Two New Species of Lophozia (Marchantiophyta) from the Sino-Himalaya and the Taxonomic Diversity of East Asian Lophozia
Abstract
1. Introduction
2. Results
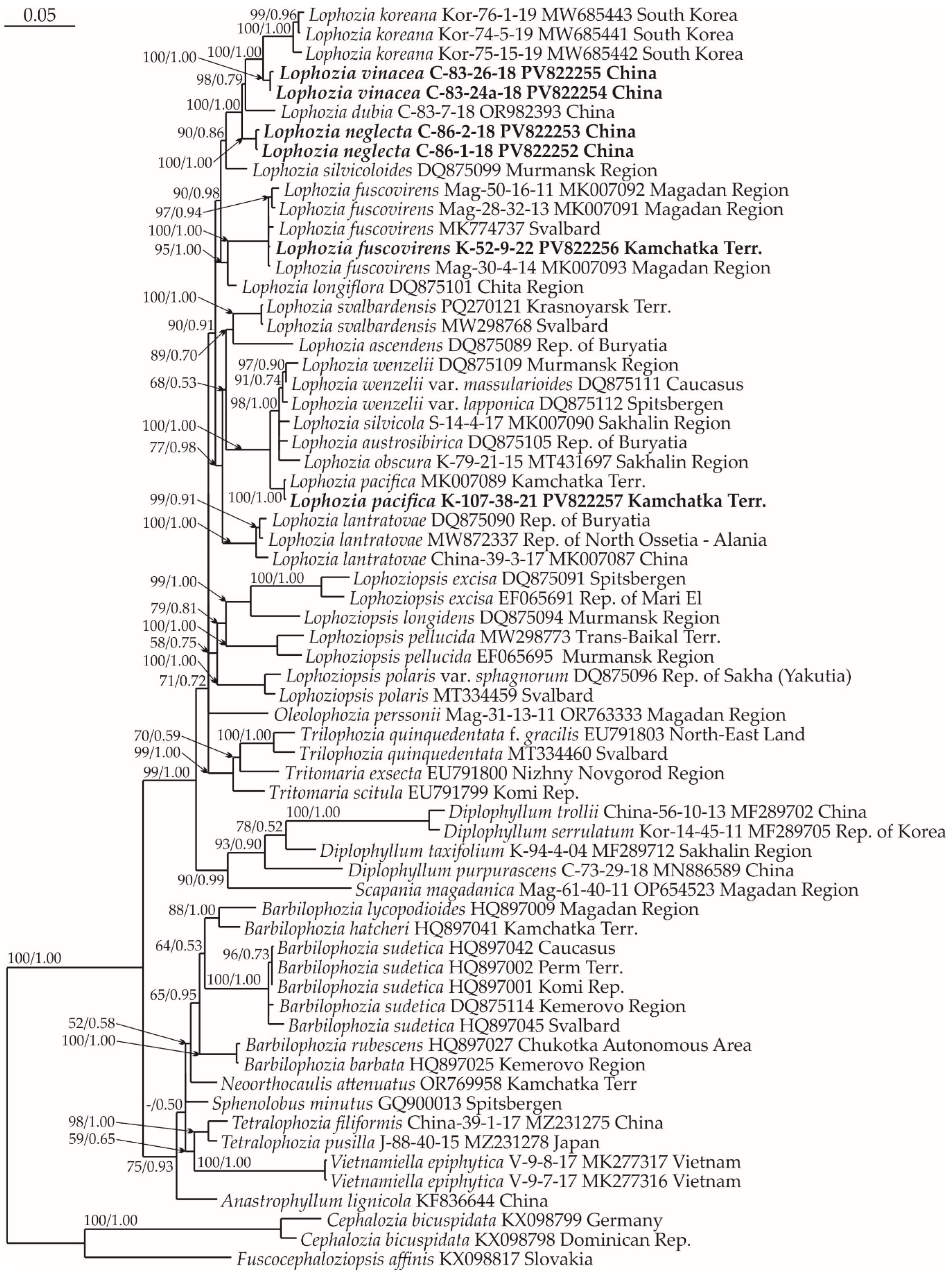
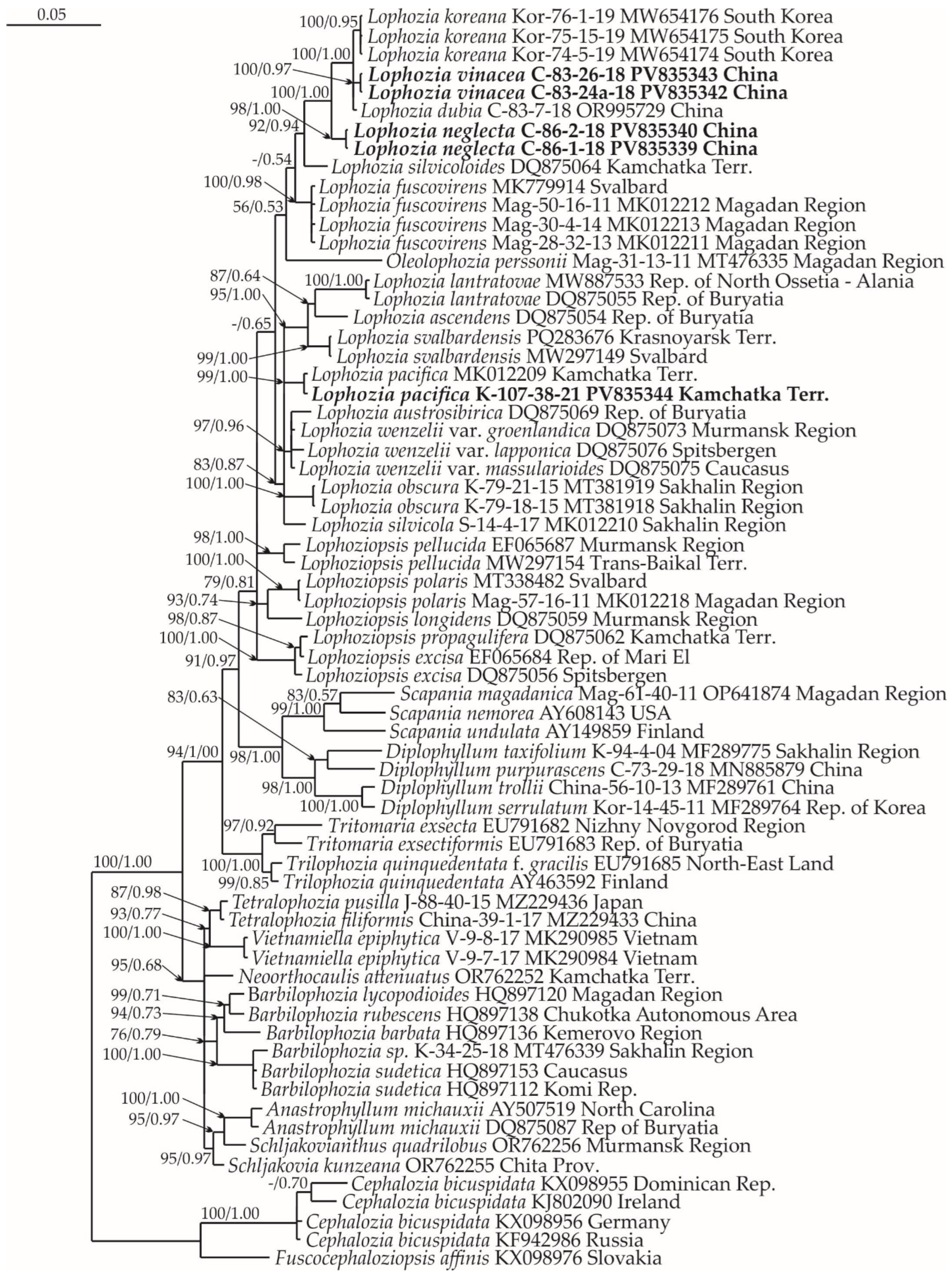
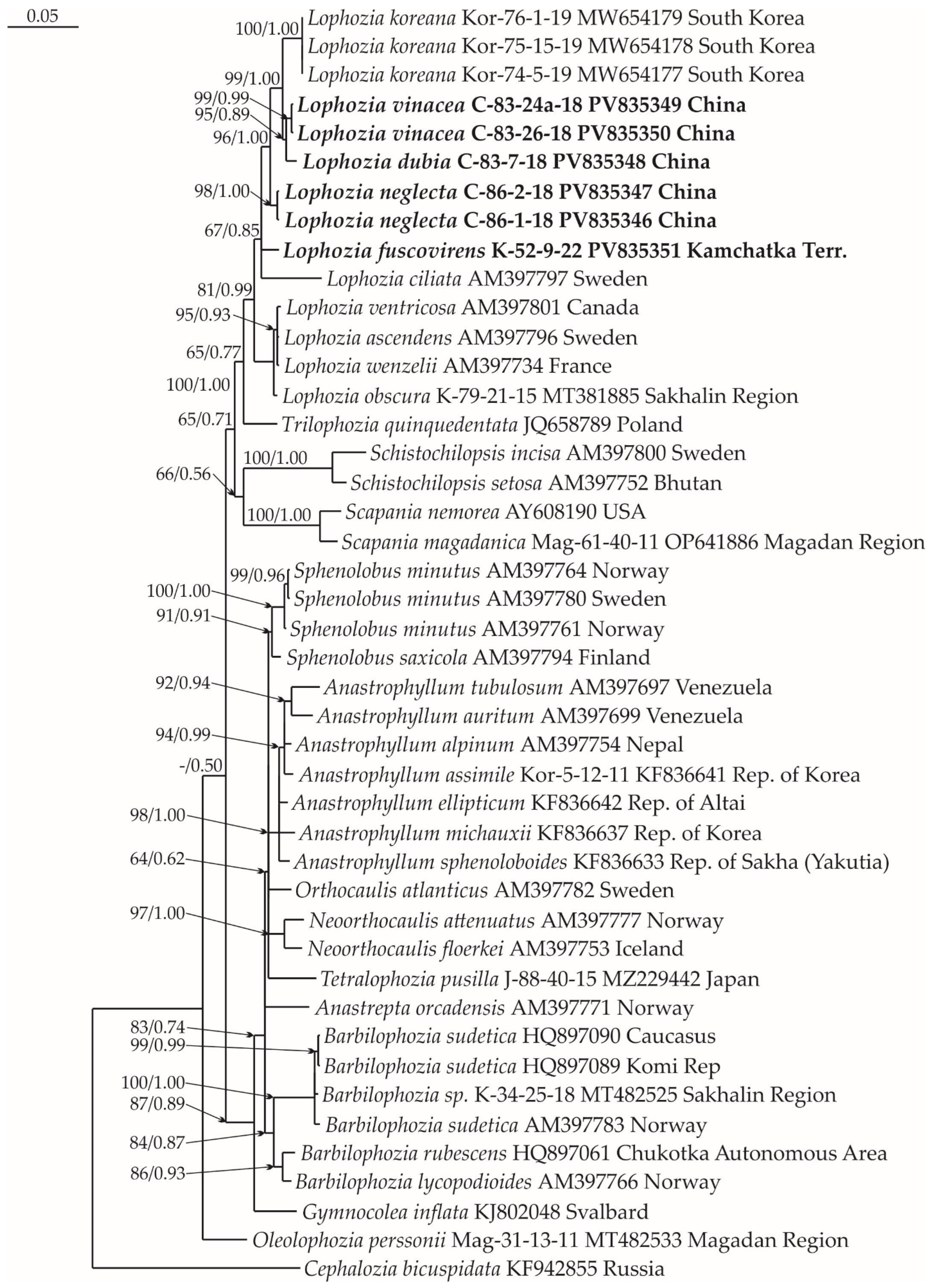
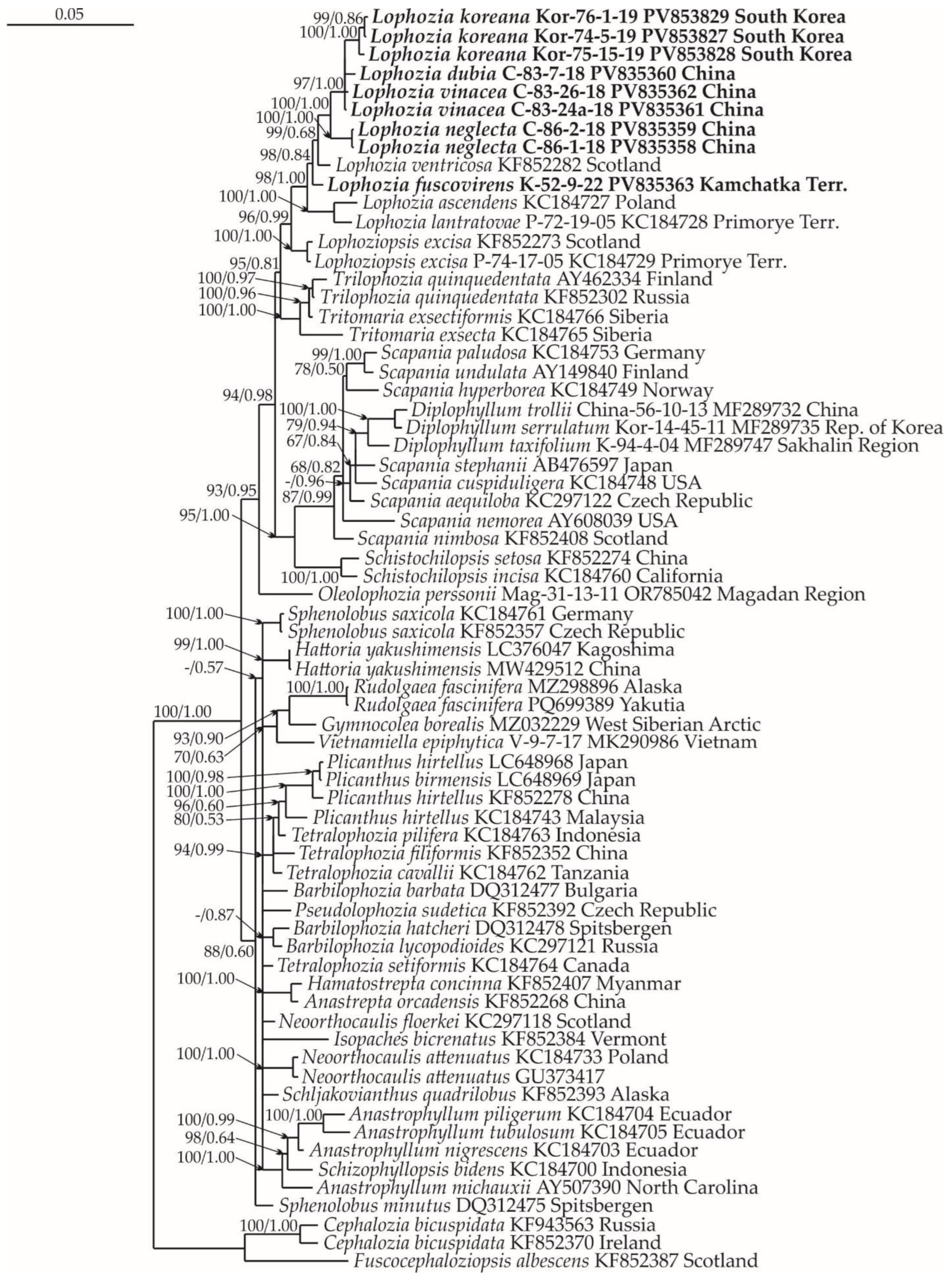
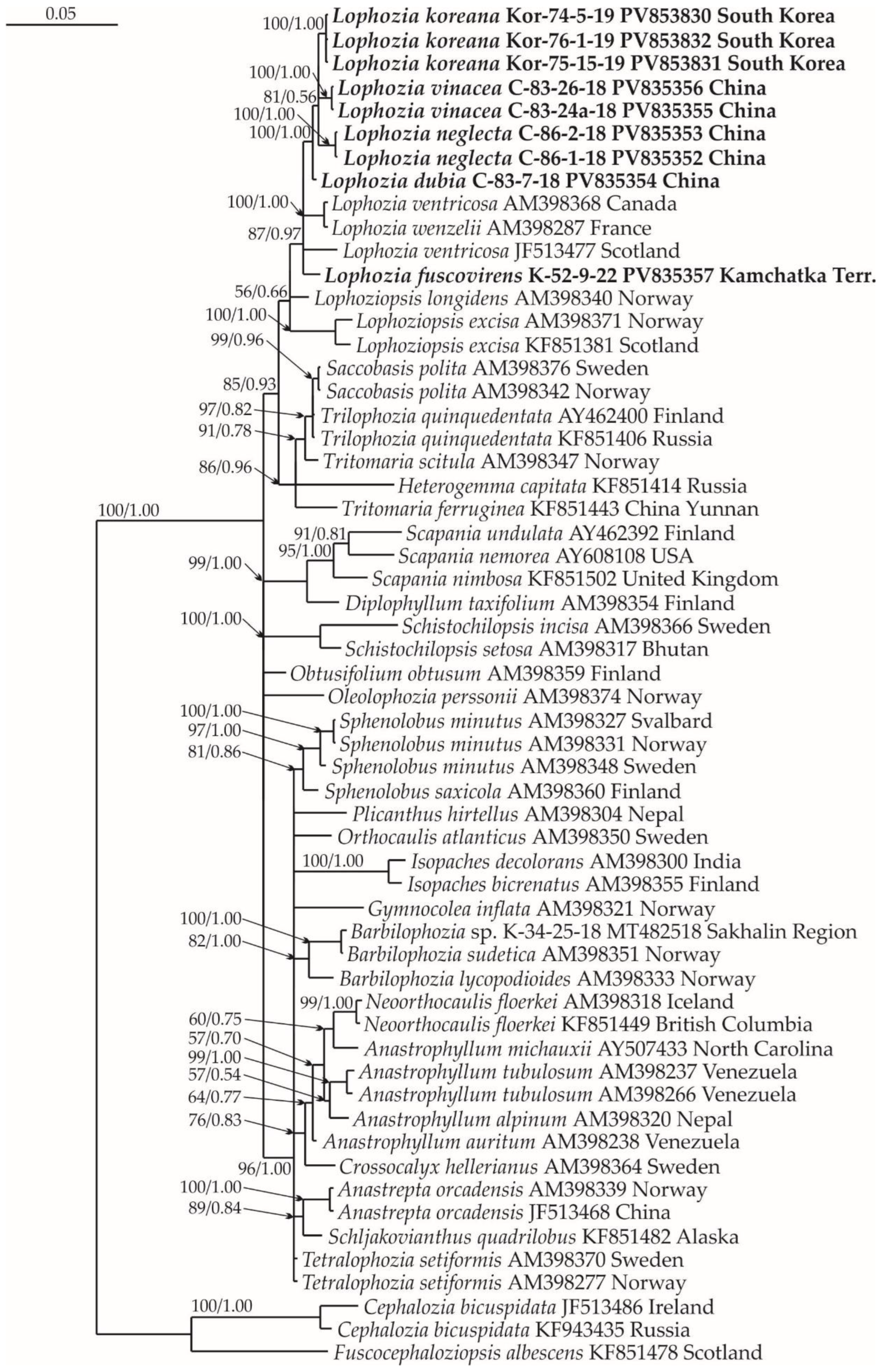
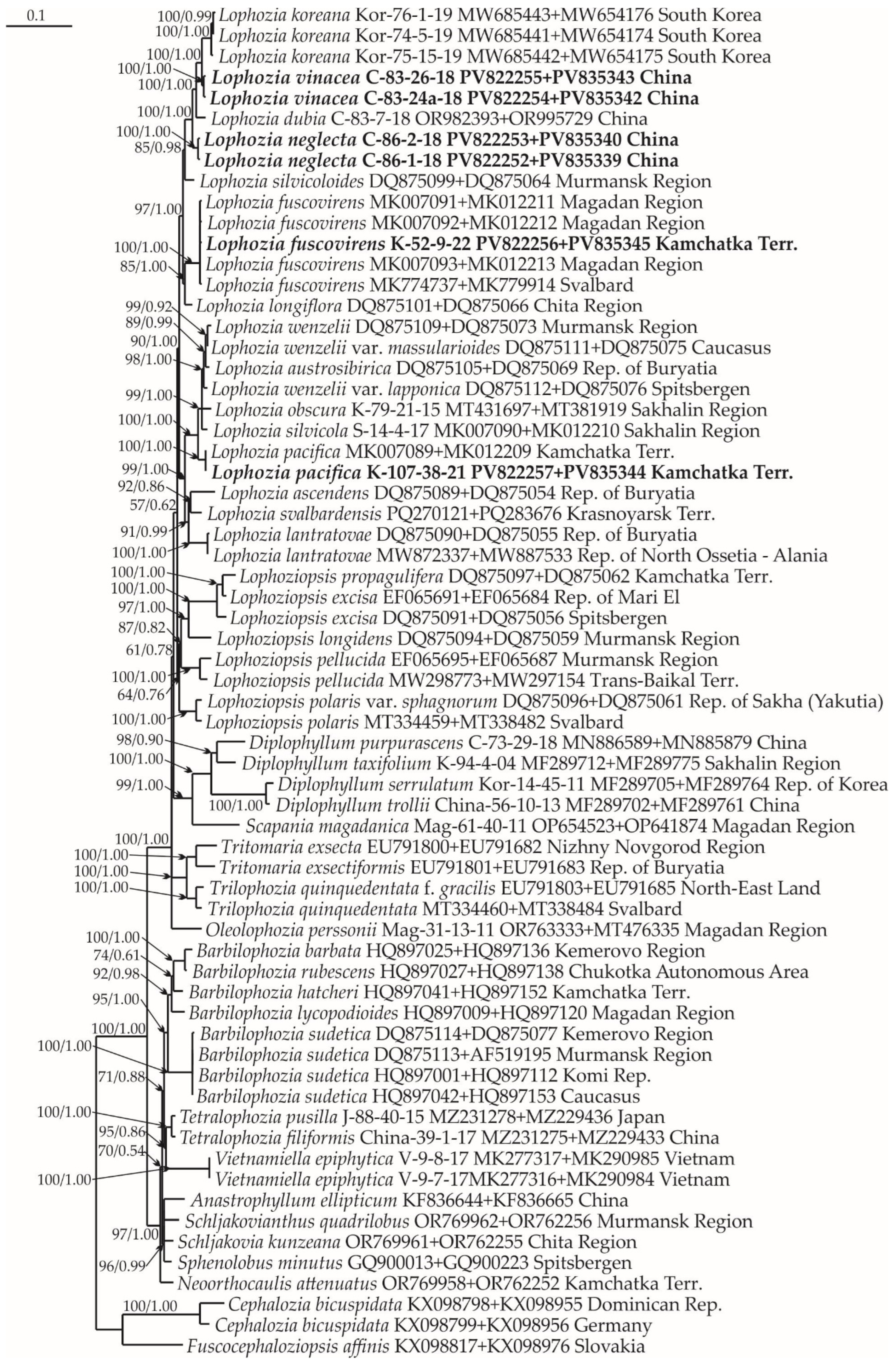
2.1. Molecular Phylogenetic Reconstruction
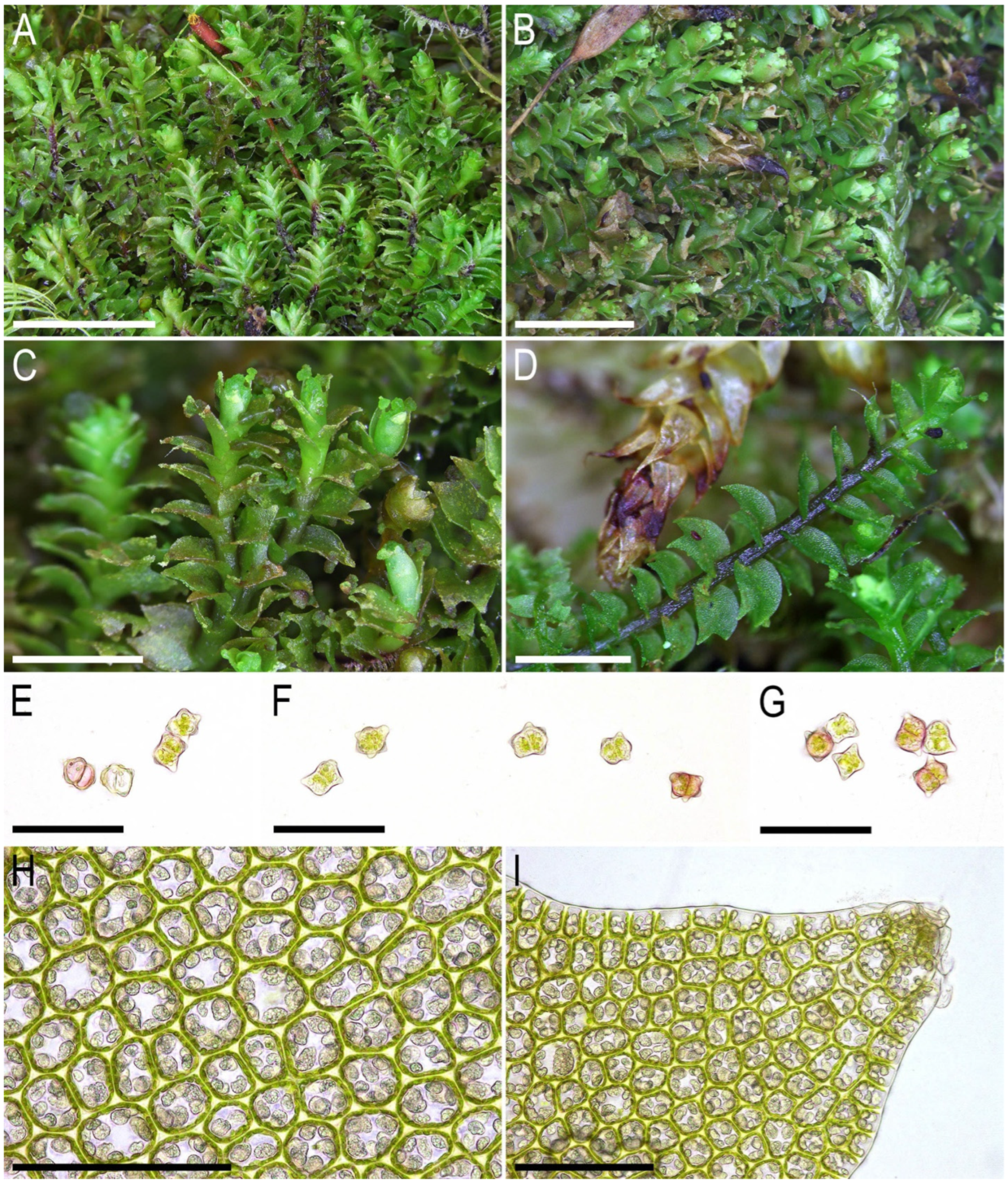
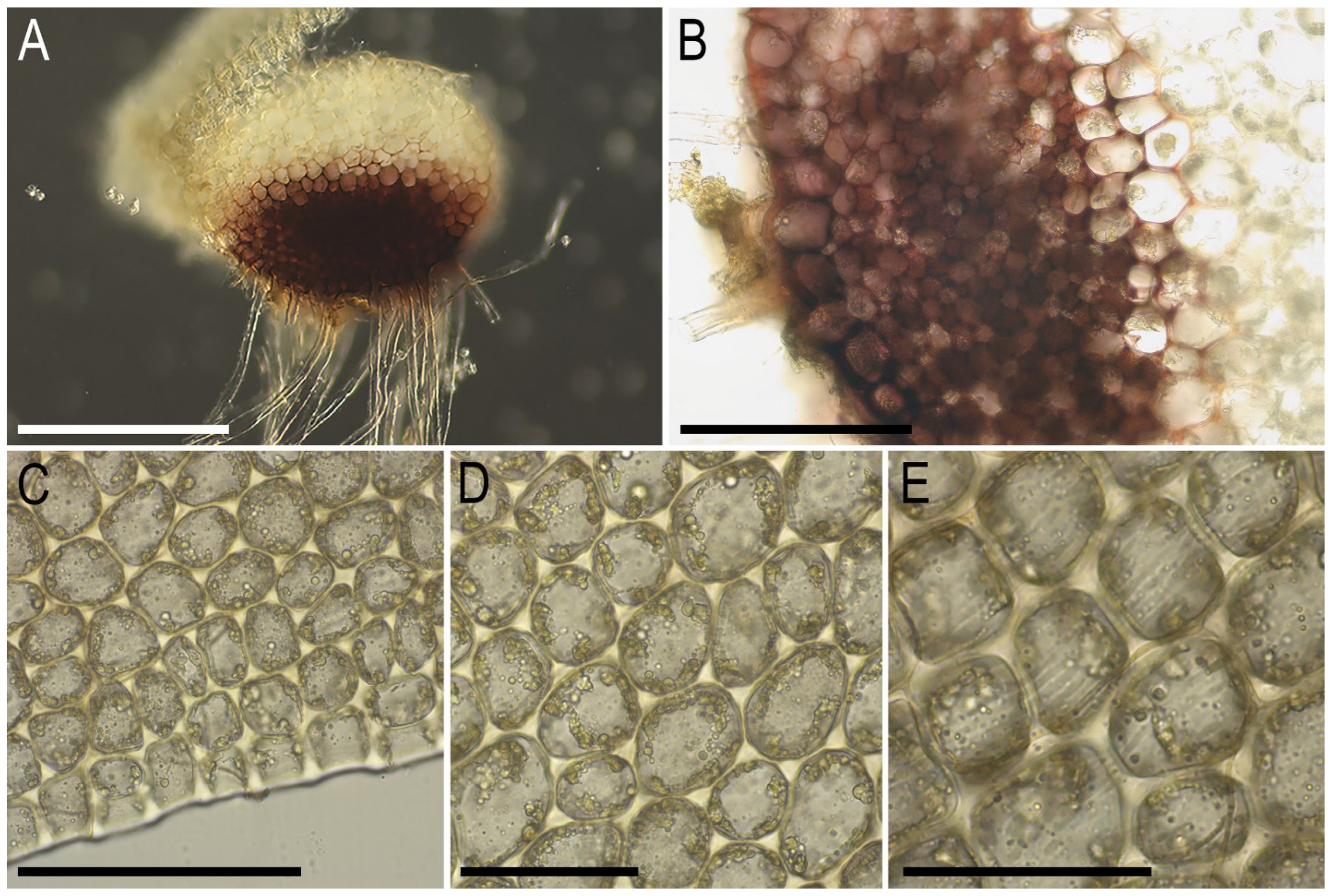
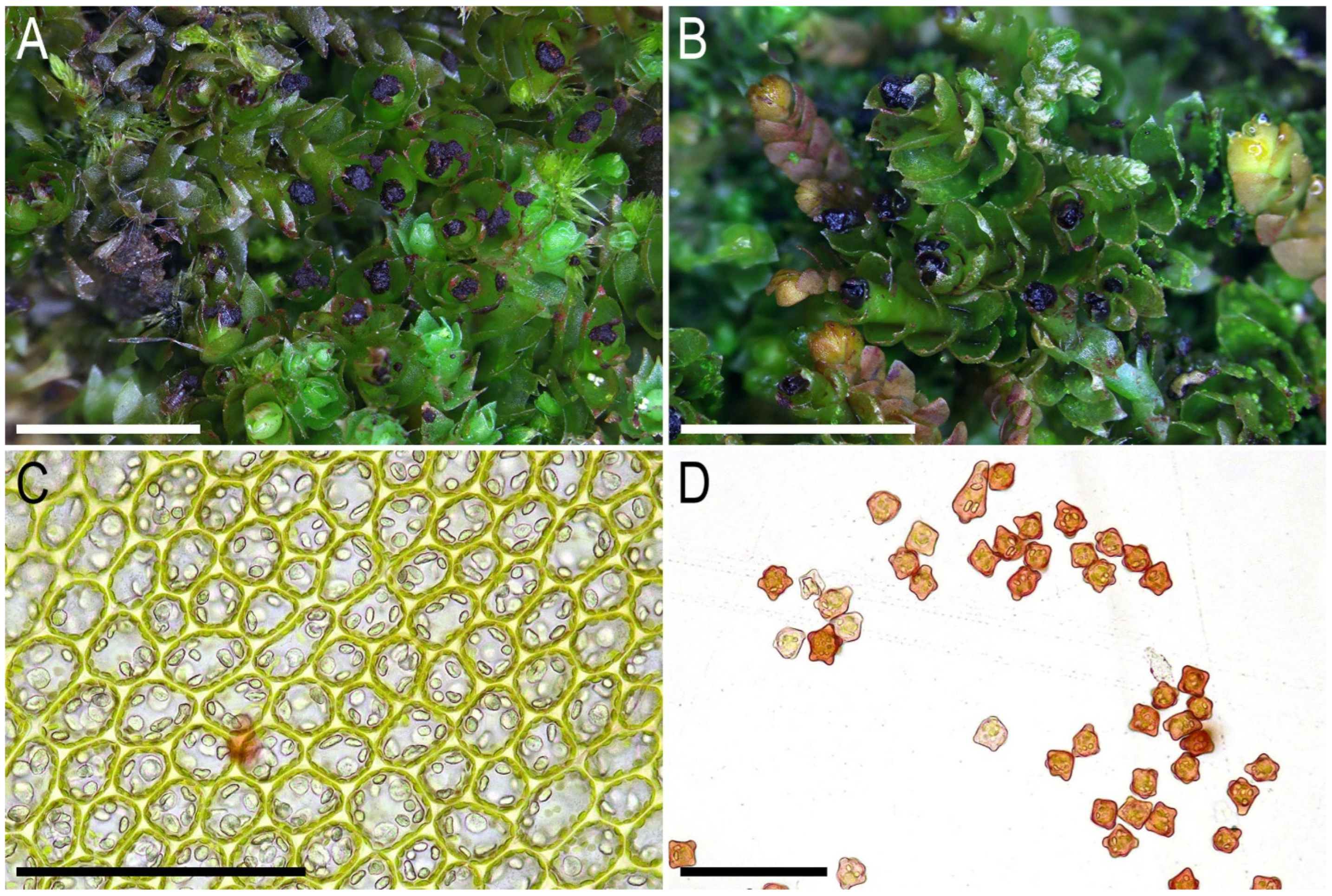
2.2. Taxonomy
- Leaves have almost equal lobes (versus, as a rule, clearly unequal);
- Leaf cells exhibit large trigones (versus small ones in L. koreana);
- A well-defined microcellous layer on the cross section of the stem is present (Figure 6B) (in L. koreana, this layer is distinguishable only owing to color, but the size of the cells is almost the same as that in other parts of the stem cross section).
- Plants are larger, exceeding 1.5 mm (versus 1.1–1.4 mm in width in L. koreana);
- Coarsely papillose oil bodies fill the cell lumen (versus finely papillose oil bodies that do not fill the lumen of the cell in L. koreana);
- The habitat is decaying wood (versus mineral soil in deep crevices for L. koreana);
- Moreover, the new species has presumable Sino–Himalayan distribution (versus the mountainous Korean–Japanese distribution in L. koreana).
- Dark vinaceous-purple gemmae, which are not present in the aforementioned Tritomaria;
- Unicellular gemmae (versus mostly bicellular, although in T. mexicana, they are also unicellular);
- More equal leaf lobes and wider leaves.
- Smaller cells varying within 15–20(–30) × 15–20(–25) μm (the distance from L. koreana in terms of cell size is less than that from L. neglecta);
- Vinaceous-purple gemmae (not present in other taxa);
- A stem in cross section with a distinct microcellous layer (a difference from L. koreana only);
- Unequally lobed leaves (a difference only from L. neglecta);
- Gemmae are located among the uppermost leaves, as is often the case in Tritomaria (but not in clusters at the tips of the lobes, as is often the case in Lophozia s. str., including L. neglecta and L. koreana). Among Lophozia s. str. taxa, this gemmae distribution is also characteristic of L. fuscovirens, where the gemmae are brown and the species’ geographic distribution is much more northern.
- The gemmae are two-celled and 10–15 × 13–18 μm in L. nepalensis versus one-celled in L. vinacea, where they are only 10–12 μm in diameter;
- Elongate shoots with gemmae are present in L. nepalensis but absent in L. vinacea;
- The cells in the midleaf of L. nepalensis are 20–26 × 22–28 μm, whereas those in L. vinacea are 15–20(–30) × 15–20(–25) μm.
2.3. Diversity of Lophozia in East Asia
3. Discussion
4. Materials and Methods
4.1. Specimen Collection
4.2. Specimen Analysis
4.2.1. Taxon Sampling
4.2.2. DNA Isolation, Amplification, and Sequencing
4.2.3. Phylogenetic Analyses
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| # | GenBank Name | Accepted Name | Specimen Voucher | ITS1-2 nrDNA | trnL-F cpDNA | trnG | rbcL | rps4 |
|---|---|---|---|---|---|---|---|---|
| 1 | Anastrepta orcadensis (Hook.) Schiffn. | Anastrepta orcadensis (Hook.) Schiffn. | China, Yunnan, Long D.G. 34711 (E) | KF852268 | JF513468 | |||
| 2 | Anastrepta orcadensis (Hook.) Schiffn. | Anastrepta orcadensis (Hook.) Schiffn. | Norway, L. Soederstroem, Soederstroem 2003/017 (BOL) | AM397771 | AM398339 | |||
| 3 | Anastrophyllum alpinum Steph. | Anastrophyllum alpinum Steph. | Nepal, Long 30460 (E) | AM397754 | AM398320 | |||
| 4 | Anastrophyllum assimile (Mitt.) Steph. | Anastrophyllum assimile (Mitt.) Steph. | South Korea, V.A. Bakalin, Kor-5-12-11 (VBGI) | KF836641 | ||||
| 5 | Anastrophyllum auritum (Lehm.) Steph. | Anastrophyllum auritum (Lehm.) Steph. | Venezuela, L. Soederstroem, Soederstroem 2004/065 (BOL) | AM397699 | AM398238 | |||
| 6 | Anastrophyllum bidens (Reinw., Blume et Nees) Steph. | Schizophyllopsis bidens (Reinw., Blume et Nees) Váňa et L. Söderstr. | Indonesia, Gradstein 12067 (GOET) | KC184700 | ||||
| 7 | Anastrophyllum ellipticum Inoue | Anastrophyllum ellipticum Inoue | Russia, Altai Territory, Yu. Mamontov, 330/2 (KPABG) | KF836642 | ||||
| 8 | Anastrophyllum lignicola D.B. Schill et D.G. Long | Anastrophyllum ellipticum Inoue | China, D. Long, 24067 (KPABG) | KF836644 | ||||
| 9 | Anastrophyllum michauxii (F. Weber) H. Buch | Anastrophyllum michauxii (F. Weber) H. Buch | USA, North Carolina, Clingman’s Dome, M. Sargent s n (ABSH) | AY507519 | AY507390 | AY507433 | ||
| 10 | Anastrophyllum michauxii (F. Weber) H. Buch | Anastrophyllum michauxii (F. Weber) H. Buch | South Korea, S.-S. Choi, Hepaticae Korea Exsiccatae F.II #54, 115520 (KPABG) | KF836637 | ||||
| 11 | Anastrophyllum minutum (Schreb.) R.M. Schust. | Sphenolobus minutus (Schreb.) Berggr. | Norway, Soederstroem 2004/271 (BOL) | AM397764 | AM398331 | |||
| 12 | Anastrophyllum minutum (Schreb.) R.M. Schust. | Sphenolobus minutus (Schreb.) Berggr. | Norway, Svalbard, L. Soederstroem, Soederstroem 2004/327 (BOL) | AM397761 | AM398327 | |||
| 13 | Anastrophyllum minutum (Schreb.) R.M. Schust. var. minutum | Sphenolobus minutus (Schreb.) Berggr. | Sweden, L. Soederstroem et P. Manyanga, Soederstroem 2003/054 (BOL) | AM397780 | AM398348 | |||
| 14 | Anastrophyllum nigrescens (Mitt.) Steph. | Anastrophyllum nigrescens (Mitt.) Steph. | Ecuador, Schaefer-Verwimp et al. 24444 (GOET) | KC184703 | ||||
| 15 | Anastrophyllum piligerum (Nees) Steph. | Anastrophyllum piligerum (Nees) Steph. | Ecuador, Schaefer-Verwimp et al. 24271 (GOET) | KC184704 | ||||
| 16 | Anastrophyllum saxicola (Schrad.) R.M. Schust. | Sphenolobus saxicola (Schrad.) Steph. | Finland, L. Soederstroem & P. Manyanga, Soederstroem 2003/099 (BOL) | AM397794 | AM398360 | |||
| 17 | Anastrophyllum sphenoloboides R.M. Schust. | Schizophyllopsis sphenoloboides (R.M. Schust.) Váňa et L. Söderstr. | Russia, Yakutiya Rep., V.A. Bakalin, 101592 (KPABG) | KF836633 | ||||
| 18 | Anastrophyllum tubulosum (Nees) Grolle | Anastrophyllum tubulosum (Nees) Grolle | Ecuador, Schaefer-Verwimp et al. 24464 (GOET) | KC184705 | ||||
| 19 | Anastrophyllum tubulosum (Nees) Grolle | Anastrophyllum tubulosum (Nees) Grolle | Venezuela, L. Soederstroem, Soederstroem 2004/030 (BOL) | AM397697 | AM398237 | |||
| 20 | Anastrophyllum tubulosum (Nees) Grolle | Anastrophyllum tubulosum (Nees) Grolle | Venezuela, L. Soederstroem, Soederstroem 2004/120 (BOL) | AM398266 | ||||
| 21 | Barbilophozia attenuata (Mart.) Loeske | Neoorthocaulis attenuatus (Mart.) L. Söderstr., De Roo et Hedd. | Norway, L. Soederstroem, Soederstroem 2003/020 (BOL) | AM397777 | ||||
| 22 | Barbilophozia attenuata (Mart.) Loeske | Neoorthocaulis attenuatus (Mart.) L. Söderstr., De Roo et Hedd. | 4230531 (H) | GU373417 | ||||
| 23 | Barbilophozia atlantica (Kaal.) Müll. Frib. | Orthocaulis atlanticus (Kaal.) H. Buch | Sweden, L. Soederstroem & P. Manyanga, Soederstroem 2003/057 (BOL) | AM398350 | ||||
| 24 | Barbilophozia barbata (Schmidel ex Schreb.) Loeske | Barbilophozia barbata (Schmidel ex Schreb.) Loeske | Russia, Kemerovskaya Prov., N.A. Konstantinova, 122-1-00 (KPABG) | HQ897025 | HQ897136 | |||
| 25 | Barbilophozia barbata (Schmidel ex Schreb.) Loeske | Barbilophozia barbata (Schmidel ex Schreb.) Loeske | Bulgaria, Hentschel Bryo 0753 (GOET) | DQ312477 | ||||
| 26 | Barbilophozia floerkei (F. Weber et D. Mohr) Loeske | Neoorthocaulis floerkei (F. Weber et D. Mohr) L. Söderstr., De Roo et Hedd. | Iceland, A. Seneca & L. Soederstroem, Soederstroem 2004/457 (BOL) | AM397753 | AM398318 | |||
| 27 | Barbilophozia hatcheri (A. Evans) Loeske | Barbilophozia hatcheri (A. Evans) Loeske | Russia, Commander Island, V.A. Bakalin, K-1-2-02-VB (KPABG) | HQ897041 | ||||
| 28 | Barbilophozia hatcheri (A. Evans) Loeske | Barbilophozia hatcheri (A. Evans) Loeske | Norway, Spitsbergen, Hentschel Bryo 0492 | DQ312478 | ||||
| 29 | Barbilophozia lycopodioides (Wallr.) Loeske | Barbilophozia lycopodioides (Wallr.) Loeske | Russia, Magadanskaya Prov., O. Mochalova, 106872 (KPABG) | HQ897009 | HQ897120 | |||
| 30 | Barbilophozia lycopodioides (Wallr.) Loeske | Barbilophozia lycopodioides (Wallr.) Loeske | Russia, N.A. Konstantinova 20 September 1994 (F) | KC297121 | ||||
| 31 | Barbilophozia lycopodioides (Wallr.) Loeske | Barbilophozia lycopodioides (Wallr.) Loeske | Norway, L. Soederstroem, Soederstroem 2003/019 (BOL) | AM397766 | AM398333 | |||
| 32 | Barbilophozia rubescens (R.M. Schust. et Damsh.) Kartt. et L. Söderstr. | Barbilophozia rubescens (R.M. Schust. et Damsh.) Kartt. et L. Söderstr. | Russia, Chukotka A.O., E. Kuzmina, 100171 (KPABG) | HQ897027 | HQ897138 | HQ897061 | ||
| 33 | Barbilophozia sudetica (Nees ex Huebener) L. Söderstr., De Roo et Hedd. | Pseudolophozia sudetica (Nees ex Huebener) Konstant. et Vilnet | Czech Republic, B. Shaw 12991 (DUKE) | KF852392 | ||||
| 34 | Cephalozia bicuspidata (L.) Dumort. | Cephalozia bicuspidata (L.) Dumort. | Dominican Republic, Province San Jose de Ocoa, Schaefer-Verwimp et Verwimp 26879/A (M) | KX098798 | KX098955 | |||
| 35 | Cephalozia bicuspidata (L.) Dumort. | Cephalozia bicuspidata (L.) Dumort. | Germany, Baden-Wuerttemberg, Schaefer-Verwimp et Verwimp 27426 (M) | KX098799 | KX098956 | |||
| 36 | Cephalozia bicuspidata (L.) Dumort. | Cephalozia bicuspidata (L.) Dumort. | Russia, N.A. Konstantinova (F) | KF942986 | KF942855 | KF943563 | KF943435 | |
| 37 | Cephalozia bicuspidata (L.) Dumort. | Cephalozia bicuspidata (L.) Dumort. | Ireland, Stotler et C.-Stotler s.n. (ABSH) | KJ802090 | KF852370 | JF513486 | ||
| 38 | Anastrophyllum hellerianum (Nees ex Lindenb.) R.M. Schust. | Crossocalyx hellerianus (Nees ex Lindenb.) Meyl. | Sweden, Soederstroem 2003/081 (BOL) | AM398364 | ||||
| 39 | Diplophyllum purpurascens Bakalin et Vilnet | Diplophyllum purpurascens Bakalin et Vilnet | China, Yunnan Province, Dali Prefecture, V.A. Bakalin & W.Z. Ma, C-73-29-18 | MN886589 | MN885879 | |||
| 40 | Diplophyllum serrulatum (Müll. Frib.) Steph. | Diplophyllum serrulatum (Müll. Frib.) Steph. | Republic of Korea, Jeonnam Province, V.A. Bakalin, Kor-14-45-11 | MF289705 | MF289764 | MF289735 | ||
| 41 | Diplophyllum taxifolium (Wahlenb.) Dumort. | Diplophyllum taxifolium (Wahlenb.) Dumort. | Russia, Sakhalin Prov., Kuril Islands, Paramushir Isl., V.A. Bakalin, K-94-4-04 | MF289712 | MF289775 | MF289747 | ||
| 42 | Diplophyllum taxifolium (Wahlenb.) Dumort. | Diplophyllum taxifolium (Wahlenb.) Dumort. | Finland, Soederstroem 2003/098 (BOL) | AM398354 | ||||
| 43 | Diplophyllum trollii Grolle | Diplophyllum trollii Grolle | China, Guizhou Prov., V.A. Bakalin, China-56-10-13 (VBGI) | MF289702 | MF289761 | MF289732 | ||
| 44 | Fuscocephaloziopsis affinis (Lindb. ex Steph.) Váňa et L. Söderstr. | Fuscocephaloziopsis affinis (Lindb. ex Steph.) Váňa et L. Söderstr. | Slovakia, Banskobystricky Kraj, Vana 6 (Vana) | KX098817 | KX098976 | |||
| 45 | Fuscocephaloziopsis albescens (Hook.) Váňa et L. Söderstr. | Fuscocephaloziopsis albescens (Hook.) Váňa et L. Söderstr. | United Kingdom, Banff, Scotland, D.G. Long & G.P. Rothero 36976 (E) | KF852387 | KF851478 | |||
| 46 | Gymnocolea borealis (Frisvoll et Moen) R.M. Schust. | Rudolgaea borealis (Frisvoll et Moen) Potemkin et Vilnet | Russia, West Siberian Arctic, Gydansky Peninsula, vicinities of Yambuto Lake, E.I. Troeva, G1-138 (LE) | MZ032229 | ||||
| 47 | Rudolgaea fascinifera (Potemkin) Potemkin et Vilnet | Rudolgaea fascinifera (Potemkin) Potemkin et Vilnet | Russia, Republic of Sakha, Yakutia, Kytalyk National Park, E.D. Lapshina, 019E-6-23 126373 (KPABG) | PQ699389 | ||||
| 48 | Gymnocolea fascinifera Potemkin | Rudolgaea fascinifera (Potemkin) Potemkin et Vilnet | USA, Alaska, A.D. Potemkin, 92-9701 (LE) | MZ298896 | ||||
| 49 | Gymnocolea inflata (Huds.) Dumort. | Gymnocolea inflata (Huds.) Dumort. | Svalbard, Spitsbergen, N.A. Konstantinova & A.N. Savchenko, 118-1-04 (F) | KJ802048 | ||||
| 50 | Gymnocolea inflata (Huds.) Dumort. | Gymnocolea inflata (Huds.) Dumort. | Norway, A. Seneca & L. Soederstroem, Soederstroem 2004/223 (BOL) | AM398321 | ||||
| 51 | Hamatostrepta concinna Váňa et D.G. Long | Hamatostrepta concinna Váňa et D.G. Long | Myanmar, Long, D. 34854 (DUKE) | KF852407 | ||||
| 52 | Hattoria yakushimensis (Horik.) R.M. Schust. | Hattoria yakushimensis (Horik.) R.M. Schust. | Japan, Kagoshima, Yakushima Island, T. Katagiri 4281 (NICH) | LC376047 | ||||
| 53 | Hattoria yakushimensis (Horik.) R.M. Schust. | Hattoria yakushimensis (Horik.) R.M. Schust. | China, Xia Tang et Rui-Ping Shi, July 2017, 20170706-94, COMPLETE CHLOROPLAST GENOME | MW429512 | ||||
| 54 | Heterogemma capitata (Hook.) Konstant. et Vilnet | Heterogemma capitata (Hook.) Konstant. et Vilnet | Russia, N. Konstantinova 121-03 (F) | KF851414 | ||||
| 55 | Isopaches bicrenatus (Schmidel ex Hoffm.) H. Buch | Isopaches bicrenatus (Schmidel ex Hoffm.) H. Buch | USA, Vermont, B. Shaw 6970 (DUKE) | KF852384 | ||||
| 56 | Isopaches decolorans (Limpr.) H. Buch | Isopaches decolorans (Limpr.) H. Buch | India, D. Long, 1995, Long 22566 (E) | AM398300 | ||||
| 57 | Lophozia bicrenata (Schmidel ex Hoffm.) Dumort. | Isopaches bicrenatus (Schmidel ex Hoffm.) H. Buch | Finland, L. Soederstroem & P. Manyanga, Soederstroem 2003/100 (BOL) | AM398355 | ||||
| 58 | Lophozia ascendens (Warnst.) R.M. Schust. | Lophozia ascendens (Warnst.) R.M. Schust. | Russia, Buryatiya, N. Konstantinova, 109-3-01 (KPABG) | DQ875089 | DQ875054 | |||
| 59 | Lophozia ascendens (Warnst.) R.M. Schust. | Lophozia ascendens (Warnst.) R.M. Schust. | Sweden, L. Soederstroem & P. Manyanga, Soederstroem 2003/077 (BOL) | AM397796 | ||||
| 60 | Lophozia ascendens (Warnst.) R.M. Schust. | Lophozia ascendens (Warnst.) R.M. Schust. | Poland, Klama et al. 171 (GOET) | KC184727 | ||||
| 61 | Lophozia austrosibirica Bakalin | Lophozia austrosibirica Bakalin | Russia, Buryatiya, V. Bakalin, 15-9-99 (KPABG) | DQ875105 | DQ875069 | |||
| 62 | Lophozia ciliata Damsh., L. Söderstr. et H. Weibull | Lophozia ciliata Damsh., L. Söderstr. et H. Weibull | Sweden, L. Soederstroem & P. Manyanga, Soederstroem 2003/084 (BOL) | AM397797 | ||||
| 63 | Lophozia excisa (Dicks.) Dumort. | Lophoziopsis excisa (Dicks.) Konstant. et Vilnet | Svalbard, Spitsbergen, N.A. Konstantinova, 104-1-04 (KPABG) | DQ875091 | DQ875056 | |||
| 64 | Lophozia excisa (Dicks.) Dumort. | Lophoziopsis excisa (Dicks.) Konstant. et Vilnet | Russia, Maryi-El, N.A. Konstantinova, K437-2-04 (KPABG) | EF065691 | EF065684 | |||
| 65 | Lophozia excisa (Dicks.) Dumort. | Lophoziopsis excisa (Dicks.) Konstant. et Vilnet | Norway, L. Soederstroem & P. Manyanga, Soederstroem 2003/030 (BOL) | AM398371 | ||||
| 66 | Lophozia fuscovirens Bakalin et Vilnet | Lophozia fuscovirens Bakalin et Vilnet | Russia, Russian Far East, Kamchatka Territory, V.A. Bakalin, K-52-9-22 | PV822256 | PV835345 | PV835351 | PV835363 | PV835357 |
| 67 | Lophozia fuscovirens Bakalin et Vilnet | Lophozia fuscovirens Bakalin et Vilnet | Norway, Svalbard, A. Savchenko, CA16-12-1c (KPABG) | MK774737 | MK779914 | |||
| 68 | Lophozia fuscovirens Bakalin et Vilnet | Lophozia fuscovirens Bakalin et Vilnet | Russia, Magadan Prov., V.A. Bakalin, Mag-28-32-13 (KPABG) | MK007091 | MK012211 | |||
| 69 | Lophozia fuscovirens Bakalin et Vilnet | Lophozia fuscovirens Bakalin et Vilnet | Russia, Magadan Prov., V.A. Bakalin, Mag-30-4-14 (KPABG) | MK007093 | MK012213 | |||
| 70 | Lophozia fuscovirens Bakalin et Vilnet | Lophozia fuscovirens Bakalin et Vilnet | Russia, Magadan Prov., V.A. Bakalin, Mag-50-16-11 (KPABG) | MK007092 | MK012212 | |||
| 71 | Lophozia incisa (Schrad.) Dumort. | Schistochilopsis incisa (Schrad.) Konstant. | Sweden, L. Soederstroem & P. Manyanga, Soederstroem 2003/079 (BOL) | AM397800 | AM398366 | |||
| 72 | Lophozia koreana (Bakalin, S.S. Choi et B.Y. Sun) Maltseva, Vilnet et Bakalin | Lophozia koreana (Bakalin, S.S. Choi et B.Y. Sun) Maltseva, Vilnet et Bakalin | Republic of Korea, South Korea, Jeollabuk-do, V.A. Bakalin & S.S. Choi, Kor-74-5-19 | MW685441 | MW654174 | MW654177 | PV853827 | PV853830 |
| 73 | Lophozia koreana (Bakalin, S.S. Choi et B.Y. Sun) Maltseva, Vilnet et Bakalin | Lophozia koreana (Bakalin, S.S. Choi et B.Y. Sun) Maltseva, Vilnet et Bakalin | Republic of Korea, South Korea, Jeollabuk-do, V.A. Bakalin & S.S. Choi, Kor-75-15-19 | MW685442 | MW654175 | MW654178 | PV853828 | PV853831 |
| 74 | Lophozia koreana (Bakalin, S.S. Choi et B.Y. Sun) Maltseva, Vilnet et Bakalin | Lophozia koreana (Bakalin, S.S. Choi et B.Y. Sun) Maltseva, Vilnet et Bakalin | Republic of Korea, South Korea, Jeollabuk-do, V.A. Bakalin & S.S. Choi, Kor-76-1-19 | MW685443 | MW654176 | MW654179 | PV853829 | PV853832 |
| 75 | Lophozia lantratovae Bakalin | Lophozia lantratovae Bakalin | Russia, Buryatiya, V.A. Bakalin, 76-7-01 (KPABG) | DQ875090 | DQ875055 | |||
| 76 | Lophozia lantratovae Bakalin | Lophozia lantratovae Bakalin | China, Sichuan Prov., V.A. Bakalin & K.G. Klimova, China-39-3-17 (VBGI) | MK007087 | ||||
| 77 | Lophozia lantratovae Bakalin | Lophozia lantratovae Bakalin | Russia, North Ossetia, Alanya Rep., A.V. Rumyantseva, 123153 (KPABG) | MW872337 | MW887533 | |||
| 78 | Lophozia lantratovae Bakalin | Lophozia lantratovae Bakalin | Russia, Russian Far East, V.A. Bakalin, P-72-19-05 | KC184728 | ||||
| 79 | Lophozia longidens (Lindb.) Macoun | Lophoziopsis longidens (Lindb.) Konstant. et Vilnet | Norway, L. Soederstroem, Soederstroem 2003/016 (BOL) | AM398340 | ||||
| 80 | Lophozia obscura (Bakalin) A.V. Troitsky, Bakalin et Fedosov | Lophozia obscura (Bakalin) A.V. Troitsky, Bakalin et Fedosov | Russia, Far East, Sakhalin Province, V.A. Bakalin, K-79-21-15 Sc21/0 (VBGI) | MT431697 | MT381919 | MT381885 | ||
| 81 | Lophozia obtusa (Lindb.) A. Evans | Obtusifolium obtusum (Lindb.) S.W. Arnell | Finland, Soederstroem 2003/094 (BOL) | AM398359 | ||||
| 82 | Lophozia pacifica Bakalin | Lophozia pacifica Bakalin | Russia, Russian Far East, Kamchatka Territory, V.A. Bakalin & K.G. Klimova, K-107-38-21 | PV822257 | PV835344 | |||
| 83 | Lophozia pacifica Bakalin | Lophozia pacifica Bakalin | Russia, Kamchatka Terr., V.A. Bakalin, K-16-2-02-VB (KPABG) | MK007089 | MK012209 | |||
| 84 | Lophozia dubia Schiffn. | Lophozia dubia Schiffn. | China, Yunnan Province, Diqing Prefecture, V.A. Bakalin & W.Z. Ma, C-83-7-18 | OR982393 | OR995729 | PV835348 | PV835360 | PV835354 |
| 85 | Lophozia pellucida R.M. Schust. | Lophoziopsis pellucida (R.M. Schust.) Konstant. et Vilnet | Russia, Murmanskaya Prov., N.A. Konstantinova, 39-2a-03 (KPABG) | EF065695 | EF065687 | |||
| 86 | Lophozia perssonii H. Buch et S.W. Arnell | Oleolophozia perssonii (H. Buch et S.W. Arnell) L. Söderstr., De Roo et Hedd. | Norway, Soederstroem 2003/036 (BOL) | AM398374 | ||||
| 87 | Lophozia propagulifera (Gottsche) Steph. | Lophoziopsis propagulifera (Gottsche) Konstant. et Vilnet | Russia, Kamchatskaya Prov., V.A. Bakalin, K-53-6-02-VB (KPABG) | DQ875062 | ||||
| 88 | Lophozia setosa (Mitt.) Steph. | Schistochilopsis setosa (Mitt.) Konstant. | Bhutan, D. Long 28644 (E) | AM397752 | AM398317 | |||
| 89 | Lophozia silvicola H. Buch | Lophozia silvicola H. Buch | Russia, Sakhalin Prov., Sakhalin Isl., V.A. Bakalin, S-14-4-17 (VBGI) | MK007090 | MK012210 | |||
| 90 | Lophozia silvicoloides N. Kitag. | Lophozia silvicoloides N. Kitag. | Russia, Murmanskaya Prov., N.A. Konstantinova, 356-4-00 (KPABG) | DQ875099 | DQ875064 | |||
| 91 | Lophozia sp. | Lophozia neglecta Bakalin, Maltseva, Klimova, S.S. Choi, W.Z. Ma sp. nov. | China, Yunnan Province, Diqing Prefecture, V.A. Bakalin & W.Z. Ma, C-86-1-18 | PV822252 | PV835339 | PV835346 | PV835358 | PV835352 |
| 92 | Lophozia sp. | Lophozia neglecta Bakalin, Maltseva, Klimova, S.S. Choi, W.Z. Ma sp. nov. | China, Yunnan Province, Diqing Prefecture, V.A. Bakalin & W.Z. Ma, C-86-2-18 | PV822253 | PV835340 | PV835347 | PV835359 | PV835353 |
| 93 | Lophozia sudetica (Nees ex Huebener) Grolle | Barbilophozia sudetica (Nees ex Huebener) L. Söderstr., De Roo et Hedd. | Russia, Murmanskaya Prov., V.A. Bakalin, 4.06.1998 (KPABG) | |||||
| 94 | Lophozia sudetica (Nees ex Huebener) Grolle | Barbilophozia sudetica (Nees ex Huebener) L. Söderstr., De Roo et Hedd. | Russia, Kemerovskaya Prov., N.A. Konstantinova, 90-7-00 (KPABG) | DQ875114 | ||||
| 95 | Lophozia sudetica (Nees ex Huebener) Grolle | Barbilophozia sudetica (Nees ex Huebener) L. Söderstr., De Roo et Hedd. | Norway, L. Soederstroem & P. Manyanga, Soederstroem 2003/049 (BOL) | AM397783 | AM398351 | |||
| 96 | Lophozia svalbardensis Konstant., Vilnet et Mamontov | Lophozia svalbardensis Konstant., Vilnet et Mamontov | Russia, Krasnoyarsk Territory, Kara Sea, Izvestii Tsik Islands, Sverdrup Island, 10 September 2021, I.V. Czenyadjeva, 28-21, LE-B0028582 | PQ270121 | PQ283676 | |||
| 97 | Lophozia svalbardensis Konstant., Vilnet et Mamontov | Lophozia svalbardensis Konstant., Vilnet et Mamontov | Norway, Svalbard, N.A. Konstantinova, K-135-3a-07 (KPABG) | MW298768 | MW297149 | |||
| 98 | Lophozia ventricosa (Dicks.) Dumort. | Lophozia ventricosa (Dicks.) Dumort. | United Kingdom, Scotland, Long, D.G. 31226 (E) | KF852282 | JF513477 | |||
| 99 | Lophozia ventricosa var. confertifolia (Schiffn.) Husn. | Lophozia wenzelii (Nees) Steph. | France, Vaňa s.n. (BOL) | AM397734 | AM398287 | |||
| 100 | Lophozia ventricosa var. confusa R.M. Schust. | Lophozia ventricosa (Dicks.) Dumort. | Canada, Hedderson 5008 (BOL) | AM397801 | AM398368 | |||
| 101 | Lophozia ventricosa var. longiflora (Nees) Macoun | Lophozia longiflora (Nees) Schiffn. | Russia, Chitinskaya Prov., V.A. Bakalin, 11-5-00 (KPABG) | DQ875101 | ||||
| 102 | Lophozia wenzelii var. groenlandica (Nees) Bakalin | Lophozia wenzelii (Nees) Steph. | Russia, Murmanskaya Prov., N.A. Konstantinova, 9329 (KPABG) | DQ875109 | DQ875073 | |||
| 103 | Lophozia wenzelii var. lapponica H. Buch et S.W. Arnell | Lophozia wenzelii var. lapponica H. Buch et S.W. Arnell | Svalbard, Spitsbergen, N. Konstantinova, 124-2-04 | DQ875112 | DQ875076 | |||
| 104 | Lophozia wenzelii var. massularioides Bakalin | Lophozia wenzelii var. massularioides Bakalin | Russia, Caucasus, Onipchenko, 31.08.83 (MHA) | DQ875111 | DQ875075 | |||
| 105 | Lophoziopsis excisa (Dicks.) Konstant. et Vilnet | Lophoziopsis excisa (Dicks.) Konstant. et Vilnet | United Kingdom, Scotland, Long, D.G. 35611 (E) | KF852273 | KF851381 | |||
| 106 | Lophoziopsis excisa (Dicks.) Konstant. et Vilnet | Lophoziopsis excisa (Dicks.) Konstant. et Vilnet | Russia, Russian Far East, V.A. Bakalin, P-74-17-05 | KC184729 | ||||
| 107 | Lophoziopsis longidens (Lindb.) Konstant. et Vilnet | Lophoziopsis longidens (Lindb.) Konstant. et Vilnet | Russia, Murmanskaya Prov., N.A. Konstantinova, 360-2-00 (KPABG) | DQ875094 | DQ875059 | |||
| 108 | Lophoziopsis pellucida (R.M. Schust.) Konstant. et Vilnet | Lophoziopsis pellucida (R.M. Schust.) Konstant. et Vilnet | Russia, Trans-Baikal Terr., Yu.S. Mamontov, 356-3-6 (KPABG) | MW298773 | MW297154 | |||
| 109 | Lophoziopsis polaris (R.M. Schust.) Konstant. et Vilnet | Lophoziopsis polaris (R.M. Schust.) Konstant. et Vilnet | Norway, Svalbard, N.A. Konstantinova, A.N. Savchenko, K129-07 (KPABG) | MT334459 | MT338482 | |||
| 110 | Lophoziopsis polaris var. sphagnorum (R.M. Schust.) Konstant. et Vilnet | Lophoziopsis polaris var. sphagnorum (R.M. Schust.) Konstant. et Vilnet | Russia, Yakutiya, V.A. Bakalin, 23-11-00 (KPABG) | DQ875096 | ||||
| 111 | Neoorthocaulis attenuatus (Mart.) L. Söderstr., De Roo et Hedd. | Neoorthocaulis attenuatus (Mart.) L. Söderstr., De Roo et Hedd. | Russia, Kamchatka Terr., V.A. Bakalin, 66-1-01-VB (KPABG) | OR769958 | OR762252 | |||
| 112 | Neoorthocaulis attenuatus (Mart.) L. Söderstr., De Roo et Hedd. | Neoorthocaulis attenuatus (Mart.) L. Söderstr., De Roo et Hedd. | Poland, Strebel 226 (GOET) | KC184733 | ||||
| 113 | Neoorthocaulis floerkei (F. Weber et D. Mohr) L. Söderstr., De Roo et Hedd. | Neoorthocaulis floerkei (F. Weber et D. Mohr) L. Söderstr., De Roo et Hedd. | Canada, British Columbia, B Shaw F699 (DUKE) | KF851449 | ||||
| 114 | Neoorthocaulis floerkei (F. Weber et D. Mohr) L. Söderstr., De Roo et Hedd. | Neoorthocaulis floerkei (F. Weber et D. Mohr) L. Söderstr., De Roo et Hedd. | United Kingdom, Scotland, Berwickshire, near Raecleugh, Long 31215 (E) | KC297118 | ||||
| 115 | Neoorthocaulis floerkei (F. Weber et D. Mohr) L. Söderstr., De Roo et Hedd. | Neoorthocaulis floerkei (F. Weber et D. Mohr) L. Söderstr., De Roo et Hedd. | Drehwald s.n. (GOET) | |||||
| 116 | Oleolophozia perssonii (H. Buch et S.W. Arnell) L. Söderstr., De Roo et Hedd. | Oleolophozia perssonii (H. Buch et S.W. Arnell) L. Söderstr., De Roo et Hedd. | Russia, Russian Far East, Magadan Province, Kolyma Upland, V.A. Bakalin, Mag-31-13-11 (VBGI) | OR763333 | MT476335 | MT482533 | OR785042 | |
| 117 | Plicanthus birmensis (Steph. ex Schiffn.) R.M. Schust. | Plicanthus birmensis (Steph. ex Schiffn.) R.M. Schust. | Japan, Hiroshima, Ryuzu gorge, K. Amamoto 13 (HIRO) | LC648969 | ||||
| 118 | Plicanthus hirtellus (F. Weber) R.M. Schust. | Plicanthus hirtellus (F. Weber) R.M. Schust. | China, Yunnan, Long, D.G. 34407 (E) | KF852278 | ||||
| 119 | Plicanthus hirtellus (F. Weber) R.M. Schust. | Plicanthus hirtellus (F. Weber) R.M. Schust. | Gradstein 10388 (GOET) | KC184743 | ||||
| 120 | Plicanthus hirtellus (F. Weber) R.M. Schust. | Plicanthus hirtellus (F. Weber) R.M. Schust. | Japan, Gifu, Mt. Kinka, K. Amamoto 10 (HIRO) | LC648968 | ||||
| 121 | Plicanthus hirtellus (F. Weber) R.M. Schust. | Plicanthus hirtellus (F. Weber) R.M. Schust. | Nepal, D. Long, Long 30335 (E) | AM398304 | ||||
| 122 | Pseudolophozia debiliformis (R.M. Schust. et Damsh.) Konstant. et Vilnet | Barbilophozia sudetica (Nees ex Huebener) L. Söderstr., De Roo et Hedd. | Svalbard, Spitsbergen, N.A. Konstantinova, K 91-3-06 (KPABG) | HQ897045 | ||||
| 123 | Pseudolophozia sp. | Barbilophozia sp. | Russia, Russian Far East, Sakhalin Province, V.A. Bakalin & K.G. Klimova, K-34-25-18 | MT476339 | MT482525 | MT482518 | ||
| 124 | Pseudolophozia sudetica (Nees ex Huebener) Konstant. et Vilnet | Barbilophozia sudetica (Nees ex Huebener) L. Söderstr., De Roo et Hedd. | Russia, Caucasus, N.A. Konstantinova, K 334/10-08 (KPABG) | HQ897042 | HQ897153 | HQ897090 | ||
| 125 | Pseudolophozia sudetica (Nees ex Huebener) Konstant. et Vilnet | Barbilophozia sudetica (Nees ex Huebener) L. Söderstr., De Roo et Hedd. | Russia, Komi Rep., M.V. Dulin, 110215 (KPABG) | HQ897001 | HQ897112 | HQ897089 | ||
| 126 | Pseudolophozia sudetica (Nees ex Huebener) Konstant. et Vilnet | Barbilophozia sudetica (Nees ex Huebener) L. Söderstr., De Roo et Hedd. | Russia, Perm Territory, N.A. Konstantinova, 108221 (KPABG) | HQ897002 | ||||
| 127 | Scapania aequiloba (Schwägr.) Dumort. | Scapania aequiloba (Schwägr.) Dumort. | Czech Republic, B. Shaw 12990 (DUKE) | KC297122 | ||||
| 128 | Scapania cuspiduligera (Nees) Müll. Frib. | Scapania cuspiduligera (Nees) Müll. Frib. | USA, Long 13996 (GOET) | KC184748 | ||||
| 129 | Scapania hyperborea Jørg. | Scapania hyperborea Jørg. | Norway, Hentschel Bryo 03230 (GOET) | KC184749 | ||||
| 130 | Scapania magadanica S.S. Choi, Bakalin et B.Y. Sun | Scapania magadanica S.S. Choi, Bakalin et B.Y. Sun | Russia, Russian Far East, Magadan Province, V.A. Bakalin, Mag-61-40-11 (VBGI) | OP654523 | OP641874 | OP641886 | ||
| 131 | Scapania nemorosa (L.) Dumort. | Scapania nemorea (L.) Grolle | USA, Davis 124 (DUKE) | AY608143 | AY608190 | AY608039 | AY608108 | |
| 132 | Scapania nimbosa Taylor ex Lehm. | Scapania nimbosa Taylor ex Lehm. | United Kingdom, Scotland, Long, D.G & Flagmeier, M. 37028 (DUKE) | KF852408 | KF851502 | |||
| 133 | Scapania paludosa (Müll. Frib.) Müll. Frib. | Scapania paludosa (Müll. Frib.) Müll. Frib. | Germany, Schaefer-Verwimp 19613 (GOET) | KC184753 | ||||
| 134 | Scapania stephanii Müll. Frib. | Scapania stephanii Müll. Frib. | Japan, Honshu, Hiroshima, H. Matsuda 26 (HIRO) | AB476597 | ||||
| 135 | Scapania undulata (L.) Dumort. | Scapania undulata (L.) Dumort. | Finland, Nuuksio National Park, 2000 He-Nygren & Piippo 1468 | AY149859 | AY149840 | AY462392 | ||
| 136 | Schistochilopsis incisa (Schrad.) Konstant. | Schistochilopsis incisa (Schrad.) Konstant. | USA, California, Shevock 27788 (GOET) | KC184760 | ||||
| 137 | Schistochilopsis setosa (Mitt.) Konstant. | Schistochilopsis setosa (Mitt.) Konstant. | China, Yunnan, Long, D.G. 34712 (E) | KF852274 | ||||
| 138 | Schljakovia kunzeana (Huebener) Konstant. et Vilnet | Schljakovia kunzeana (Huebener) Konstant. et Vilnet | Russia, Chita Prov., V.A. Bakalin, 38-1-00 (KPABG) | OR762255 | ||||
| 139 | Schljakovianthus quadrilobus (Lindb.) Konstant. et Vilnet | Schljakovianthus quadrilobus (Lindb.) Konstant. et Vilnet | Russia, Murmansk Prov., Yu. Mamontov, YuSM-412-3-1, 400334 (INEP) | OR762256 | ||||
| 140 | Schljakovianthus quadrilobus (Lindb.) Konstant. et Vilnet | Schljakovianthus quadrilobus (Lindb.) Konstant. et Vilnet | USA, Alaska, B Shaw F982b/8 DUKE | KF852393 | KF851482 | |||
| 141 | Sphenolobus minutus (Schreb.) Berggr. | Sphenolobus minutus (Schreb.) Berggr. | Norway, Spitsbergen, Hentschel Bryo421 (GOET) | GQ900013 | DQ312475 | |||
| 142 | Sphenolobus saxicola (Schrad.) Steph. | Sphenolobus saxicola (Schrad.) Steph. | Czech Republic, N. Bohemia, B. Buryova 26.9.1995 (DUKE) | KF852357 | ||||
| 143 | Sphenolobus saxicola (Schrad.) Steph. | Sphenolobus saxicola (Schrad.) Steph. | Germany, Heinrichs et al. JH3734 (GOET) | KC184761 | ||||
| 144 | Tetralophozia cavallii (Gola) Váňa | Tetralophozia cavallii (Gola) Váňa | Tanzania, Pocs & Ochyra 88152/C (GOET) | KC184762 | ||||
| 145 | Tetralophozia filiformis (Steph.) Urmi | Tetralophozia filiformis (Steph.) Urmi | China, Yunnan, B. Shaw 5790 (DUKE) | KF852352 | ||||
| 146 | Tetralophozia filiformis (Steph.) Urmi | Tetralophozia filiformis (Steph.) Urmi | China, Sichuan Prov., Bakalin & Klimova, China-39-1-17 (VBGI) | MZ231275 | MZ229433 | |||
| 147 | Tetralophozia pilifera (Steph.) R.M. Schust. | Tetralophozia pilifera (Steph.) R.M. Schust. | Indonesia, Gradstein 11015 (GOET) | KC184763 | ||||
| 148 | Tetralophozia pusilla (Steph.) Bakalin et Vilnet | Tetralophozia pusilla (Steph.) Bakalin et Vilnet | Japan, Yamanashi Pref., V.A. Bakalin, J-88-40-15 (VBGI) (Tl1+2/12+13) | MZ231278 | MZ229436 | MZ229442 | ||
| 149 | Tetralophozia setiformis (Ehrh.) Schljakov | Tetralophozia setiformis (Ehrh.) Schljakov | Canada, Faubert 268.3 (GOET) | KC184764 | ||||
| 150 | Tetralophozia setiformis (Ehrh.) Schljakov | Tetralophozia setiformis (Ehrh.) Schljakov | Sweden, L. Soederstroem & P. Manyanga, Soederstroem 2003/056 (BOL) | AM398370 | ||||
| 151 | Tetralophozia setiformis (Ehrh.) Schljakov | Tetralophozia setiformis (Ehrh.) Schljakov | Norway, Soederstroem 2004/195 (BOL) | AM398277 | ||||
| 152 | Trilophozia quinquedentata (Huds.) Bakalin | Trilophozia quinquedentata (Huds.) Bakalin | Norway, Svalbard, N.A. Konstantinova & A.N. Savchenko, K70-08 (KPABG) | MT334460 | ||||
| 153 | Tritomaria exsecta (Schmidel) Schiffn. ex Loeske | Tritomaria exsecta (Schmidel) Schiffn. ex Loeske | Russia, Nijegorodskaya Prov., N.A. Konstantinova, 103-1-03 (KPABG) | EU791800 | EU791682 | |||
| 154 | Tritomaria exsecta (Schmidel) Schiffn. ex Loeske | Tritomaria exsecta (Schmidel) Schiffn. ex Loeske | Russia, Siberia, Schaefer-Verwimp & Verwimp 21816 (GOET) | KC184765 | ||||
| 155 | Tritomaria exsectiformis (Breidl.) Schiffn. ex Loeske | Lophozia vinacea Bakalin, Maltseva, Klimova, S.S. Choi, W.Z. Ma sp. nov. | China, Yunnan Province, Diqing Prefecture, V.A. Bakalin & W.Z. Ma, C-83-24a-18 | PV822254 | PV835342 | PV835349 | PV835361 | PV835355 |
| 156 | Tritomaria exsectiformis (Breidl.) Schiffn. ex Loeske | Lophozia vinacea Bakalin, Maltseva, Klimova, S.S. Choi, W.Z. Ma sp. nov. | China, Yunnan Province, Diqing Prefecture, V.A. Bakalin & W.Z. Ma, C-83-26-18 | PV822255 | PV835343 | PV835350 | PV835362 | PV835356 |
| 157 | Tritomaria exsectiformis (Breidl.) Schiffn. ex Loeske | Tritomaria exsectiformis (Breidl.) Schiffn. ex Loeske | Russia, Rep. Buryatiya, N.A. Konstantinova, 83-4-01 (KPABG) | EU791683 | ||||
| 158 | Tritomaria exsectiformis (Breidl.) Schiffn. ex Loeske | Tritomaria exsectiformis (Breidl.) Schiffn. ex Loeske | Russia, Siberia, Schaefer-Verwimp & Verwimp 21849/A (GOET) | KC184766 | ||||
| 159 | Tritomaria ferruginea (Grolle) Váňa | Tritomaria ferruginea (Grolle) Váňa | China, Yunnan, B. Shaw 5763 (DUKE) | KF851443 | ||||
| 160 | Tritomaria polita (Nees) Jørg. | Saccobasis polita (Nees) H. Buch | Norway, L. Soederstroem & P. Manyanga, Soederstroem 2003/037 (BOL) | AM398342 | ||||
| 161 | Tritomaria polita (Nees) Jørg. | Saccobasis polita (Nees) H. Buch | Sweden, L. Soederstroem, Soederstroem 2003/063 (BOL) | AM398376 | ||||
| 162 | Tritomaria quinquedentata (Huds.) H. Buch | Trilophozia quinquedentata (Huds.) Bakalin | Poland, 41204 (POZW) | JQ658789 | ||||
| 163 | Tritomaria quinquedentata (Huds.) H. Buch | Trilophozia quinquedentata (Huds.) Bakalin | Russia, N.A. Konstantinova & A.N. Savchenko 515-3-05 (F) | KF852302 | KF851406 | |||
| 164 | Tritomaria quinquedentata (Huds.) H. Buch | Trilophozia quinquedentata (Huds.) Bakalin | Finland, Lammi, 2003 He-Nygren & Piippo 1474 | AY463592 | AY462334 | AY462400 | ||
| 165 | Tritomaria quinquedentata f. gracilis R.M. Schust. | Trilophozia quinquedentata f. gracilis (R.M. Schust.) Konstant. | Svalbard, North-East Land, N.A. Konstantinova, K 72-2-06 (KPABG) | EU791803 | EU791685 | |||
| 166 | Tritomaria scitula (Taylor) Jørg. | Tritomaria scitula (Taylor) Jørg. | Russia, Komi Rep., M. Dulin, N.A. Konstantinova, G101301 (KPABG) | EU791799 | ||||
| 167 | Tritomaria scitula (Taylor) Jørg. | Tritomaria scitula (Taylor) Jørg. | Norway, L. Soederstroem & P. Manyanga, Soederstroem 2003/028 (BOL) | AM398347 | ||||
| 168 | Vietnamiella epiphytica Bakalin et Vilnet | Vietnamiella epiphytica Bakalin et Vilnet | Vietnam, Lao Cai Prov., V.A. Bakalin & K.G. Klimova, V-9-7-17 (VBGI) | MK277316 | MK290984 | MK290986 | ||
| 169 | Vietnamiella epiphytica Bakalin et Vilnet | Vietnamiella epiphytica Bakalin et Vilnet | Vietnam, Lao Cai Prov., V.A. Bakalin & K.G. Klimova, V-9-8-17 (VBGI) | MK277317 | MK290985 |
Appendix B
References
- Rimington, W.R.; Duckett, J.G.; Field, K.R.; Bidartondo, M.I.; Pressel, S. The distribution and evolution of fungal symbioses in ancient lineages of land plants. Mycorrhiza 2000, 30, 23–49. [Google Scholar] [CrossRef]
- Söderström, L.; Váňa, J.; Hagborg, A.; Von Konrat, M. Notes on Early Land Plants Today. 35. Notes on Lophoziaceae (Marchantiophyta). Phytotaxa 2013, 97, 27–35. [Google Scholar] [CrossRef]
- Bakalin, V.A.; Maltseva, Y.D.; Troitsky, A.V. The Position of Lophozia dubia (Lophoziaceae, Marchantiophyta) in the Phylogenetic System of Lophozia and the Distribution of Lophozia in Southeast Eurasia, Extending to Indonesia. Plants 2024, 13, 367. [Google Scholar] [CrossRef]
- Schuster, R.M. The Hepaticae and Anthocerotae of North America East of the Hundredth Meridian. II; Columbia University Press: New York, NY, USA, 1969; 1062p. [Google Scholar]
- Bakalin, V.A. Notes on Lophozia III. Some taxonomic problems in Lophozia sect. Lophozia. Arctoa 2001, 10, 207–218. [Google Scholar] [CrossRef]
- Saukel, J. Zum Merkmalsbestang einiger. Mitteleuropдischer Arten der Lebermoosgattung. Lophozia (Dum.) Dum. (Sektion Lophozia). Stapfia 1985, 14, 149–185. [Google Scholar]
- Cao, T.; Sun, J.; Yu, J.; Song, G.-Y. Scapania gaochii, a new species of Hepaticae from Yunnan, China. Acta Bot. Yunnan. 2003, 25, 541–543. [Google Scholar]
- Cao, T.; Gao, C.; Sung, J.; Yu, J.; Song, G.-Y.; Zuo, B.-R. Scapania macroparaphyllia, a new species of Scapania (Scapaniaceae) from Xizang, China. Acta Phytotax. Sin. 2004, 42, 180–182. [Google Scholar]
- Mamontov, Y.S.; Vilnet, A.A.; Potemkin, A.D. Scapania marsupelloides sp. nov. (Scapaniaceae, Marchantiophyta), a remarkable new species near the base of Scapania phylogeny. Phytotaxa 2018, 385, 55–66. [Google Scholar] [CrossRef]
- Potemkin, A.D. Three new species of Scapania (Hepaticae) collected by Dr. David G. Long in Nepal. Arctoa 2000, 9, 115–122. [Google Scholar] [CrossRef]
- Potemkin, A.D. Three new species of Scapania (Hepaticae) from India and China. Ann. Bot. Fenn. 2001, 38, 83–89. [Google Scholar]
- Potemkin, A.D.; Vilnet, A.A.; Mamontov, Y.S. Gymnomitrion blankae (Marchantiophyta), a new species From Yunnan Province, China. Arctoa 2022, 31, 111–123. [Google Scholar] [CrossRef]
- Potemkin, A.D.; Mamontov, Y.S.; Gamova, N.S. Gymnomitrion fissum (Gymnomitriaceae, Marchantiophyta)—A new species with fissured leaf surface from China. Novosti Sist. Nizsh. Rast. 2017, 51, 274–280. [Google Scholar] [CrossRef]
- Konstantinova, N.A.; Long, D.G.; Mamontov, Y.S.; Vilnet, A.A. Gymnomitrion schusteranum (Gymnomitricaceae), a new species from the Sino-Himalaya. Arctoa 2021, 30, 149–158. [Google Scholar] [CrossRef]
- Schuster, R.M. The Hepaticae and Anthocerotae of North America East of the Hundredth Meridian. III; Columbia University Press: New York, NY, USA, 1974; 880p. [Google Scholar]
- Ellis, L.T.; Afonina, O.M.; Czernyadjeva, I.V.; Ivchenko, T.G.; Kholod, S.S.; Kotkova, V.M.; Kuzmina, E.Y.; Potemkin, A.D.; Sergeeva, Y.M.; Asthana, A.K.; et al. New national and regional bryophyte records, 61. J. Bryol. 2019, 41, 364–384. [Google Scholar] [CrossRef]
- Bakalin, V.A.; Maltseva, Y.D.; Vilnet, A.A.; Choi, S.S. The transfer of Tritomaria koreana to Lophozia has led to recircumscription of the genus and shown convergence in Lophoziaceae (Hepaticae). Phytotaxa 2021, 512, 41–56. [Google Scholar] [CrossRef]
- Takhtadzhan, A.L.; Crovello, T.J. Floristic Regions of the World; University of California Press: Berkeley/Los Angeles, CA, USA, 1986; pp. 1–522. [Google Scholar]
- Chen, Y.-S.; Deng, T.; Zhou, Z.; Sun, H. Is the East Asian flora ancient or not? Natl. Sci. Rev. 2018, 5, 920–932. [Google Scholar] [CrossRef]
- Bakalin, V.A.; Xiong, Y. Lophozia silvicola (Scapaniaceae, Hepaticae)—An Unexpected Record of a Boreal Species in the Subtropical Forest of Guizhou (Southern China). Herzogia 2015, 28, 44–49. [Google Scholar] [CrossRef]
- Kitagawa, N. A revision of the family Lophoziaceae of Japan and its adjacent regions. I. J. Hattori Bot. Lab. 1965, 28, 239–291. [Google Scholar] [CrossRef]
- Ager, T.A. Late Quaternary vegetation development following deglaciation of northwestern Alexander Archi-pelago, Alaska. Front. Earth Sci. 2019, 7, 104. [Google Scholar] [CrossRef]
- Beck, H.E.; McVicar, T.R.; Vergopolan, N.; Berg, A.; Lutsko, N.J.; Dufour, A.; Zeng, Z.; Jiang, X.; van Dijk, A.I.J.M.; Miralles, D.G. High-resolution (1 km) Köppen–Geiger maps for 1901–2099 based on constrained CMIP6 projections. Sci. Data 2023, 10, 724. [Google Scholar] [CrossRef]
- GloH2O. Köppen-Geiger Global 1-km Climate Classification Maps. 2024. Available online: https://www.gloh2o.org/koppen/ (accessed on 26 November 2024).
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1 km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Worldclim, 2020–2022. Available online: https://www.worldclim.org/ (accessed on 24 June 2023).
- Heinrichs, J.; Hentschel, J.; Wilson, R.; Feldberg, K.; Schneider, H. Evolution of leafy liverworts (Jungermanniidae, Marchantiophyta): Estimating divergence times from chloroplast DNA sequences using penalized likelihood with integrated fossil evidence. Taxon 2007, 56, 31–44. [Google Scholar] [CrossRef]
- Feldberg, K.; Heinrichs, J.; Schmidt, A.R.; Váňa, J.; Schneider, H. Exploring the impact of fossil constraints on the divergence time estimates of derived liverworts. Pl. Syst. Evol. 2013, 299, 585–601. [Google Scholar] [CrossRef]
- Konstantinova, N.A.; Vilnet, A.A.; Mamontov, Y.S. How many taxa are in the genus Saccobasis H. Buch? Evidence from integrative taxonomy. Arctoa 2022, 31, 166–180. [Google Scholar] [CrossRef]
- Bakalin, V.A.; Maltseva, Y.D.; Klimova, K.G.; Ma, W.Z. Bazzania calypogeoides (Marchantiophyta)—A peculiar new taxon from Eastern Hengduan (China). Phytotaxa 2024, 645, 214–231. [Google Scholar] [CrossRef]
- Friedl, T. Evolution of the polyphyletic genus Pleurastrum (Chlorophyta): Inferences from nuclear-encoded ribosomal DNA sequences and motile cell ultrastructure. Phycologia 1996, 35, 456–469. [Google Scholar] [CrossRef]
- Milyutina, I.A.; Goryunov, D.V.; Ignatov, M.S.; Ignatova, E.A.; Troitsky, A.V. The phylogeny of Schistidium (Bryophyta, Grimmiaceae) based on the primary and secondary structure of nuclear rDNA internal transcribed spacers. Mol. Biol. 2010, 44, 883–897. [Google Scholar] [CrossRef]
- Feldberg, K.; Váňa, J.; Krusche, J.; Kretschmann, J.; Patzak, S.D.F.; Pérez-Escobar, O.A.; Rudolf, N.R.; Seefelder, N.; Schäfer-Verwimp, A.; David, G.L.; et al. A phylogeny of Cephaloziaceae (Jungermanniopsida) based on nuclear and chloroplast DNA markers. Org. Divers. Evol. 2016, 16, 727–742. [Google Scholar] [CrossRef]
- Pacak, A.; Szweykowska-Kuliñska, Z. Molecular data concerning alloploid character and the origin of chloroplast and mitochondrial genomes in the liverwort species Pellia borealis. J. Plant Biotech. 2000, 2, 101–108. [Google Scholar]
- Matveeva, T.V.; Pavlova, O.A.; Bogomaz, D.I.; Demkovich, A.E.; Lutova, L.A. Molecular markers for plant species identification and phylogenetics. Ecol. Genet. 2011, 9, 32–43. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Felsenstein, J. Maximum Likelihood and Minimum-Steps Methods for Estimating Evolutionary Trees from Data on Discrete Characters. Syst. Biol. 1973, 22, 240–249. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 2001, 294, 2310–2314. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]






| Locus | Number of Sequences in the Alignment | Total Sites in the Alignment | Conservative Sites | Variable Sites | Parsimony-Informative Sites | Base Frequencies | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Base Pairs | % | Base Pairs | % | Base Pairs | % | A | C | G | T | |||
| ITS1–ITS2 | 65 | 890 | 415 | 46.63 | 453 | 50.90 | 341 | 38.31 | 0.174 | 0.291 | 0.338 | 0.197 |
| trnL–trnF | 68 | 484 | 288 | 59.50 | 177 | 36.57 | 142 | 29.34 | 0.347 | 0.152 | 0.173 | 0.328 |
| trnG | 44 | 640 | 352 | 55.00 | 255 | 39.84 | 139 | 21.72 | 0.363 | 0.134 | 0.127 | 0.376 |
| rbcL | 69 | 1131 | 843 | 74.54 | 288 | 25.46 | 201 | 17.77 | 0.289 | 0.172 | 0.218 | 0.322 |
| rps4 | 58 | 741 | 468 | 63.16 | 273 | 36.84 | 186 | 25.10 | 0.358 | 0.149 | 0.174 | 0.320 |
| ITS1–2 + trnL–F | 65 | 1367 | 686 | 50.18 | 631 | 46.16 | 481 | 35.19 | 0.237 | 0.241 | 0.278 | 0.245 |
| trnG + rps4 | 44 | 1253 | 753 | 60.10 | 463 | 36.95 | 266 | 21.23 | 0.360 | 0.143 | 0.152 | 0.346 |
| rbcL + rps4 | 34 | 1842 | 1333 | 72.37 | 507 | 27.52 | 331 | 17.97 | 0.317 | 0.162 | 0.201 | 0.320 |
| Locus | ML Analysis | Bayesian Analysis | |
|---|---|---|---|
| Run 1 | Run 2 | ||
| ITS1–ITS2 | −7467.54 | −7524.67 | −7523.95 |
| trnL–trnF | −3199.90 | −3265.75 | −3264.95 |
| trnG | −3325.28 | −3362.99 | −3362.03 |
| rbcL | −5521.40 | −5600.09 | −5603.52 |
| rps4 | −3988.17 | −4048.25 | −4044.00 |
| ITS1–2 + trnL–F | −11,246.90 | −11,293.19 | −11,297.05 |
| trnG + rps4 | −6694.954 | −6755.03 | −6756.83 |
| rbcL + rps4 | −7840.28 | −7876.37 | −7878.80 |
| № | Taxa | Infraspecific p-Distances, ITS1–ITS2/trnL–trnF/trnG/rbcL/rps4, % | Interspecific p-Distances, ITS1–ITS2/trnL–trnF/trnG/rbcL/rps4, % | |||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |||
| 1 | Lophozia koreana | 0.52/0/0/0.18/0 | ||||
| 2 | Lophozia vinacea | 0.26/0/0/0.11/0 | 2.8/0.45/1.57/0.76/0.85 | |||
| 3 | Lophozia dubia | n/c/n/c/n/c/n/c/n/c | 5.10/0.26/2.08/1.02/0.42 | 3.48/0.25/0.87/0.38/0.69 | ||
| 4 | Lophozia neglecta | 0.13/0/0/0/0 | 4.19/2.05/2.60/1.61/1.13 | 2.79/2.04/1.57/1.29/1.38 | 3.17/2.04/2.08/1.37/0.97 | |
| 5 | Lophozia fuscovirens | 0.41/0/n/c/n/c/n/c | 7.09/3.9/3.84/1.82/1.56 | 5.67/3.89/2.46/1.14/1.8 | 6.32/3.34/2.97/1.58/1.11 | 5.22/3.20/2.27/1.44/1.81 |
| Locus | Sequence (5′-3′) | Direction | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|
| trnL–trnF cpDNA | CGAAATTGGTAGACGCTGCG | forward | 62 | [17] |
| trnL–trnF cpDNA | TGCCAGAAACCAGATTTGAAC | reverse | 58 | [17] |
| trnT–F cpDNA | GAGGTCCTCGATAACGNGACATAA | forward | 70–72 | [30] |
| ITS1–ITS2 nrDNA | ACCTGCGGAAGGATCATTG | forward | 58 | [31] |
| ITS1–ITS2 nrDNA | GATATGCTTAAACTCAGCGG | reverse | 58 | [32] |
| ITS1–ITS2 nrDNA | CGTTGTGAGAAGTTCATTAAACC | reverse | 64 | [33] |
| trnG-intron cpDNA | CGGGTAGCGGGAATCGAAC | forward | 62 | present study |
| trnG-intron cpDNA | GCGGGTATAGTTTAGTGG | reverse | 54 | [34] |
| rbcL cpDNA | ATGTCACCACAAACAGAGACTAAAGC | forward | 50 | [35] |
| rbcL cpDNA | GCAGCAGCTAAMTCRGGACTCCA | reverse | 50 | [30] |
| rps4 cpDNA | GATGGTTGAGTGGTYTAAGAT | forward | 58–60 | present study |
| rps4 cpDNA | ATGTCCCGTTATCGAGG | reverse | 52 | present study |
| Step | Time | Temperature, °C | Cycles |
|---|---|---|---|
| Initial denaturation | 3 min | 95 (ITS1-2, trnL-F, trnG-intron), 94 (rbcL, rps4) | 35–40 |
| Denaturation | 30 s (ITS1-2, trnL-F, trnG-intron), 40 s (rbcL, rps4) | 94 | |
| Annealing | 20 s (trnL-F), 30 s (ITS1-2, trnG-intron), 90 s (rbcL, rps4) | 50 (rbcL, rps4), 56 (trnG-intron), 58 (ITS1-2, trnL-F) | |
| Elongation | 30 s (ITS1-2, trnL-F, trnG-intron), 2 min (rbcL, rps4) | 72 | |
| Final elongation | 5 min | 72 | 1 |
| Step | Time | Temperature, °C | Cycles |
|---|---|---|---|
| Initial denaturation | 3 min | 95 | 35 |
| Denaturation | 30 s | 95 | |
| Annealing | 70 s | 60 | |
| Elongation | 90 s | 72 | |
| Final elongation | 5 min | 72 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakalin, V.A.; Maltseva, Y.D.; Klimova, K.G.; Ma, W.; Choi, S.S. Two New Species of Lophozia (Marchantiophyta) from the Sino-Himalaya and the Taxonomic Diversity of East Asian Lophozia. Plants 2025, 14, 2997. https://doi.org/10.3390/plants14192997
Bakalin VA, Maltseva YD, Klimova KG, Ma W, Choi SS. Two New Species of Lophozia (Marchantiophyta) from the Sino-Himalaya and the Taxonomic Diversity of East Asian Lophozia. Plants. 2025; 14(19):2997. https://doi.org/10.3390/plants14192997
Chicago/Turabian StyleBakalin, Vadim A., Yulia D. Maltseva, Ksenia G. Klimova, Wenzhang Ma, and Seung Se Choi. 2025. "Two New Species of Lophozia (Marchantiophyta) from the Sino-Himalaya and the Taxonomic Diversity of East Asian Lophozia" Plants 14, no. 19: 2997. https://doi.org/10.3390/plants14192997
APA StyleBakalin, V. A., Maltseva, Y. D., Klimova, K. G., Ma, W., & Choi, S. S. (2025). Two New Species of Lophozia (Marchantiophyta) from the Sino-Himalaya and the Taxonomic Diversity of East Asian Lophozia. Plants, 14(19), 2997. https://doi.org/10.3390/plants14192997








