COP9 Signalosome’s Role in Plant Defense Mechanisms
Abstract
1. Introduction
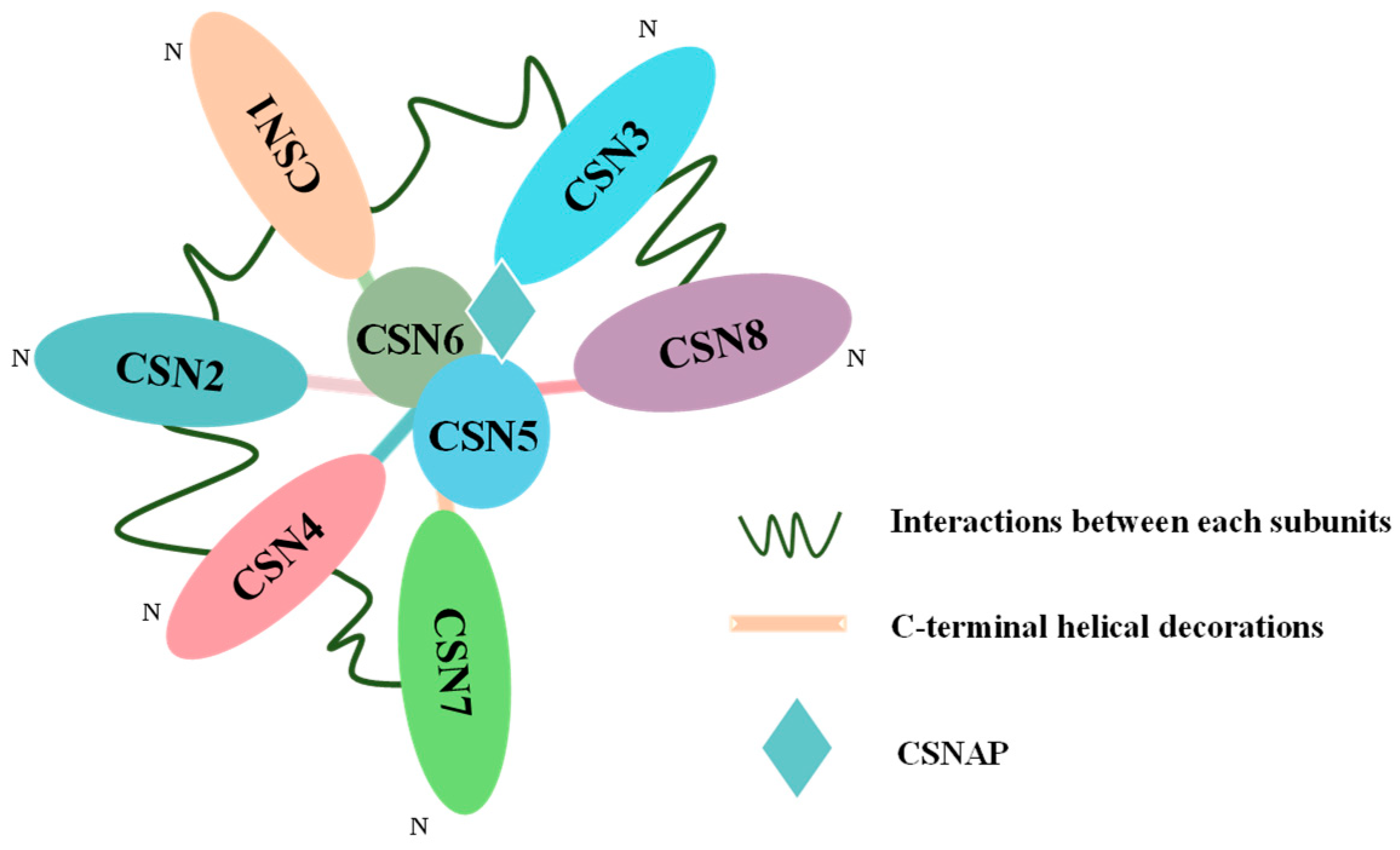
2. Structure and Function of COP9
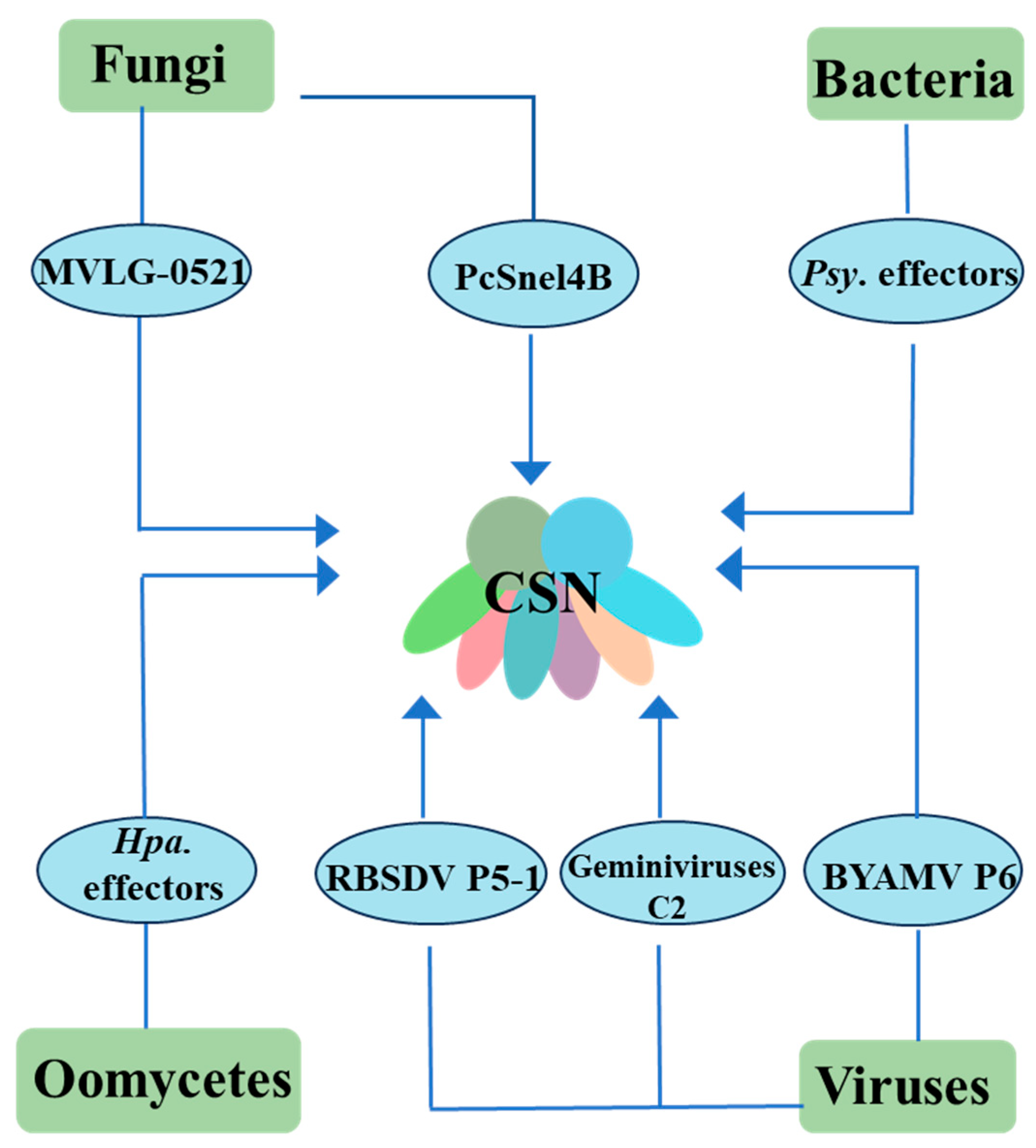
3. Various Pathogenic Microorganisms Target COP9
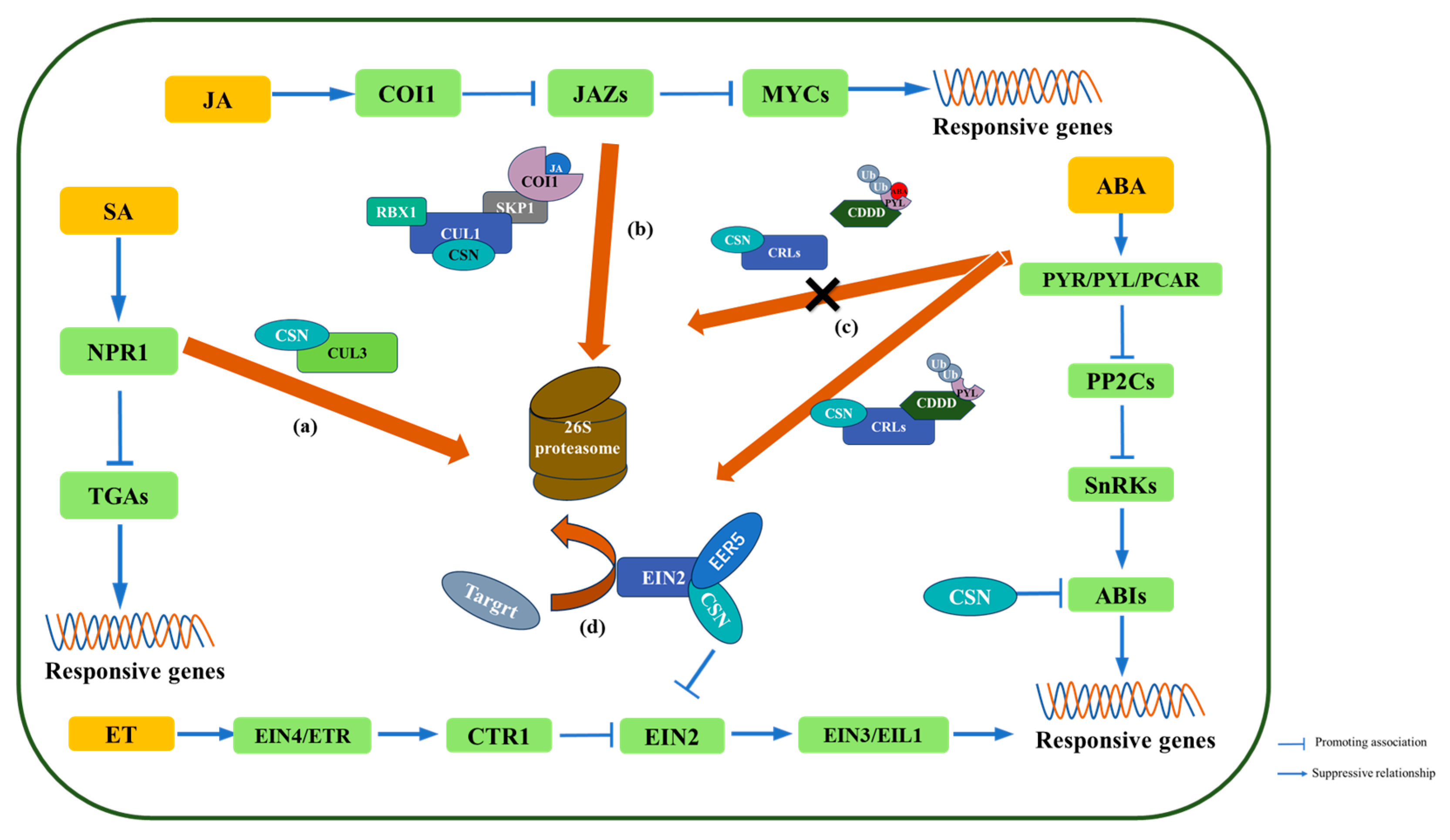
4. COP9 Regulates Hormone-Mediated Resistance Signaling Pathways
4.1. Salicylic Acid
4.2. Jasmonate
4.3. Abscisic Acid
4.4. Ethylene
5. COP9 and Immune-Related Secondary Metabolites
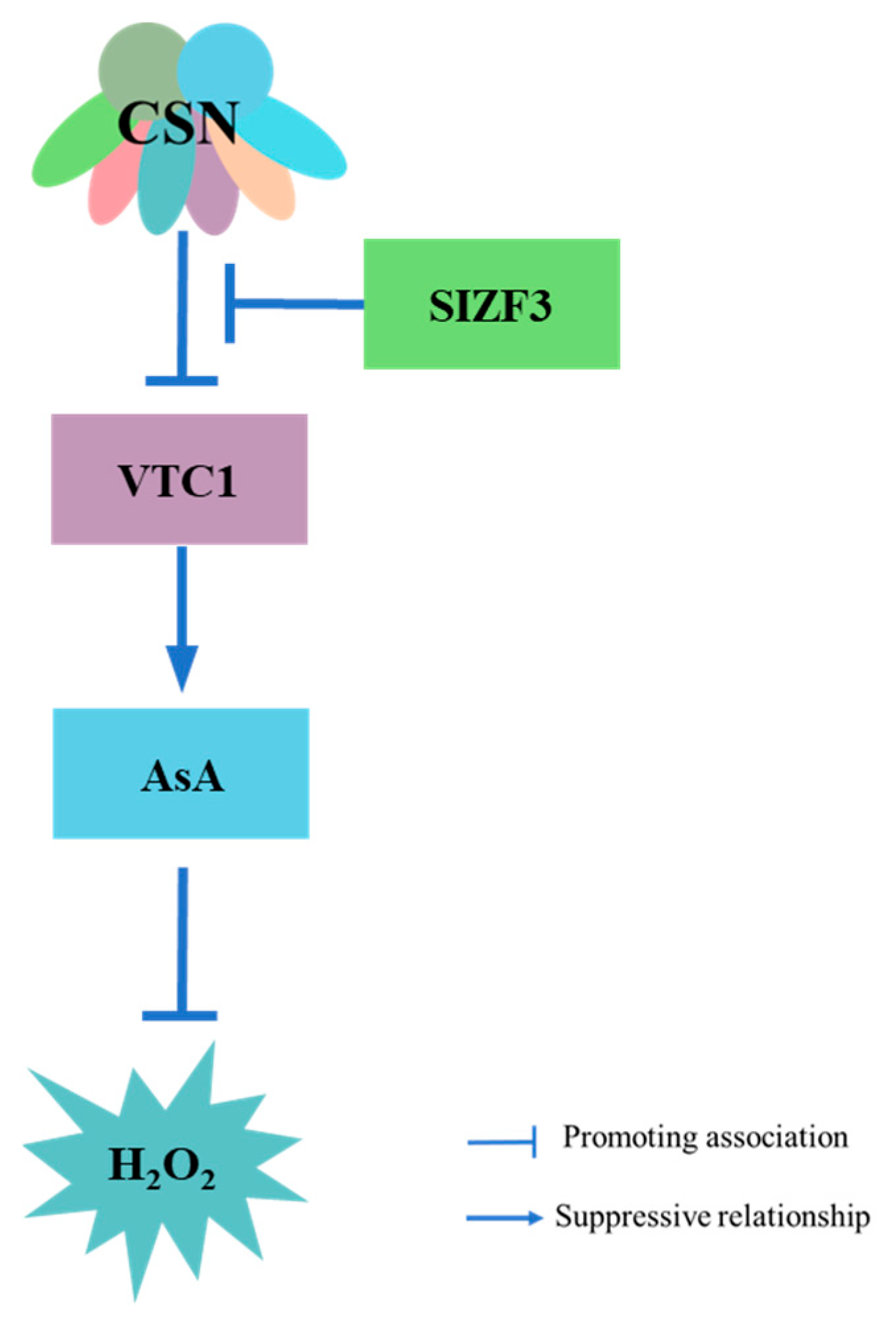
6. COP9 Regulation of ROS in Plant Immunity
7. COP9 Mediate Regulation of R Gene Homeostasis and Function
8. Concluding Remarks and Future Perspective
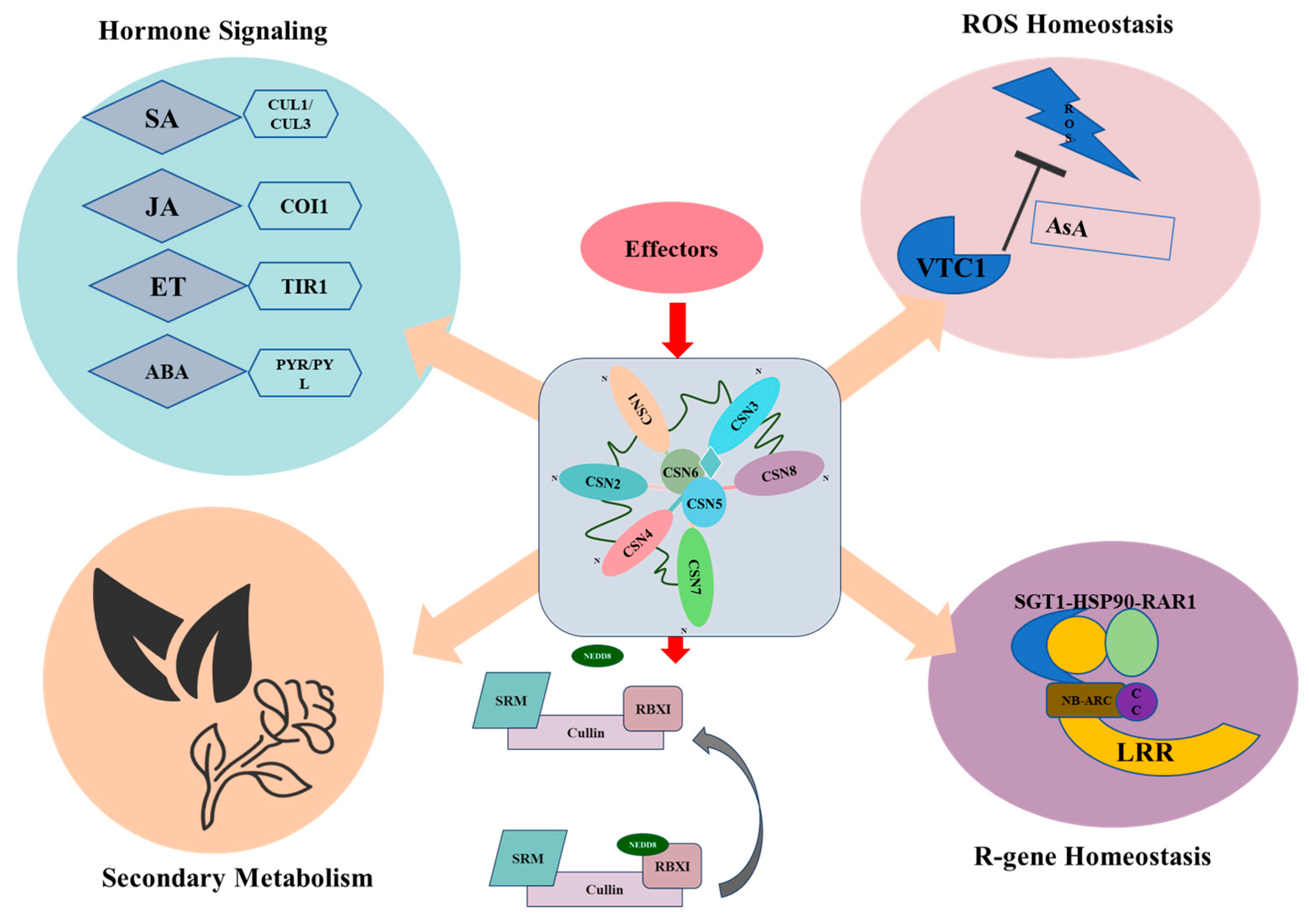
8.1. An Integrated Model for CSN in Plant Immunity: The Central Regulatory Hub
8.2. Functional Specialization and Coordination of COP9 Subunits
8.3. COP9 as a Central Regulator of the Growth-Defense Trade-Off
8.4. Future Perspectives and Experimental Roadmap
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| AMPs | Antimicrobial peptides |
| AsA | Ascorbic acid |
| BYSMV | Barley yellow striate mosaic virus |
| COI1 | Coronatine Insensitive 1 |
| CNL | CC-NB-LRR |
| CRLs | Cullin-RING E3 ubiquitin ligases |
| CSN | COP9 signalosome |
| CSNAP | COP9 signalosome-associated acidic protein |
| ET | Ethylene |
| ETI | Effector-triggered immunity |
| HR | Hypersensitive response |
| JA | Jasmonate |
| JAZ | Jasmonate-ZIM |
| LC-MS | Liquid chromatography–mass spectrometry |
| MAPK | Mitogen-activated protein kinase |
| NLR | Nucleotide-binding leucine-rich repeat |
| NPR1 | Nonexpressor Of PR genes 1 |
| PAMPs | Pathogen-associated molecular patterns |
| PCD | Programmed cell death |
| PCI | Proteasome-COP9-eIF3 |
| PR | Pathogenesis-related |
| PRRs | Pattern recognition receptors |
| PTI | Pattern-triggered immunity |
| RBSDV | Rice black -streaked dwarf virus |
| ROS | Reactive oxygen species |
| SA | Salicylic acid |
| SAR | Systemic acquired resistance |
| SLY1 | Sleepy 1 |
| SM | Secondary metabolism |
| TIR1 | Transport inhibitor response 1 |
| TNL | TIR-NB-LRR |
| UFO | Unusual flower organs |
| UPS | Ubiquitin-proteasome |
References
- Ngou, B.P.M.; Ding, P.; Jones, J.D.G. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 2022, 34, 1447–1478. [Google Scholar] [CrossRef] [PubMed]
- Nürnberger, T.; Brunner, F.; Kemmerling, B.; Piater, L. Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 2004, 198, 249–266. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Klessig, D.F. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001, 6, 520–527. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.-L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Wang, C.; Tang, R.-J.; Kou, S.; Xu, X.; Lu, Y.; Rauscher, K.; Voelker, A.; Luan, S. Mechanisms of calcium homeostasis orchestrate plant growth and immunity. Nature 2024, 627, 382–388. [Google Scholar] [CrossRef]
- Wang, C.; Luan, S. Calcium homeostasis and signaling in plant immunity. Curr. Opin. Plant Biol. 2023, 77, 102485. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Jones, J.D.; Ding, P. Plant immune networks. Trends Plant Sci. 2022, 27, 255–273. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Czymmek, K.; Tallóczy, Z.; Levine, B.; Dinesh-Kumar, S. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef]
- Hayward, A.P.; Dinesh-Kumar, S. What can plant autophagy do for an innate immune response? Annu. Rev. Phytopathol. 2011, 49, 557–576. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2008, 69, 473–488. [Google Scholar] [CrossRef]
- Gao, H.; Guo, M.; Song, J.; Ma, Y.; Xu, Z. Signals in systemic acquired resistance of plants against microbial pathogens. Mol. Biol. Rep. 2021, 48, 3747–3759. [Google Scholar] [CrossRef]
- Wei, N.; Deng, X.W. The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 2003, 19, 261–286. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Füzesi-Levi, M.G.; Ben-Nissan, G.; Mizrachi, L.; Gabashvili, A.; Levin, Y.; Ben-Dor, S.; Eisenstein, M.; Sharon, M. CSNAP Is a stoichiometric subunit of the COP9 signalosome. Cell Rep. 2015, 13, 585–598. [Google Scholar] [CrossRef]
- Wei, N.; Chamovitz, D.A.; Deng, X.-W. Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 1994, 78, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Chamovitz, D.A.; Wei, N.; Osterlund, M.T.; von Arnim, A.G.; Staub, J.M.; Matsui, M.; Deng, X.-W. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 1996, 86, 115–121. [Google Scholar] [CrossRef]
- Qin, N.; Xu, D.; Li, J.; Deng, X.W. COP9 signalosome: Discovery, conservation, activity, and function. J. Integr. Plant Biol. 2020, 62, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Franciosini, A.; Lombardi, B.; Iafrate, S.; Pecce, V.; Mele, G.; Lupacchini, L.; Rinaldi, G.; Kondou, Y.; Gusmaroli, G.; Aki, S.; et al. The Arabidopsis COP9 SIGNALOSOME INTERACTING F-BOX KELCH 1 protein forms an SCF ubiquitin ligase and regulates hypocotyl elongation. Mol. Plant 2013, 6, 1616–1629. [Google Scholar] [CrossRef]
- Wang, X.; Feng, S.; Nakayama, N.; Crosby, W.L.; Irish, V.; Deng, X.W.; Wei, N. The COP9 signalosome interacts with SCF UFO and participates in Arabidopsis flower development. Plant Cell 2003, 15, 1071–1082. [Google Scholar] [CrossRef]
- Feng, S.; Ma, L.; Wang, X.; Xie, D.; Dinesh-Kumar, S.P.; Wei, N.; Deng, X.W. The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 2003, 15, 1083–1094. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Serino, G.; Callis, J.; Crosby, W.L.; Lyapina, S.; Deshaies, R.J.; Gray, W.M.; Estelle, M.; Deng, X.-W. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCF TIR1 in mediating auxin response. Science 2001, 292, 1379–1382. [Google Scholar] [CrossRef]
- Dohmann, E.M.N.; Nill, C.; Schwechheimer, C. DELLA proteins restrain germination and elongation growth in Arabidopsis thaliana COP9 signalosome mutants. Eur. J. Cell Biol. 2010, 89, 163–168. [Google Scholar] [CrossRef]
- Zhang, C.; Fang, L.; He, F.; You, X.; Wang, M.; Zhao, T.; Hou, Y.; Xiao, N.; Li, A.; Yang, J.; et al. Ubiquitination of OsCSN5 by OsPUB45 activates immunity by modulating the OsCUL3a-OsNPR1 module. Sci. Adv. 2025, 11, eadr2441. [Google Scholar] [CrossRef]
- Wei, N.; Deng, X.W. COP9: A new genetic locus involved in light-regulated development and gene expression in arabidopsis. Plant Cell 1992, 4, 1507–1518. [Google Scholar] [PubMed]
- Zhang, H.; Wang, X.; Giroux, M.J.; Huang, L. A wheat COP9 subunit 5-like gene is negatively involved in host response to leaf rust. Mol. Plant Pathol. 2016, 18, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Cui, K.-C.; Liu, M.; Ke, G.-H.; Zhang, X.-Y.; Mu, B.; Zhou, M.; Hu, Y.; Wen, Y.-Q. Transient silencing of VvCSN5 enhances powdery mildew resistance in grapevine (Vitis vinifera). Plant Cell Tissue Organ Cult. (PCTOC) 2021, 146, 621–633. [Google Scholar] [CrossRef]
- Schwechheimer, C. The COP9 signalosome (CSN): An evolutionary conserved proteolysis regulator in eukaryotic development. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2004, 1695, 45–54. [Google Scholar] [CrossRef]
- Lingaraju, G.M.; Bunker, R.D.; Cavadini, S.; Hess, D.; Hassiepen, U.; Renatus, M.; Fischer, E.S. Crystal structure of the human COP9 signalosome. Nature 2014, 512, 161–165. [Google Scholar] [CrossRef]
- Tsuge, T.; Matsui, M.; Wei, N. The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J. Mol. Biol. 2001, 305, 1–9. [Google Scholar] [CrossRef]
- Tran, H.J.T.T.; Allen, M.D.; Löwe, J.; Bycroft, M. Structure of the Jab1/MPN domain and its implications for proteasome function. Biochemistry 2003, 42, 11460–11465. [Google Scholar] [CrossRef]
- Choi, H.H.; Gully, C.; Su, C.-H.; Velazquez-Torres, G.; Chou, P.-C.; Tseng, C.; Zhao, R.; Phan, L.; Shaiken, T.; Chen, J.; et al. COP9 signalosome subunit 6 stabilizes COP1, which functions as an E3 ubiquitin ligase for 14-3-3σ. Oncogene 2011, 30, 4791–4801. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rozen, S.; Tieri, A.; Ridner, G.; Stark, A.-K.; Schmaler, T.; Ben-Nissan, G.; Dubiel, W.; Sharon, M. Exposing the subunit diversity within protein complexes: A mass spectrometry approach. Methods 2013, 59, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Rockel, B.; Schmaler, T.; Huang, X.; Dubiel, W. Electron microscopy and in vitro deneddylation reveal similar architectures and biochemistry of isolated human and Flag-mouse COP9 signalosome complexes. Biochem. Biophys. Res. Commun. 2014, 450, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Birol, M.; Enchev, R.I.; Padilla, A.; Stengel, F.; Aebersold, R.; Betzi, S.; Yang, Y.; Hoh, F.; Peter, M.; Dumas, C.; et al. Structural and biochemical characterization of the Cop9 signalosome CSN5/CSN6 heterodimer. PLoS ONE 2014, 9, e105688. [Google Scholar] [CrossRef]
- Gusmaroli, G.; Feng, S.; Deng, X.W. The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell 2004, 16, 2984–3001. [Google Scholar] [CrossRef]
- Enchev, R.I.; Schulman, B.A.; Peter, M. Protein neddylation: Beyond cullin–RING ligases. Nat. Rev. Mol. Cell Biol. 2014, 16, 30–44. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Skaar, J.R.; Pagan, J.K.; Pagano, M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Lyapina, S.; Cope, G.; Shevchenko, A.; Serino, G.; Tsuge, T.; Zhou, C.; Wolf, D.A.; Wei, N.; Deshaies, R.J. Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science 2001, 292, 1382–1385. [Google Scholar] [CrossRef]
- Enchev, R.I.; Scott, D.C.; da Fonseca, P.C.; Schreiber, A.; Monda, J.K.; Schulman, B.A.; Peter, M.; Morris, E.P. Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep. 2012, 2, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Niemand, E.; Naumann, M. The COP9 signalosome: A versatile regulatory hub of Cullin-RING ligases. Trends Biochem. Sci. 2022, 48, 82–95. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Shen, Y.; Feng, S.; Wang, X.; Chitteti, B.N.; Vierstra, R.D.; Deng, X.W. Evidence for a physical association of the COP9 signalosome, the proteasome, and specific SCF E3 ligases in vivo. Curr. Biol. 2003, 13, R504–R505. [Google Scholar] [CrossRef]
- Huang, X.; Hetfeld, B.K.J.; Seifert, U.; Kähne, T.; Kloetzel, P.; Naumann, M.; Bech-Otschir, D.; Dubiel, W. Consequences of COP9 signalosome and 26S proteasome interaction. FEBS J. 2005, 272, 3909–3917. [Google Scholar] [CrossRef]
- Arkinson, C.; Dong, K.C.; Gee, C.L.; Martin, A. Mechanisms and regulation of substrate degradation by the 26S proteasome. Nat. Rev. Mol. Cell Biol. 2024, 26, 104–122. [Google Scholar] [CrossRef]
- Peng, Z.; Serino, G.; Deng, X.W. Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell 2001, 13, 2393–2407. [Google Scholar] [CrossRef][Green Version]
- Malec, P.; Chamovitz, D.A. Characterization and Purification of Kinase Activities against Arabidopsis COP9 Signalosome Subunit 7. Isr. J. Chem. 2006, 46, 239–246. [Google Scholar] [CrossRef]
- Chamovitz, D.A. Revisiting the COP9 signalosome as a transcriptional regulator. EMBO Rep. 2009, 10, 352–358. [Google Scholar] [CrossRef]
- Menon, S.; Chi, H.; Zhang, H.; Deng, X.W.; Flavell, R.A.; Wei, N. COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor–induced entry into the cell cycle from quiescence. Nat. Immunol. 2007, 8, 1236–1245. [Google Scholar] [CrossRef]
- Tsai, M.-C.; Barati, M.T.; Kuppireddy, V.S.; Beckerson, W.C.; Long, G.; Perlin, M.H. Characterization of Microbotryum lychnidis-dioicae Secreted Effector Proteins, Their Potential Host Targets, and Localization in a Heterologous Host Plant. J. Fungi 2024, 10, 262. [Google Scholar] [CrossRef]
- Gao, H.; Guo, Y.; Ren, M.; Tang, L.; Gao, W.; Tian, S.; Shao, G.; Peng, Q.; Gu, B.; Miao, J.; et al. Phytophthora RxLR effector PcSnel4B promotes degradation of resistance protein AtRPS2. Plant Physiol. 2023, 193, 1547–1560. [Google Scholar] [CrossRef]
- He, L.; Chen, X.; Yang, J.; Zhang, T.; Li, J.; Zhang, S.; Zhong, K.; Zhang, H.; Chen, J.; Yang, J. Rice black-streaked dwarf virus-encoded P5-1 regulates the ubiquitination activity of SCF E3 ligases and inhibits jasmonate signaling to benefit its infection in rice. New Phytol. 2019, 225, 896–912. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Durán, R.; Rosas-Díaz, T.; Gusmaroli, G.; Luna, A.P.; Taconnat, L.; Deng, X.W.; Bejarano, E.R. Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 2011, 23, 1014–1032. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.-M.; Zhang, Z.-J.; Qiao, J.-H.; Gao, Q.; Zang, Y.; Xu, W.-Y.; Xie, L.; Fang, X.-D.; Ding, Z.-H.; Yang, Y.-Z.; et al. A rhabdovirus accessory protein inhibits jasmonic acid signaling in plants to attract insect vectors. Plant Physiol. 2022, 190, 1349–1364. [Google Scholar] [CrossRef]
- Mukhtar, M.S.; Carvunis, A.-R.; Dreze, M.; Epple, P.; Steinbrenner, J.; Moore, J.; Tasan, M.; Galli, M.; Hao, T.; Nishimura, M.T.; et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 2011, 333, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Guo, H.; Batool, W.; Lin, L.; Cao, J.; An, Q.; Aliyu, S.R.; Bao, J.; Wang, Z.; Norvienyeku, J. Translocon Subunits of the COP9 Signalosome Complex Are a Central Hub for Regulating Multiple Photoresponsive Processes and Autophagic Flux in Magnaporthe oryzae. J. Agric. Food Chem. 2024, 72, 22015–22034. [Google Scholar] [CrossRef]
- Chen, A.; Ren, Y.; Han, X.; Liu, C.; Zhou, Y.; Xu, C.; Qi, H.; Ma, Z.; Chen, Y. The COP9 signalosome complex regulates fungal development and virulence in the wheat scab fungus Fusarium graminearum. Front. Microbiol. 2023, 14, 1179676. [Google Scholar] [CrossRef]
- Meister, C.; Thieme, K.G.; Thieme, S.; Köhler, A.M.; Schmitt, K.; Valerius, O.; Braus, G.H. COP9 Signalosome Interaction with UspA/Usp15 Deubiquitinase Controls VeA-Mediated Fungal Multicellular Development. Biomolecules 2019, 9, 238. [Google Scholar] [CrossRef]
- Wang, D.; Musazade, E.; Wang, H.; Liu, J.; Zhang, C.; Liu, W.; Liu, Y.; Guo, L. Regulatory Mechanism of the Constitutive Photomorphogenesis 9 Signalosome Complex in Response to Abiotic Stress in Plants. J. Agric. Food Chem. 2022, 70, 2777–2788. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Penninckx, I.A.; Cammue, B.P.; Broekaert, W.F. The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 2001, 13, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Chamovitz, D.A. Role of Cop9 Signalosome Subunits in the Environmental and Hormonal Balance of Plant. Biomolecules 2019, 9, 224. [Google Scholar] [CrossRef]
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef]
- Gu, X.Y.; Liu, Y.; Liu, L.J. Progress on the biosynthesis and signal transduction of phytohormone salicylic acid. Yi Chuan 2020, 42, 858–869. [Google Scholar] [CrossRef]
- Bai, X.; Huang, X.; Tian, S.; Peng, H.; Zhan, G.; Goher, F.; Guo, J.; Kang, Z.; Guo, J. RNAi-mediated stable silencing of TaCSN5 confers broad-spectrum resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 2021, 22, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Fonseca, S.; Chico, J.M.; Solano, R. The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 2009, 12, 539–547. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Mayer, R.; Raventos, D.; Chua, N.-H. det1, cop1, and cop9 mutations cause inappropriate expression of several gene sets. Plant Cell 1996, 8, 1951. [Google Scholar] [CrossRef][Green Version]
- Hind, S.R.; Pulliam, S.E.; Veronese, P.; Shantharaj, D.; Nazir, A.; Jacobs, N.S.; Stratmann, J.W. The COP9 signalosome controls jasmonic acid synthesis and plant responses to herbivory and pathogens. Plant J. 2011, 65, 480–491. [Google Scholar] [CrossRef]
- Zou, J.-P.; Zhao, Q.-F.; Yang, T.; Shang, Y.-F.; Ahammed, G.J.; Zhou, J. The E3 ubiquitin ligase RING1 interacts with COP9 Signalosome Subunit 4 to positively regulate resistance to root-knot nematodes in Solanum lycopersicum L. Plant Sci. 2022, 322, 111344. [Google Scholar] [CrossRef]
- Antoni, R.; Rodriguez, L.; Gonzalez-Guzman, M.; Pizzio, G.A.; Rodriguez, P.L. News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling. Curr. Opin. Plant Biol. 2011, 14, 547–553. [Google Scholar] [CrossRef]
- Mohr, P.G.; Cahill, D.M. Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. tomato. Funct. Integr. Genom. 2006, 7, 181–191. [Google Scholar] [CrossRef]
- Xie, K.; Li, L.; Zhang, H.; Wang, R.; Tan, X.; He, Y.; Hong, G.; Li, J.; Ming, F.; Yao, X.; et al. Abscisic acid negatively modulates plant defence against rice black-streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant Cell Environ. 2018, 41, 2504–2514. [Google Scholar] [CrossRef]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Lee, J.-H.; Yoon, H.-J.; Terzaghi, W.; Martinez, C.; Dai, M.; Li, J.; Byun, M.-O.; Deng, X.W. DWA1 and DWA2, Two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 2010, 22, 1716–1732. [Google Scholar] [CrossRef]
- Martínez, C.; Iniesto, E.; García-León, M.; García-Corredera, D.; Fonseca, S.; Santiago, C.; Yang, M.; Yu, R.; Chen, H.; Altmann, E.; et al. Hormone-mediated disassembly and inactivation of a plant E3 ubiquitin ligase complex. Cell Rep. 2024, 43, 114802. [Google Scholar] [CrossRef]
- Zheng, Q.; Zhang, L.; Zhang, Q.; Pang, Z.; Sun, Y.; Yin, Z.; Lou, Z. Discovery of Interacting Proteins of ABA Receptor PYL5 via Covalent Chemical Capture. ACS Chem. Biol. 2019, 14, 2557–2563. [Google Scholar] [CrossRef]
- Jin, D.; Wu, M.; Li, B.; Bücker, B.; Keil, P.; Zhang, S.; Li, J.; Kang, D.; Liu, J.; Dong, J.; et al. The COP9 Signalosome regulates seed germination by facilitating protein degradation of RGL2 and ABI5. PLoS Genet. 2018, 14, e1007237. [Google Scholar] [CrossRef]
- Guan, R.; Su, J.; Meng, X.; Li, S.; Liu, Y.; Xu, J.; Zhang, S. Multilayered Regulation of Ethylene Induction Plays a Positive Role in Arabidopsis Resistance against Pseudomonas syringae. Plant Physiol. 2015, 169, 299–312. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, T.; Yin, K.-Q.; Chen, Z.; Gu, H.; Qu, L.-J.; Qin, G. Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol. 2012, 195, 450–460. [Google Scholar] [CrossRef]
- An, F.; Zhao, Q.; Ji, Y.; Li, W.; Jiang, Z.; Yu, X.; Zhang, C.; Han, Y.; He, W.; Liu, Y.; et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-Box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 2010, 22, 2384–2401. [Google Scholar] [CrossRef]
- Christians, M.J.; Robles, L.M.; Zeller, S.M.; Larsen, P.B. The eer5 mutation, which affects a novel proteasome-related subunit, indicates a prominent role for the COP9 signalosome in resetting the ethylene-signaling pathway in Arabidopsis. Plant J. 2008, 55, 467–477. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Wei, S.; Li, X.; Gruber, M.Y.; Feyissa, B.A.; Amyot, L.; Hannoufa, A. COP9 signalosome subunit 5A affects phenylpropanoid metabolism, trichome formation and transcription of key genes of a regulatory tri-protein complex in Arabidopsis. BMC Plant Biol. 2018, 18, 134. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef]
- Luo, D.; Xiong, C.; Lin, A.; Zhang, C.; Sun, W.; Zhang, J.; Yang, C.; Lu, Y.; Li, H.; Ye, Z.; et al. SlBBX20 interacts with the COP9 signalosome subunit SlCSN5-2 to regulate anthocyanin biosynthesis by activating SlDFR expression in tomato. Hortic. Res. 2021, 8, 163. [Google Scholar] [CrossRef]
- Mo, X.; Zhang, M.; Zhang, Z.; Lu, X.; Liang, C.; Tian, J. Phosphate (Pi) Starvation Up-Regulated GmCSN5A/B Participates in Anthocyanin Synthesis in Soybean (Glycine max) Dependent on Pi Availability. Int. J. Mol. Sci. 2021, 22, 12348. [Google Scholar] [CrossRef]
- Nawrot, R.; Barylski, J.; Nowicki, G.; Broniarczyk, J.; Buchwald, W.; Goździcka-Józefiak, A. Plant antimicrobial peptides. Folia Microbiol. 2013, 59, 181–196. [Google Scholar] [CrossRef]
- Damon, C.; Dmitrieva, J.; Muhovski, Y.; Francis, F.; Lins, L.; Ledoux, Q.; Luwaert, W.; Markó, I.E.; Mauro, S.; Ongena, M.; et al. Interaction network of antimicrobial peptides of Arabidopsis thaliana, based on high-throughput yeast two-hybrid screening. Plant Physiol. Biochem. 2012, 58, 245–252. [Google Scholar] [CrossRef]
- Wang, R.; He, F.; Ning, Y.; Wang, G.-L. Fine-Tuning of RBOH-Mediated ROS Signaling in Plant Immunity. Trends Plant Sci. 2020, 25, 1060–1062. [Google Scholar] [CrossRef]
- Gilroy, S.; Suzuki, N.; Miller, G.; Choi, W.-G.; Toyota, M.; Devireddy, A.R.; Mittler, R. A tidal wave of signals: Calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 2014, 19, 623–630. [Google Scholar] [CrossRef]
- Wang, C.; Liu, R.; Lim, G.-H.; de Lorenzo, L.; Yu, K.; Zhang, K.; Hunt, A.G.; Kachroo, A.; Kachroo, P. Pipecolic acid confers systemic immunity by regulating free radicals. Sci. Adv. 2018, 4, eaar4509. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, C.; Li, H.; Xu, Y.; Zhang, B.; Zheng, F.; Zhao, B.; Zhang, H.; Zhao, H.; Liu, B.; et al. OsJAB1 Positively Regulates Ascorbate Biosynthesis and Negatively Regulates Salt Tolerance Due to Inhibiting Early-Stage Salt-Induced ROS Accumulation in Rice. Plants 2023, 12, 3859. [Google Scholar] [CrossRef]
- Mach, J. COP9 Signalosome-regulated proteolysis: Turning off ascorbic acid synthesis when the lights go out. Plant Cell 2013, 25, 359. [Google Scholar] [CrossRef][Green Version]
- Li, S.; Wang, J.; Yu, Y.; Wang, F.; Dong, J.; Huang, R. D27E mutation of VTC1 impairs the interaction with CSN5B and enhances ascorbic acid biosynthesis and seedling growth in Arabidopsis. Plant Mol. Biol. 2016, 92, 473–482. [Google Scholar] [CrossRef]
- Li, Y.; Chu, Z.; Luo, J.; Zhou, Y.; Cai, Y.; Lu, Y.; Xia, J.; Kuang, H.; Ye, Z.; Ouyang, B. The C2H2 zinc-finger protein SlZF3 regulates AsA synthesis and salt tolerance by interacting with CSN5B. Plant Biotechnol. J. 2017, 16, 1201–1213. [Google Scholar] [CrossRef]
- Liu, Y.; Burch-Smith, T.; Schiff, M.; Feng, S.; Dinesh-Kumar, S.P. Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 2004, 279, 2101–2108. [Google Scholar] [CrossRef]
- Leister, R.T.; Dahlbeck, D.; Day, B.; Li, Y.; Chesnokova, O.; Staskawicz, B.J. Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 2005, 17, 1268–1278. [Google Scholar] [CrossRef]
- Slootweg, E.; Roosien, J.; Spiridon, L.N.; Petrescu, A.-J.; Tameling, W.; Joosten, M.; Pomp, R.; van Schaik, C.; Dees, R.; Borst, J.W.; et al. Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 2010, 22, 4195–4215. [Google Scholar] [CrossRef]
- Austin, M.J.; Muskett, P.; Kahn, K.; Feys, B.J.; Jones, J.D.G.; Parker, J.E. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 2002, 295, 2077–2080. [Google Scholar] [CrossRef]
- Fu, D.Q.; Ghabrial, S.; Kachroo, A. GmRAR1 and GmSGT1 are required for basal, R gene-mediated and systemic acquired resistance in soybean. Mol. Plant Microbe Interact. 2009, 22, 86–95. [Google Scholar] [CrossRef]
- Azevedo, C.; Sadanandom, A.; Kitagawa, K.; Freialdenhoven, A.; Shirasu, K.; Schulze-Lefert, P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 2002, 295, 2073–2076. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Serino, G.; Deng, X.-W.; Dinesh-Kumar, S.P. Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene–mediated resistance response to Tobacco mosaic virus. Plant Cell 2002, 14, 1483–1496. [Google Scholar] [CrossRef]
- Shirasu, K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 2009, 60, 139–164. [Google Scholar] [CrossRef]
- Liu, C.; Guo, L.-Q.; Menon, S.; Jin, D.; Pick, E.; Wang, X.; Deng, X.W.; Wei, N. COP9 signalosome subunit Csn8 is involved in maintaining proper duration of the G1 phase. J. Biol. Chem. 2013, 288, 20443–20452. [Google Scholar] [CrossRef]
- Huang, Y.; Gao, J.; Ji, G.; Li, W.; Wang, J.; Wang, Q.; Shen, Y.; Guo, J.; Gao, F. COP9 SIGNALOSOME SUBUNIT 5A facilitates POLYAMINE OXIDASE 5 degradation to regulate strawberry plant growth and fruit ripening. Plant Cell 2025, 37, koaf022. [Google Scholar] [CrossRef]
- Han, S.; Yue, W.; Bao, A.; Jiao, T.; Liu, Y.; Zeng, H.; Song, K.; Wu, M.; Guo, L. OsCSN2 orchestrates Oryza sativa L. growth and development through modulation of the GA and BR pathways. Funct. Integr. Genom. 2024, 24, 39. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Z.; Li, C.; Deng, K.; Han, C.; Shan, Z.; Chen, S.; Yang, H.; Yang, Y.; Chen, H.; Hao, Q. COP9 Signalosome’s Role in Plant Defense Mechanisms. Plants 2025, 14, 3017. https://doi.org/10.3390/plants14193017
Lu Z, Li C, Deng K, Han C, Shan Z, Chen S, Yang H, Yang Y, Chen H, Hao Q. COP9 Signalosome’s Role in Plant Defense Mechanisms. Plants. 2025; 14(19):3017. https://doi.org/10.3390/plants14193017
Chicago/Turabian StyleLu, Zihua, Chao Li, Kelin Deng, Cong Han, Zhihui Shan, Shuilian Chen, Hongli Yang, Yuanxiao Yang, Haifeng Chen, and Qingnan Hao. 2025. "COP9 Signalosome’s Role in Plant Defense Mechanisms" Plants 14, no. 19: 3017. https://doi.org/10.3390/plants14193017
APA StyleLu, Z., Li, C., Deng, K., Han, C., Shan, Z., Chen, S., Yang, H., Yang, Y., Chen, H., & Hao, Q. (2025). COP9 Signalosome’s Role in Plant Defense Mechanisms. Plants, 14(19), 3017. https://doi.org/10.3390/plants14193017





