACAGCCC |
| CL16 | Pop-CL16 | FAM | GATCCGACCGTTGGATCGCGG |
| CL65 | Pop-CL65 | TAMRA | ACTCGGTGACGAGGAGGTCT |
| CL85 | Pop-CL85 | FAM | TCTGCAGCGACTCAGTTTTCG |
| CL90 | Pop-CL90 | FAM | GGTGACGAGTGGATTCGTCA |
| CL93 | Pop-CL93 | TAMRA | GGTGACCTGCCCATCTTAAA |
| CL97 | Pop-CL97 | TAMRA | CTACTGCTGGCTCAGAAACCG |
| CL100 | Pop-CL100 | TAMRA | GATCAGCACTGGCATAGTGT |
| CL109 | Pop-CL109(a) | FAM | TTCCCCAGCTGATGCCTGGTT |
| Pop-CL109(b) | FAM | GCACAATCCCTGTTGACTCG | |
| CL110 | Pop-CL110 | FAM | CAACGAAACTGCTTCCACGG |
| CL117 | Pop-CL117 | FAM | AAAACAGAAATCCTAATTACT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Wang, X.; Shen, W.; Wang, M.; Qu, G.; Dou, Q. Satellite DNA in Populus and Molecular Karyotyping of Populus xiaohei and Its Derived Double Haploids. Plants 2025, 14, 3046. https://doi.org/10.3390/plants14193046
Liu B, Wang X, Shen W, Wang M, Qu G, Dou Q. Satellite DNA in Populus and Molecular Karyotyping of Populus xiaohei and Its Derived Double Haploids. Plants. 2025; 14(19):3046. https://doi.org/10.3390/plants14193046
Chicago/Turabian StyleLiu, Bo, Xinyu Wang, Wenjie Shen, Meng Wang, Guanzheng Qu, and Quanwen Dou. 2025. "Satellite DNA in Populus and Molecular Karyotyping of Populus xiaohei and Its Derived Double Haploids" Plants 14, no. 19: 3046. https://doi.org/10.3390/plants14193046
APA StyleLiu, B., Wang, X., Shen, W., Wang, M., Qu, G., & Dou, Q. (2025). Satellite DNA in Populus and Molecular Karyotyping of Populus xiaohei and Its Derived Double Haploids. Plants, 14(19), 3046. https://doi.org/10.3390/plants14193046






