WSF: A Transformer-Based Framework for Microphenotyping and Genetic Analyzing of Wheat Stomatal Traits
Abstract
1. Introduction
2. Results
2.1. Model Performance Testing and Comparative Experiments
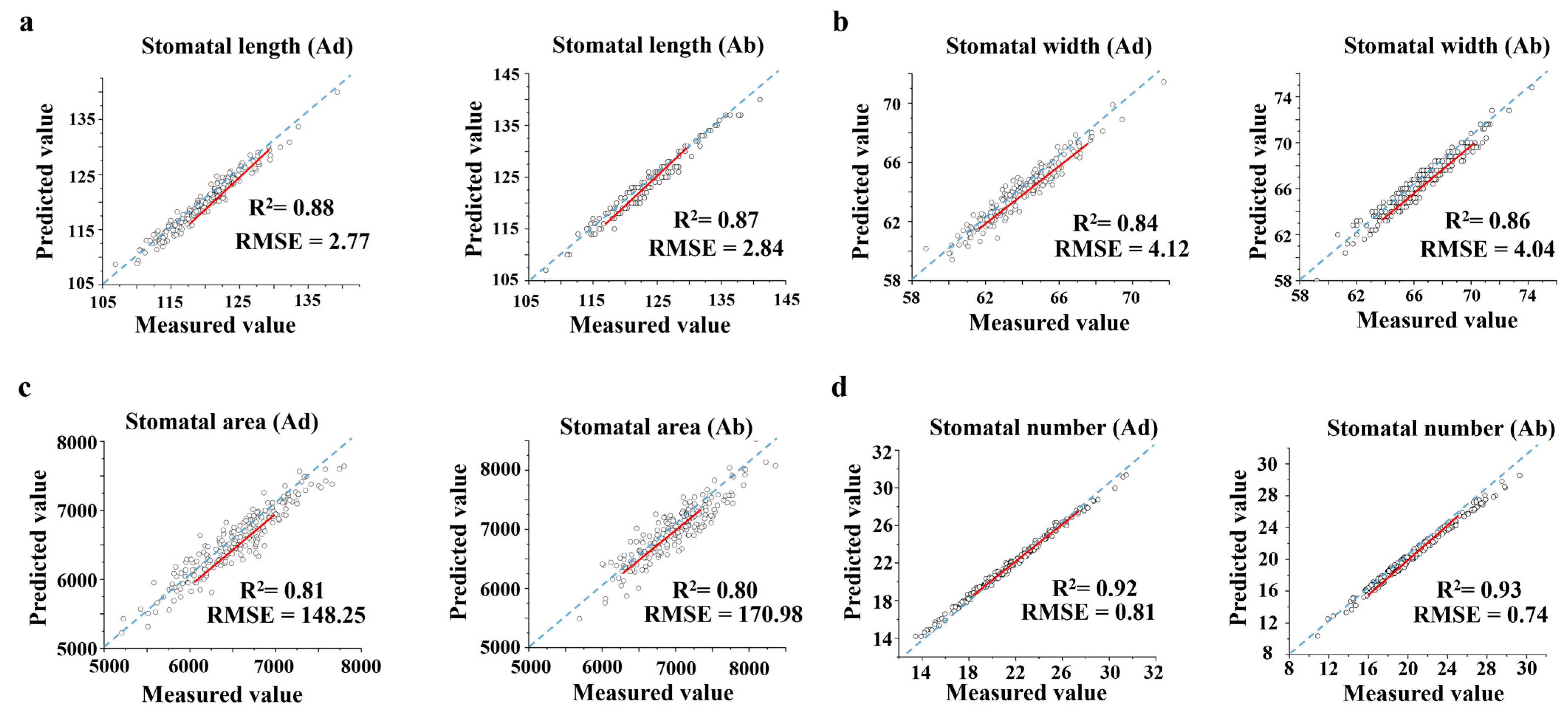
2.2. Stomatal Trait Accuracy Assessment
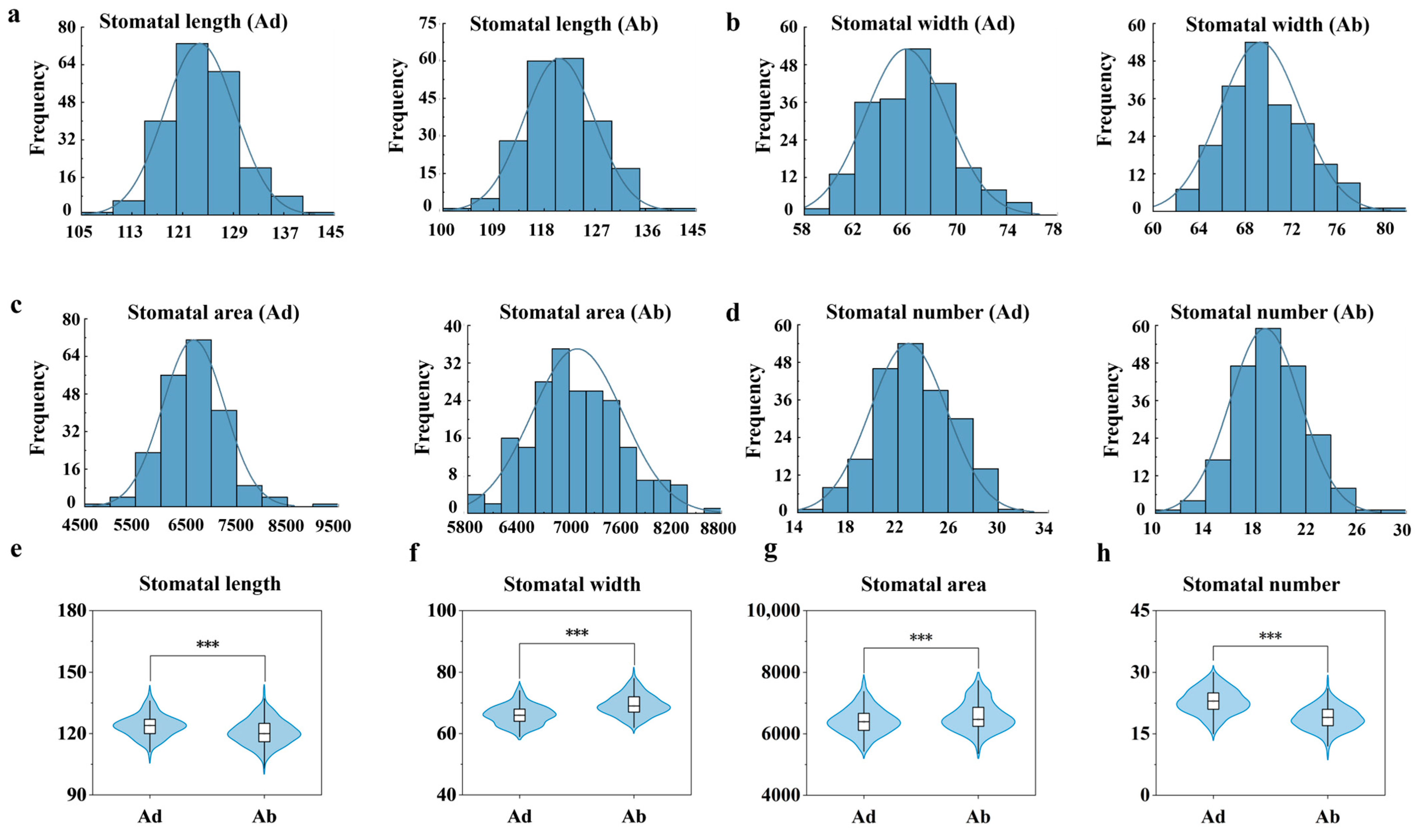
2.3. Analysis of Stomatal Differences Between the Adaxial and Abaxial Surfaces
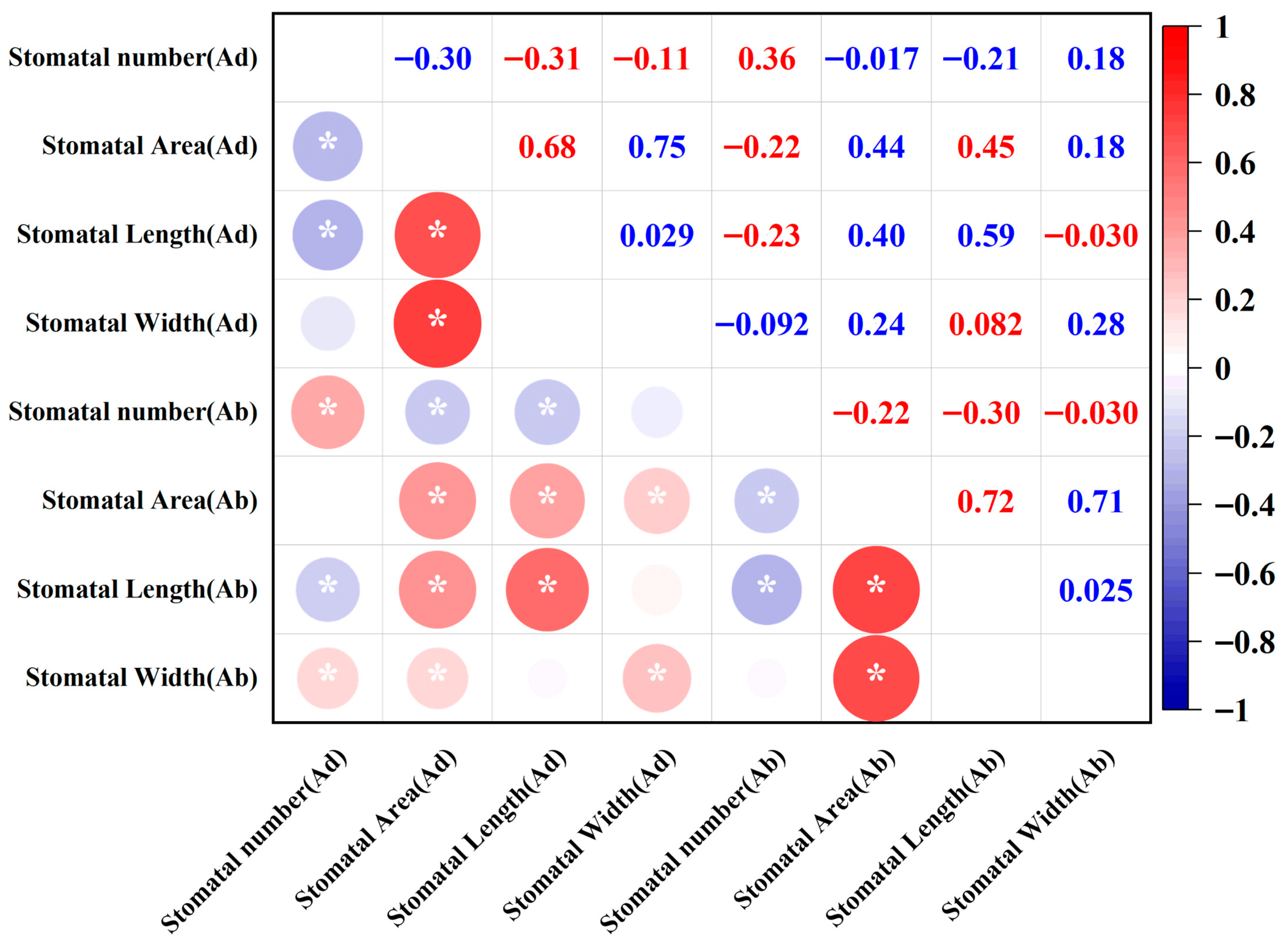
2.4. Stomatal Trait Correlation Analysis
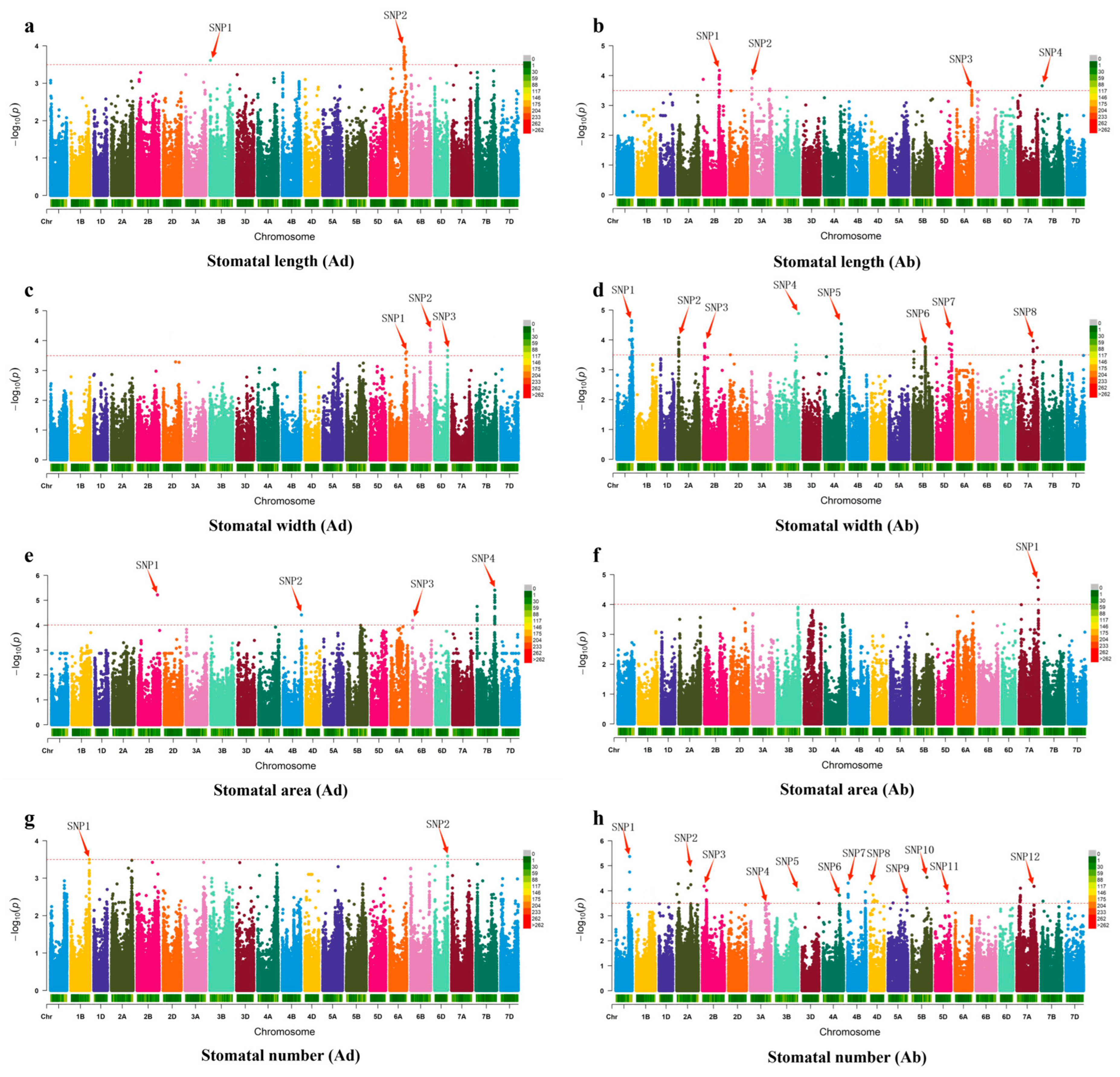
2.5. GWAS Results and Candidate Gene Identification
3. Discussion
3.1. Model Performance Testing and Comparative Experiments
3.2. Genetic Regulation of Stomatal Traits and Application Prospects of GWAS
3.3. The Generalization Ability of the WSF Model to Other Plants
3.4. Limitations of the WSF Model and Future Research Directions
4. Materials and Methods
4.1. Stomatal Data Collection for 210 Wheat Varieties
4.2. Construction of the Wheat Stomata Dataset
4.3. WSF Model Architecture
4.4. Wheat Stomatal Trait Extraction
4.5. Genotyping and GWAS of Stomatal Traits in Wheat
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Wheat Varieties Used in the Study
| Number | Variety Name | Number | Variety Name | Number | Variety Name | Number | Variety Name | Number | Variety Name |
| 1 | Aikang 58 | 43 | Jinan 16 | 85 | Luomai 21 | 127 | Shimai 22 | 169 | Yannong 15 |
| 2 | Baomai 10 | 44 | Jinan 17 | 86 | Luomai 26 | 128 | Shiyou 20 | 170 | Yannong 173 |
| 3 | Beijing 10 | 45 | Jining 16 | 87 | Luomai 7 | 129 | Shunmai 1718 | 171 | Yannong 19 |
| 4 | Boai 7023 | 46 | Ji 5265 | 88 | Machang 2 | 130 | Sumai 188 | 172 | Yannong 22 |
| 5 | Chuanmai 22 | 47 | Jimai 19 | 89 | Mianmai 37 | 131 | Sumai 3 | 173 | Yannong 836 |
| 6 | Chuanmai 36 | 48 | Jimai 1 | 90 | Mianmai 39 | 132 | Tai 10604 | 174 | Yannong 999 |
| 7 | Chuanmai 42 | 49 | Jimai 26 | 91 | Xumai 32 | 133 | Taikemai 31 | 175 | Yangmai 10 |
| 8 | Dekang 961 | 50 | Jinan 2 | 92 | Nannong 0686 | 134 | Taikemai 33 | 176 | Yangmai 13 |
| 9 | Dexuan 1 | 51 | Jinan 8 | 93 | Ningmai 13 | 135 | Taimai 1918 | 177 | Yangmai 15 |
| 10 | Emai 11 | 52 | Jinan Ai 6 | 94 | Ningmai 15 | 136 | Tainong 19 | 178 | Yangmai 158 |
| 11 | Emai 12 | 53 | Jinhe 9123 | 95 | Ningmai 22 | 137 | Taishan 1 | 179 | Yangmai 16 |
| 12 | Emai 15 | 54 | Jinmai 31 | 96 | Ningmai 24 | 138 | Taishan 27 | 180 | Yangmai 18 |
| 13 | Emai 19 | 55 | Jinmai 33 | 97 | Ningmai 26 | 139 | Taishan 28 | 181 | Yangmai 1 |
| 14 | Emai 6 | 56 | Jinmai 47 | 98 | Ningmai 9 | 140 | Taishan 4 | 182 | Yangmai 22 |
| 15 | Fan 6 | 57 | Jingdong 17 | 99 | Ningnuomai 1 | 141 | Taishan 5366 | 183 | Yangmai 5 |
| 16 | Fengde Cunmai 1 | 58 | Jingdong 18 | 100 | Nongda 211 | 142 | Taitianmai 118 | 184 | Yangmai 9 |
| 17 | Fengkang 8 | 59 | Jingdong 22 | 101 | Qida 195 | 143 | Tainong 24 | 185 | Yumai 13 |
| 18 | Gaoyou 503 | 60 | Jinghua 9 | 102 | Qimai 2 | 144 | Wanmai 0066 | 186 | Yumai 18 |
| 19 | Guomai 301 | 61 | Jingshuang 16 | 103 | Qingnong 2 | 145 | Wanmai 19 | 187 | Yumai 2 |
| 20 | Han 7086 | 62 | Keyuan 088 | 104 | Rumai 1 | 146 | Wanmai 33 | 188 | Yunhan 2233 |
| 21 | Haoyou 2018 | 63 | Laizhou 137 | 105 | Ruihuamai 520 | 147 | Wanmai 50 | 189 | Yunhan 618 |
| 22 | Henong 2063 | 64 | Laizhou 953 | 106 | Ruihuamai 523 | 148 | Wanxi Mai 0638 | 190 | Xumai 31 |
| 23 | Henong 6049 | 65 | Lianmai 2 | 107 | Shannong 12 | 149 | Wanfeng 269 | 191 | Chang 4738 |
| 24 | Henong 825 | 66 | Lianmai 6 | 108 | Shannong 15 | 150 | Wannian 2 | 192 | Zhemai 1 |
| 25 | Heng 136 | 67 | Lianmai 7 | 109 | Shannong 20 | 151 | Wennong 14 | 193 | Zhenmai 12 |
| 26 | Heng 4399 | 68 | Liangxing 66 | 110 | Shannong 205 | 152 | Wennong 5 | 194 | Zhenmai 168 |
| 27 | Yangmai 20 | 69 | Liangxing 77 | 111 | Shannong 21 | 153 | Wennong 17 | 195 | Zhenmai 3 |
| 28 | Hengguan 35 | 70 | Lin Y8012 | 112 | Shannong 22 | 154 | Xinmai 16 | 196 | Zhenmai 4 |
| 29 | Hengza 102 | 71 | Linmai 4 | 113 | Shannong 24 | 155 | Xinmai 18 | 197 | Zhenmai 9 |
| 30 | Huaimai 20 | 72 | Lukenmai 9 | 114 | Shannong 28 | 156 | Xinmai 20 | 198 | Zhengmai 004 |
| 31 | Huaimai 22 | 73 | Lumai 14 | 115 | Shannong 29 | 157 | Xinmai 9 | 199 | Zhengmai 7698 |
| 32 | Huaimai 28 | 74 | Lumai 15 | 116 | Shengxuan 3 | 158 | Xinmai 296 | 200 | Zhengmai 9023 |
| 33 | Huaimai 29 | 75 | Lumai 1 | 117 | Shengxuan 6 | 159 | Xingmai 18 | 201 | Zhongmai 175 |
| 34 | Huaimai 30 | 76 | Lumai 21 | 118 | Shiluan 02-1 | 160 | Xingmai 7 | 202 | Zhongmai 9 |
| 35 | Huaimai 33 | 77 | Lumai 22 | 119 | Shimai 26 | 161 | Xumai 30 | 203 | Zhoumai 12 |
| 36 | Huaimai 6 | 78 | Lumai 5 | 120 | Shimai 28 | 162 | Xumai 33 | 204 | Zhoumai 18 |
| 37 | Jimai 19 | 79 | Luyuan 502 | 121 | Shi 4185 | 163 | Xuzhou 24 | 205 | Zhoumai 23 |
| 38 | Jimai 20 | 80 | Lunxuan 987 | 122 | Shijiazhuang 407 | 164 | Xuzhou 438 | 206 | Zhoumai 24 |
| 39 | Jimai 21 | 81 | Luohan 11 | 123 | Shijiazhuang 8 | 165 | Xuzhou 8 | 207 | Zhoumai 26 |
| 40 | Jimai 22 | 82 | Luohan 13 | 124 | Shimai 12 | 166 | Yanfu 188 | 208 | Zhoumai 27 |
| 41 | Jimai 229 | 83 | Luohan 2 | 125 | Shimai 15 | 167 | Yannong 0428 | 209 | Zhoumai 32 |
| 42 | Jimai 23 | 84 | Luohan 9 | 126 | Shimai 18 | 168 | Yannong 1212 | 210 | Zinong 033 |
Appendix B. Significant Associations Markers Based on GWAS of Stomatal Traits
| Trait | Marker | Loc | Var | −log10 | SnpEffect_GeneID | Gene Annotation |
|---|---|---|---|---|---|---|
| Length (Ad) | SNP1 | Chr3B:24820702 | C->T | 3.50 | TraesCS3B02G048900 | oxidoreductase activity |
| Length (Ad) | SNP2 | Chr6A:522154021 | G->A | 3.86 | TraesCS6A02G290600 | cellular response to calcium ion, regulation of stomatal closure |
| Length (Ab) | SNP1 | Chr2B:579569265 | C->A | 4.01 | TraesCS2B02G407900 | integral component of membrane transmembrane transport |
| Length (Ab) | SNP2 | Chr3A:15558194 | G->A | 3.90 | TraesCS3A02G030300 | integral component of membrane |
| Length (Ab) | SNP3 | Chr6A:571935441 | A->C | 3.50 | TraesCS6A02G338600 | transaminase activity cellular amino acid metabolic process |
| Length (Ab) | SNP4 | Chr7B:52781 | G->A | 3.65 | TraesCS7B02G000200 | ATP binding, ATPase activity, coupled to transmembrane movement of substances |
| Width (Ad) | SNP1 | Chr6A:609452855 | C->A | 3.63 | TraesCS6A02G400000 | iron–sulfur cluster binding, metal ion binding |
| Width (Ad) | SNP2 | Chr6B:710007244 | C->T | 4.36 | TraesCS6B02G451100 | aspartic endopeptidase activity, intramembrane cleaving, membrane protein proteolysis |
| Width (Ad) | SNP3 | Chr6D:468806891 | A->G | 3.67 | TraesCS6D02G396300 | RNA polymerase |
| Width (Ab) | SNP1 | Chr1A:531677694 | T->C | 4.58 | TraesCS1A02G342900 | ATPase activity, coupled to transmembrane movement of substances, ATP binding |
| Width (Ab) | SNP2 | Chr2A:36021015 | G->T | 4.08 | TraesCS2A02G073500 | UDP-glycosyltransferase activity |
| Width (Ab) | SNP3 | Chr2B:54532482 | A->G | 3.75 | TraesCS2B02G094100 | protein kinase activity, ubiquitin–protein transferase activity, ATP binding, oxidoreductase activity |
| Width (Ab) | SNP4 | Chr3B:705591855 | C->T | 3.84 | TraesCS3B02G462700 | integral component of membrane |
| Width (Ab) | SNP5 | Chr4A:624015701 | T->G | 4.54 | TraesCS4A02G344100 | O-methyltransferase activity; protein dimerization activity; P-nucleus; hormone-mediated signaling pathway; methylation; Q-positive regulation of transcription, DNA-templated; |
| Width (Ab) | SNP6 | Chr5B:457122696 | T->C | 3.64 | TraesCS5B02G271400 | RNA binding; intracellular membrane-bounded organelle; RNA modification |
| Width (Ab) | SNP7 | Chr5D:549858298 | C->G | 4.23 | TraesCS5D02G536800 | plant-pathogen interaction; environmental adaptation; organismal Systems |
| Width (Ab) | SNP8 | Chr7A:548367285 | T->C | 3.68 | TraesCS7A02G375500 | calcium ion binding; RNA binding; methyltransferase activity; RNA methylation |
| Area (Ad) | SNP1 | Chr2B:703976259 | G-> T | 5.22 | TraesCS2B02G507100 | ATP binding, ADP binding, defense response |
| Area (Ad) | SNP2 | Chr4B:656816645 | G-> T | 4.41 | TraesCS4B02G370800 | cation transmembrane transporter activity |
| Area (Ad) | SNP3 | Chr6B:32330944 | C->G | 4.19 | TraesCS6B02G068800 | raffinose alpha-galactosidase activity, carbohydrate metabolic process |
| Area (Ad) | SNP4 | Chr7B:641806970 | G->T | 5.41 | TraesCS7B02G377100 | extracellular region |
| Area (Ab) | SNP1 | Chr7A:675287935 | G->T | 4.57 | TraesCS7A02G684200 | integral component of membrane |
| Number (Ad) | SNP1 | Chr1B:667223816 | G->A | 3.50 | TraesCS1B02G447600 | RNA binding, metal ion binding; nucleus |
| Number (Ad) | SNP2 | Chr6D:471016984 | G->A | 3.59 | TraesCS6D02G402800 | amRNA binding, DNA binding, metal ion binding; poly(A)+mRNA export from nucleus |
| Number (Ab) | SNP1 | Chr1A:503377978 | C->A | 4.75 | TraesCS1A02G311900 | DNA-binding transcription factor activity, RNA polymerase II-specific |
| Number (Ab) | SNP2 | Chr2A:52763048 | T->C | 4.28 | TraesCS2A02G102500 | other types of O-glycan biosynthesis, Glycan biosynthesis and metabolism; oxidoreductase activity |
| Number (Ab) | SNP3 | Chr2B:153128770 | A->G | 3.57 | TraesCS2B02G178000 | regulation of stomatal movement |
| Number (Ab) | SNP4 | Chr3A:535189608 | G->T | 3.50 | TraesCS3A02G301400 | rRNA 5′-end processing, rRNA processing |
| Number (Ab) | SNP5 | Chr3B:822681495 | C->T | 4.03 | TraesCS3B02G602700 | MAPK signaling pathway-plant |
| Number (Ab) | SNP6 | Chr4A:590590535 | C->T | 3.82 | TraesCS4A02G283700 | amino sugar and nucleotide sugar metabolism |
| Number (Ab) | SNP7 | Chr4B:21556722 | G->A | 4.30 | TraesCS4B02G029400 | amino sugar and nucleotide sugar metabolism |
| Number (Ab) | SNP8 | Chr4D:11914487 | C->T | 4.29 | TraesCS4D02G026900 | amino sugar and nucleotide sugar metabolism |
| Number (Ab) | SNP9 | Chr5A:682898348 | T->C | 3.75 | TraesCS5A02G521600 | magnesium ion binding, terpene synthase activity; terpenoid biosynthetic process |
| Number (Ab) | SNP10 | Chr5B:550156642 | C->T | 4.54 | TraesCS5B02G563800 | Aminoacyl-tRNA biosynthesis; DNA binding, sigma factor activity |
| Number (Ab) | SNP11 | Chr5D:450860102 | G->T | 3.89 | TraesCS5D02G380500 | ribosome biogenesis in eukaryotes; GTPase activity, GTP binding |
| Number (Ab) | SNP12 | Chr7A:116506563 | T->C | 3.66 | TraesCS7A02G160400 | MAPK signaling pathway-plant, Plant hormone signal transduction, Plant-pathogen interaction |
References
- Qin, X.; Zhang, F.; Liu, C.; Yu, H.; Cao, B.; Tian, S.; Liao, Y.; Siddique, K.H. Wheat yield improvements in China: Past trends and future directions. Field Crops Res. 2015, 177, 117–124. [Google Scholar] [CrossRef]
- Lawson, T.; Matthews, J. Guard cell metabolism and stomatal function. Annu. Rev. Plant Biol. 2020, 71, 273–302. [Google Scholar] [CrossRef]
- Xie, J.; Fernandes, S.B.; Mayfield-Jones, D.; Erice, G.; Choi, M.; E Lipka, A.; Leakey, A.D. Optical topometry and machine learning to rapidly phenotype stomatal patterning traits for maize QTL mapping. Plant Physiol. 2021, 187, 1462–1480. [Google Scholar] [CrossRef]
- Danila, F.R.; Quick, W.P.; White, R.G.; Furbank, R.T.; von Caemmerer, S. The metabolite pathway between bundle sheath and mesophyll: Quantification of plasmodesmata in leaves of C3 and C4 monocots. Plant Cell 2016, 28, 1461–1471. [Google Scholar] [CrossRef]
- Kim, K.W. Methanol fixation for scanning electron microscopy of plants. Appl. Microsc. 2020, 50, 10. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Cheng, Y.; Cao, L.; Wang, S.; Li, Y.; Wang, H.; Zhou, Y. Analyses of plant leaf cell size, density and number, as well as trichome number using cell counter plugin. Bio-Protocol 2014, 4, e1165. [Google Scholar] [CrossRef]
- Meeus, S.; Van den Bulcke, J.; Wyffels, F. From leaf to label: A robust automated workflow for stomata detection. Ecol. Evol. 2020, 10, 9178–9191. [Google Scholar] [CrossRef] [PubMed]
- Fetter, K.C.; Eberhardt, S.; Barclay, R.S.; Wing, S.; Keller, S.R. StomataCounter: A neural network for automatic stomata identification and counting. New Phytol. 2019, 223, 1671–1681. [Google Scholar] [CrossRef] [PubMed]
- Badgujar, C.M.; Poulose, A.; Gan, H. Agricultural object detection with You Only Look Once (YOLO) Algorithm: A bibliometric and systematic literature review. Comput. Electron. Agric. 2024, 223, 109090. [Google Scholar] [CrossRef]
- Pathoumthong, P.; Zhang, Z.; Roy, S.J.; El Habti, A. Rapid non-destructive method to phenotype stomatal traits. Plant Methods 2023, 19, 36. [Google Scholar] [CrossRef] [PubMed]
- Sultana, S.N.; Park, H.; Choi, S.H.; Jo, H.; Song, J.T.; Lee, J.-D.; Kang, Y.J. Optimizing the experimental method for stomata-profiling automation of soybean leaves based on deep learning. Plants 2021, 10, 2714. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Yang, Q.; Yang, L.; Shen, T.; Wang, R.; Fu, C. Deep learning implementation of image segmentation in agricultural applications: A comprehensive review. Artif. Intell. Rev. 2024, 57, 149. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017. [Google Scholar]
- Costa, L.; Archer, L.; Ampatzidis, Y.; Casteluci, L.; Caurin, G.A.; Albrecht, U. Determining leaf stomatal properties in citrus trees utilizing machine vision and artificial intelligence. Precis. Agric. 2021, 22, 1107–1119. [Google Scholar] [CrossRef]
- Jayakody, H.; Petrie, P.; Boer, H.J.d.; Whitty, M. A generalised approach for high-throughput instance segmentation of stomata in microscope images. Plant Methods 2021, 17, 27. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015. [Google Scholar]
- Zhu, C.; Hu, Y.; Mao, H.; Li, S.; Li, F.; Zhao, C.; Luo, L.; Liu, W.; Yuan, X. A deep learning-based method for automatic assessment of stomatal index in wheat microscopic images of leaf epidermis. Front. Plant Sci. 2021, 12, 716784. [Google Scholar] [CrossRef] [PubMed]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Visscher, P.M.; Brown, M.A.; McCarthy, M.I.; Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012, 90, 7–24. [Google Scholar] [CrossRef]
- Duan, P.; Xu, J.; Zeng, D.; Zhang, B.; Geng, M.; Zhang, G.; Huang, K.; Huang, L.; Xu, R.; Ge, S. Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 2017, 10, 685–694. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, H.; Wu, L.; Warburton, M.; Yan, J. Genome-wide association studies in maize: Praise and stargaze. Mol. Plant 2017, 10, 359–374. [Google Scholar] [CrossRef]
- Saini, D.K.; Chopra, Y.; Singh, J.; Sandhu, K.S.; Kumar, A.; Bazzer, S.; Srivastava, P. Comprehensive evaluation of mapping complex traits in wheat using genome-wide association studies. Mol. Breed. 2022, 42, 1. [Google Scholar] [CrossRef]
- Maurer, A.; Draba, V.; Pillen, K. Genomic dissection of plant development and its impact on thousand grain weight in barley through nested association mapping. J. Exp. Bot. 2016, 67, 2507–2518. [Google Scholar] [CrossRef]
- Soltabayeva, A.; Ongaltay, A.; Omondi, J.O.; Srivastava, S. Morphological, physiological and molecular markers for salt-stressed plants. Plants 2021, 10, 243. [Google Scholar] [CrossRef]
- Gibbs, J.A.; Mcausland, L.; Robles-Zazueta, C.A.; Murchie, E.H.; Burgess, A.J. A deep learning method for fully automatic stomatal morphometry and maximal conductance estimation. Front. Plant Sci. 2021, 12, 780180. [Google Scholar] [CrossRef] [PubMed]
- Qur’ania, A.; Herdiyeni, Y.; Kusuma, W.A.; Wigena, A.H.; Suhesti, S. Detection and Calculation of Stomatal Density Using YOLOv5: A Study of High-Yielding Patchouli Varieties. J. Image Graph. 2024, 12, 228–238. [Google Scholar] [CrossRef]
- Wacker, T.S.; Smith, A.G.; Jensen, S.M.; Pflüger, T.; Hertz, V.G.; Rosenqvist, E.; Liu, F.; Dresbøll, D.B. Stomata morphology measurement with interactive machine learning: Accuracy, speed, and biological relevance? Plant Methods 2025, 21, 95. [Google Scholar] [CrossRef] [PubMed]
- Lawson, T.; Blatt, M.R. Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 2014, 164, 1556–1570. [Google Scholar] [CrossRef]
- Caine, R.S.; Yin, X.; Sloan, J.; Harrison, E.L.; Mohammed, U.; Fulton, T.; Biswal, A.K.; Dionora, J.; Chater, C.C.; Coe, R.A. Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 2019, 221, 371–384. [Google Scholar] [CrossRef]
- Dunn, J.; Hunt, L.; Afsharinafar, M.; Meselmani, M.A.; Mitchell, A.; Howells, R.; Wallington, E.; Fleming, A.J.; Gray, J.E. Reduced stomatal density in bread wheat leads to increased water-use efficiency. J. Exp. Bot. 2019, 70, 4737–4748. [Google Scholar] [CrossRef]
- Hughes, J.; Hepworth, C.; Dutton, C.; Dunn, J.A.; Hunt, L.; Stephens, J.; Waugh, R.; Cameron, D.D.; Gray, J.E. Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 2017, 174, 776–787. [Google Scholar] [CrossRef]
- Wang, S.-X.; Zhu, Y.-L.; Zhang, D.-X.; Shao, H.; Liu, P.; Hu, J.-B.; Zhang, H.; Zhang, H.-P.; Chang, C.; Lu, J. Genome-wide association study for grain yield and related traits in elite wheat varieties and advanced lines using SNP markers. PLoS ONE 2017, 12, e0188662. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yu, S.; Zhang, Y.; Zhu, D.; Li, F.; Chen, B.; Mei, F.; Du, L.; Ding, L.; Chen, L. Genome-wide association study revealed TaHXK3-2A as a candidate gene controlling stomatal index in wheat seedlings. Plant Cell Environ. 2022, 45, 2306–2323. [Google Scholar] [CrossRef] [PubMed]
- Schoppach, R.; Taylor, J.D.; Majerus, E.; Claverie, E.; Baumann, U.; Suchecki, R.; Fleury, D.; Sadok, W. High resolution mapping of traits related to whole-plant transpiration under increasing evaporative demand in wheat. J. Exp. Bot. 2016, 67, 2847–2860. [Google Scholar] [CrossRef]
- MacAlister, C.A.; Ohashi-Ito, K.; Bergmann, D.C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 2007, 445, 537–540. [Google Scholar] [CrossRef]
- Hunt, L.; Gray, J.E. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 2009, 19, 864–869. [Google Scholar] [CrossRef]
- Sun, C.; Dong, Z.; Zhao, L.; Ren, Y.; Zhang, N.; Chen, F. The Wheat 660K SNP array demonstrates great potential for marker-assisted selection in polyploid wheat. Plant Biotechnol. J. 2020, 18, 1354–1360. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X. rMVP: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Ma, S.; Wang, M.; Wu, J.; Guo, W.; Chen, Y.; Li, G.; Wang, Y.; Shi, W.; Xia, G.; Fu, D. WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 2021, 14, 1965–1968. [Google Scholar] [CrossRef] [PubMed]

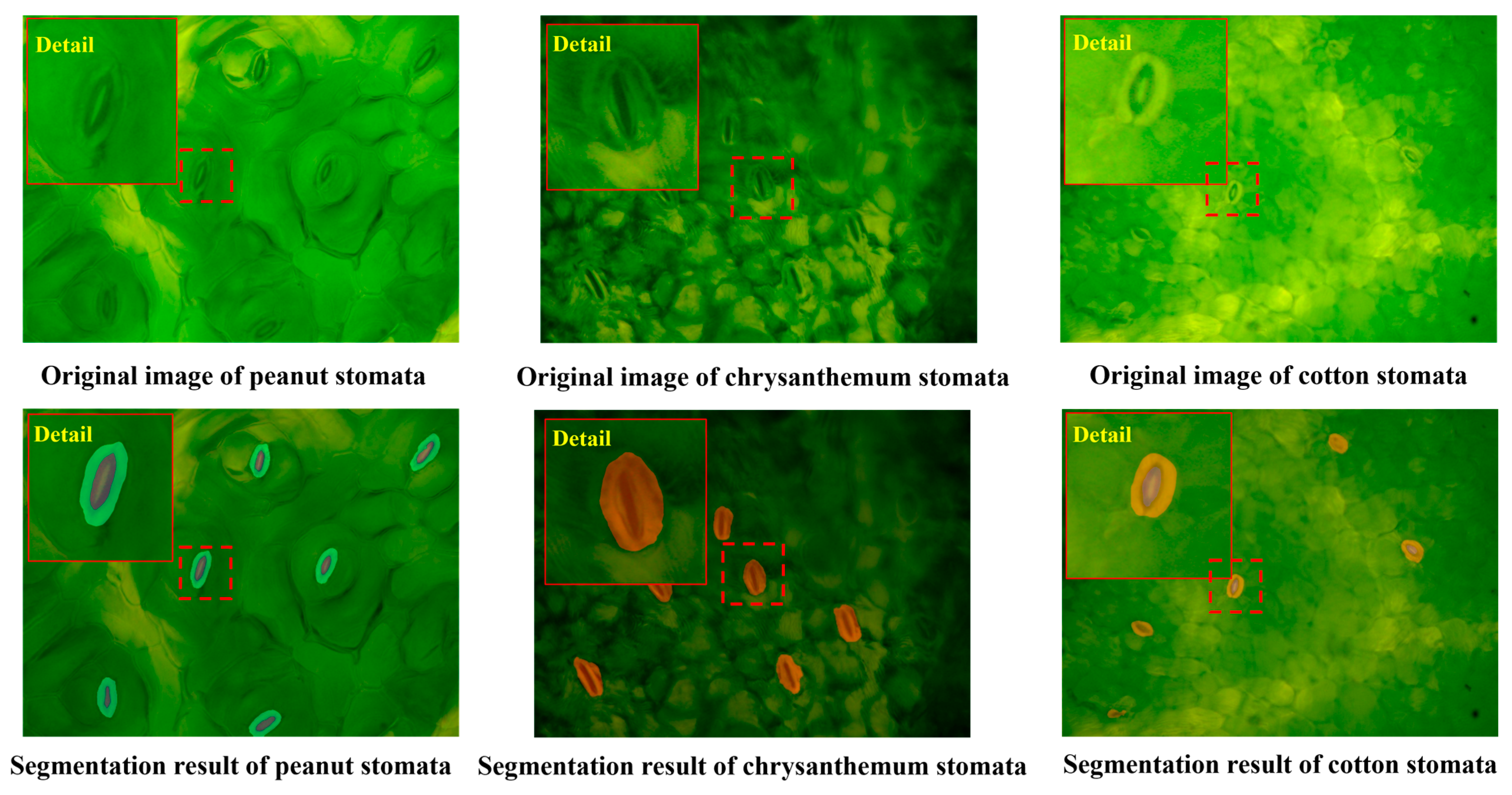

| Model Name | Precision | MIoU |
|---|---|---|
| WSF (our) | 93.6% | 91.2% |
| SegFormer | 91.7% | 90.1% |
| Deeplabv3+ | 89.5% | 86.8% |
| UNet | 87.7% | 85.2% |
| Plant Name | Precision | MIoU |
|---|---|---|
| Peanut | 81.1% | 77.5% |
| Chrysanthemum | 78.2% | 72.3% |
| Cotton | 83.6% | 79.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Min, H.; Liang, S.; Qin, B.; Sun, Q.; Pei, Z.; Pan, Q.; Wang, X.; Cai, J.; Zhou, Q.; et al. WSF: A Transformer-Based Framework for Microphenotyping and Genetic Analyzing of Wheat Stomatal Traits. Plants 2025, 14, 3016. https://doi.org/10.3390/plants14193016
Zhou H, Min H, Liang S, Qin B, Sun Q, Pei Z, Pan Q, Wang X, Cai J, Zhou Q, et al. WSF: A Transformer-Based Framework for Microphenotyping and Genetic Analyzing of Wheat Stomatal Traits. Plants. 2025; 14(19):3016. https://doi.org/10.3390/plants14193016
Chicago/Turabian StyleZhou, Honghao, Haijiang Min, Shaowei Liang, Bingxi Qin, Qi Sun, Zijun Pei, Qiuxiao Pan, Xiao Wang, Jian Cai, Qin Zhou, and et al. 2025. "WSF: A Transformer-Based Framework for Microphenotyping and Genetic Analyzing of Wheat Stomatal Traits" Plants 14, no. 19: 3016. https://doi.org/10.3390/plants14193016
APA StyleZhou, H., Min, H., Liang, S., Qin, B., Sun, Q., Pei, Z., Pan, Q., Wang, X., Cai, J., Zhou, Q., Zhong, Y., Huang, M., Jiang, D., Chen, J., & Li, Q. (2025). WSF: A Transformer-Based Framework for Microphenotyping and Genetic Analyzing of Wheat Stomatal Traits. Plants, 14(19), 3016. https://doi.org/10.3390/plants14193016








