Genome-Wide Identification and Expression Profiling of the RNA-Directed DNA Methylation Pathway Genes in Cucumis sativus L.
Abstract
1. Introduction
2. Results
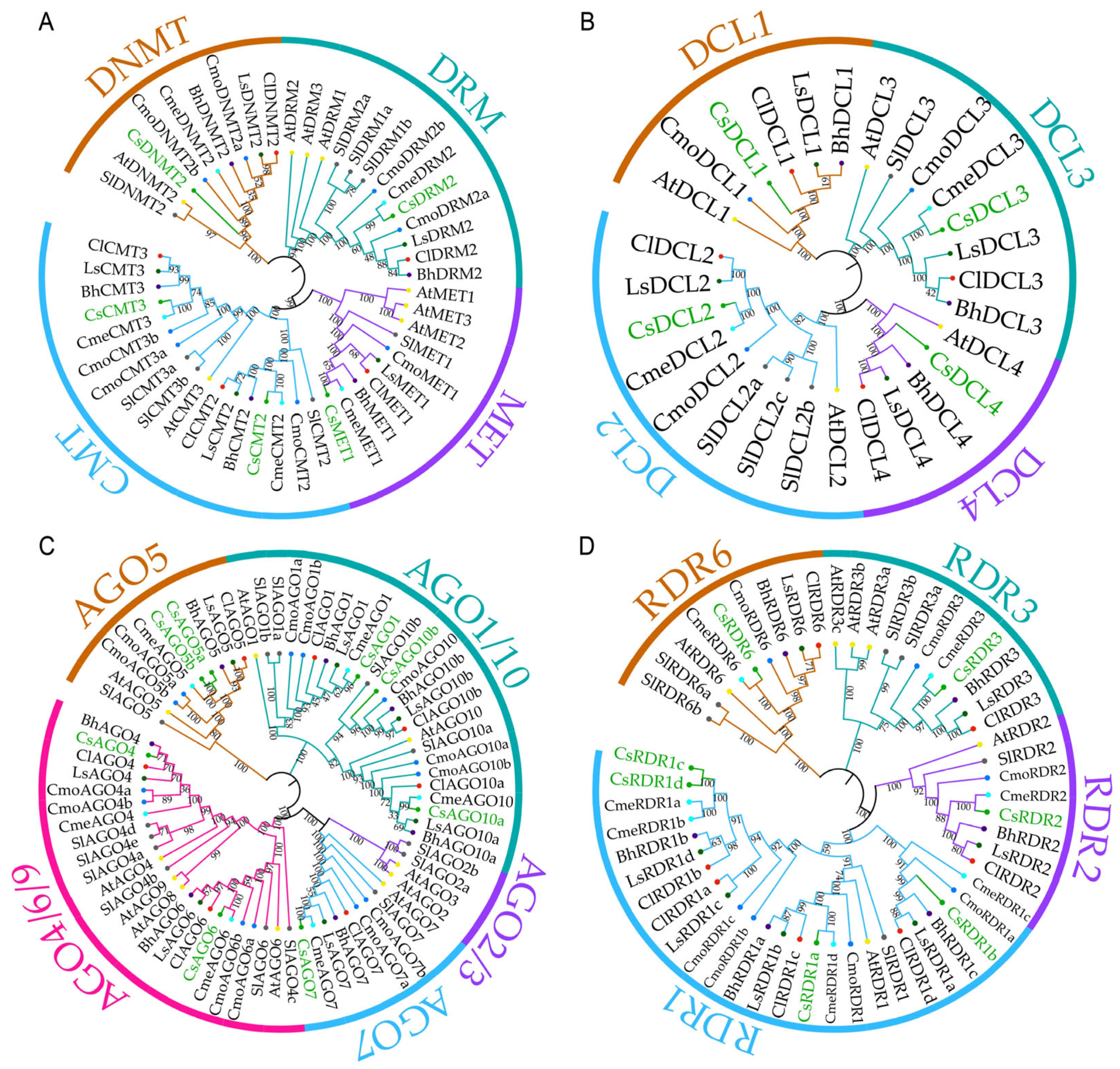
2.1. Genome-Wide Identification and Phylogenetic Analysis of CsRdDMs
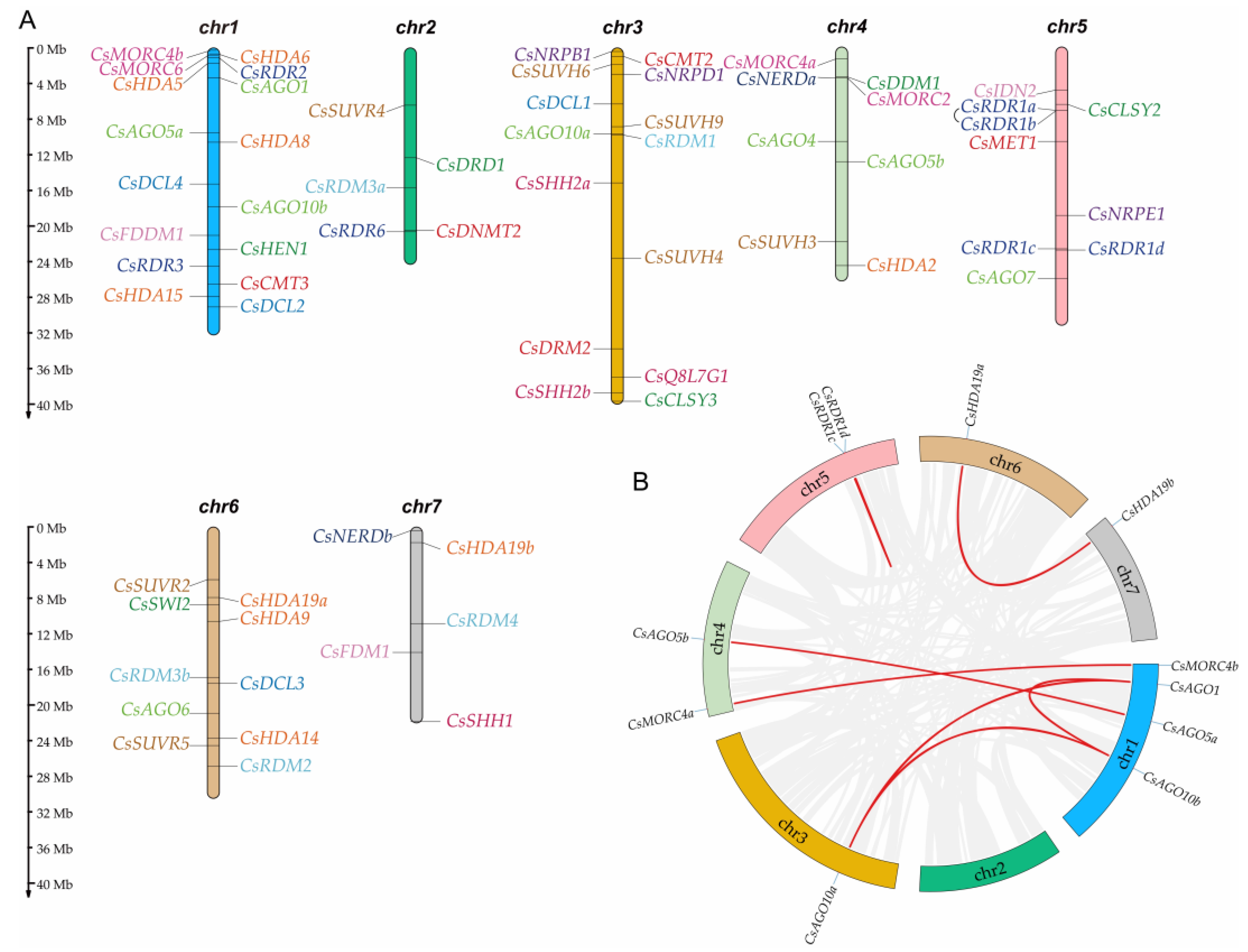
2.2. Chromosomal Location and Duplication of CsRdDMs
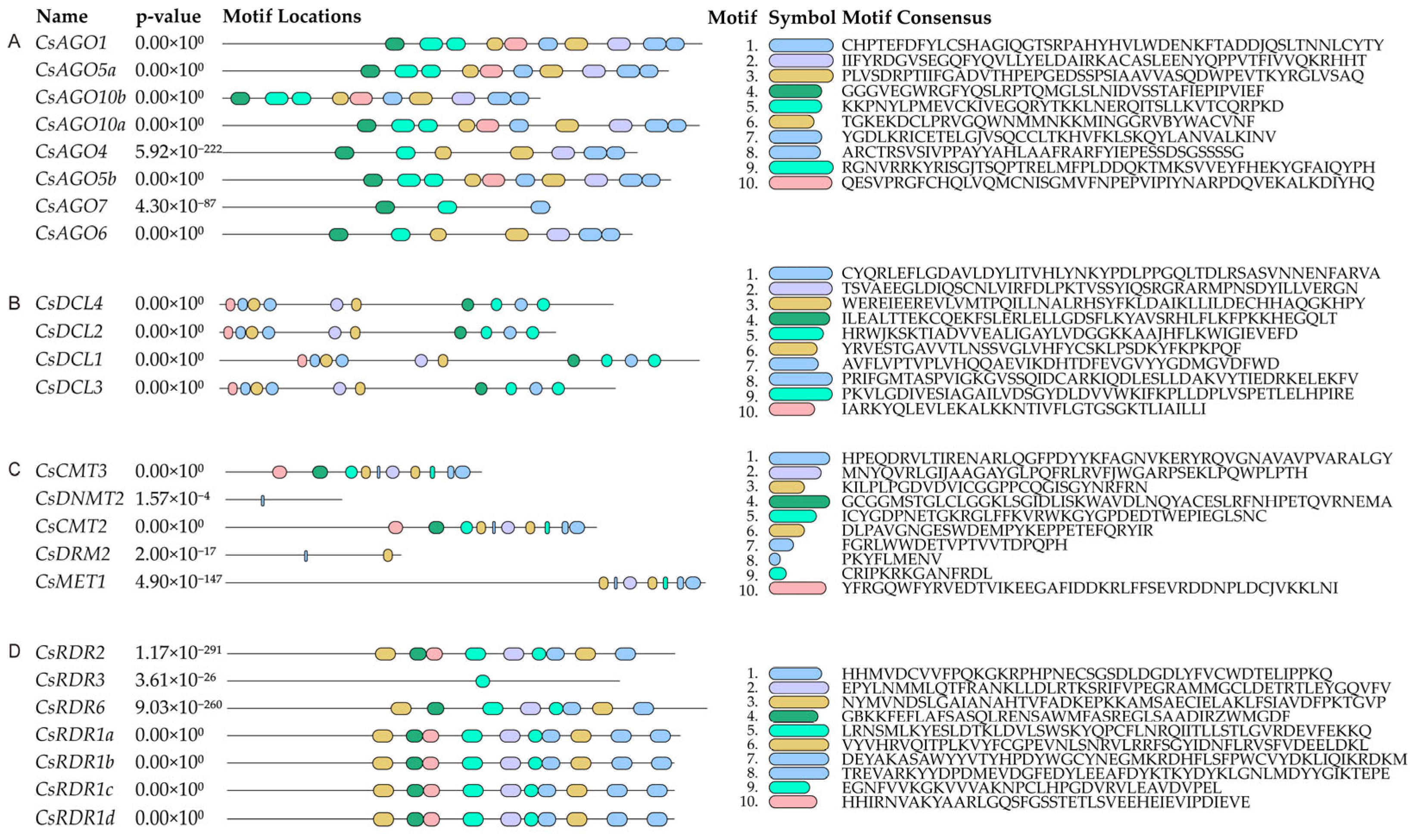
2.3. Protein Structure Analysis of CsRdDMs
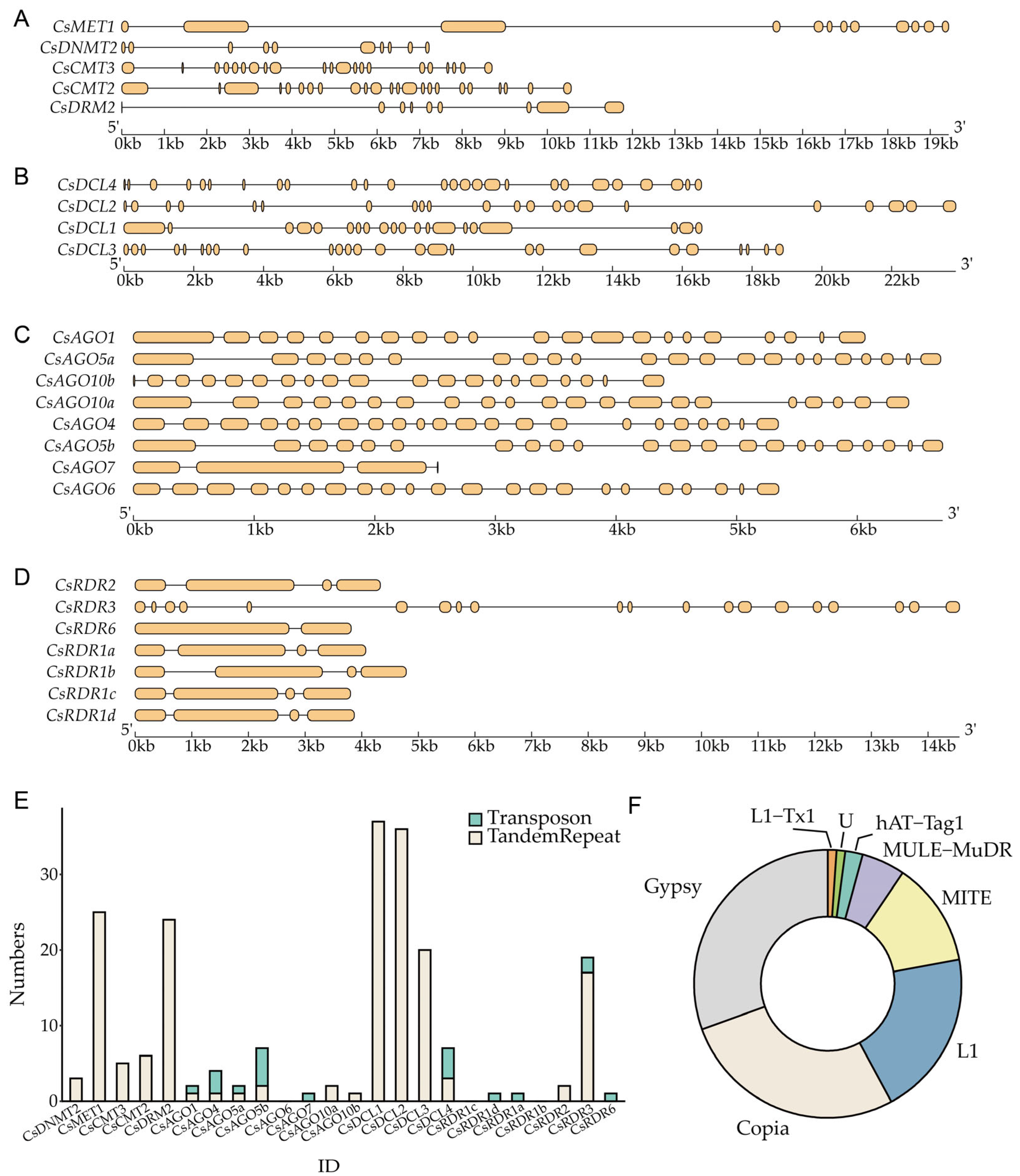
2.4. Gene Structure Analysis of CsRdDMs
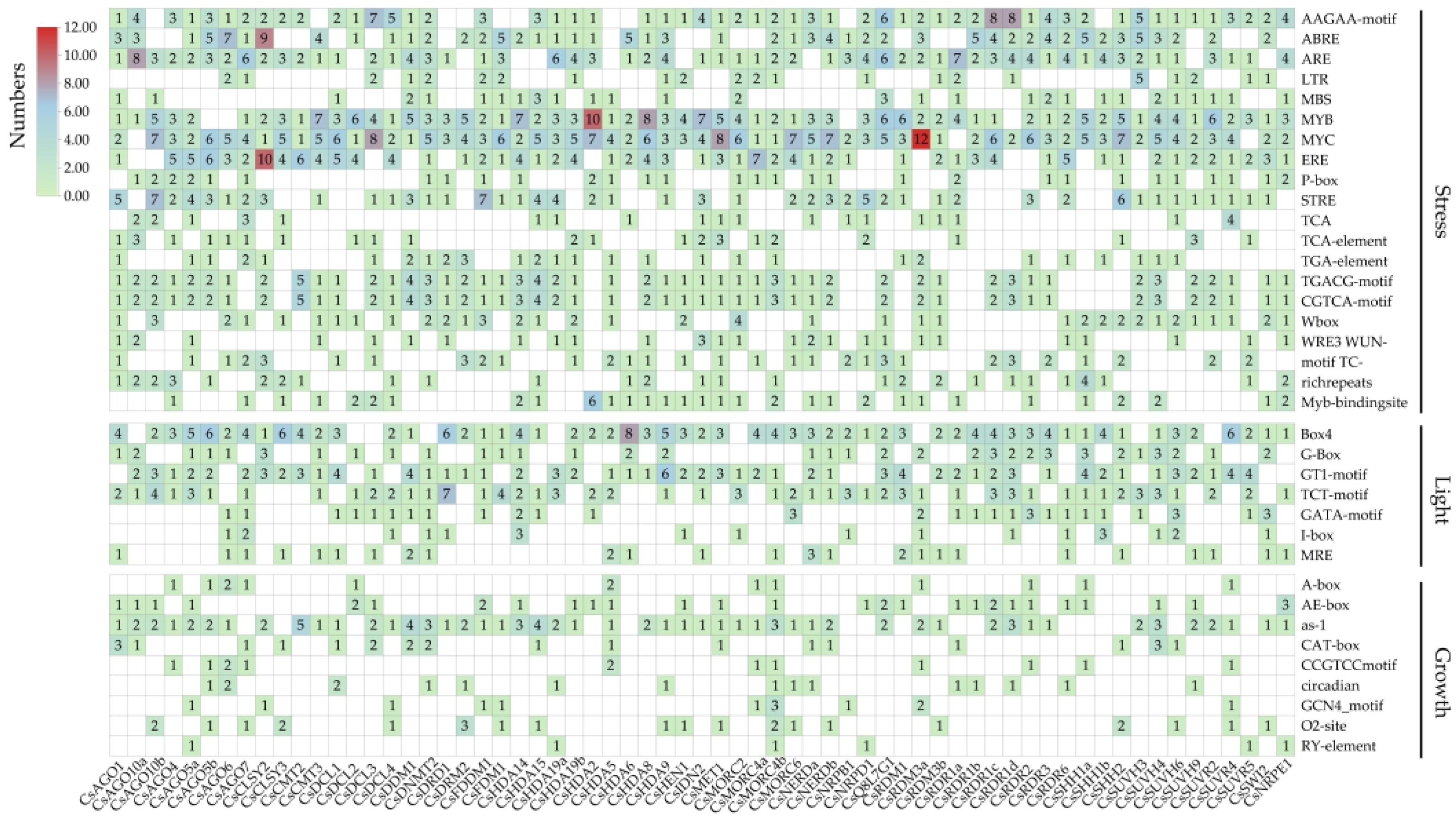
2.5. Cis-Regulatory Elements Analysis of CsRdDMs
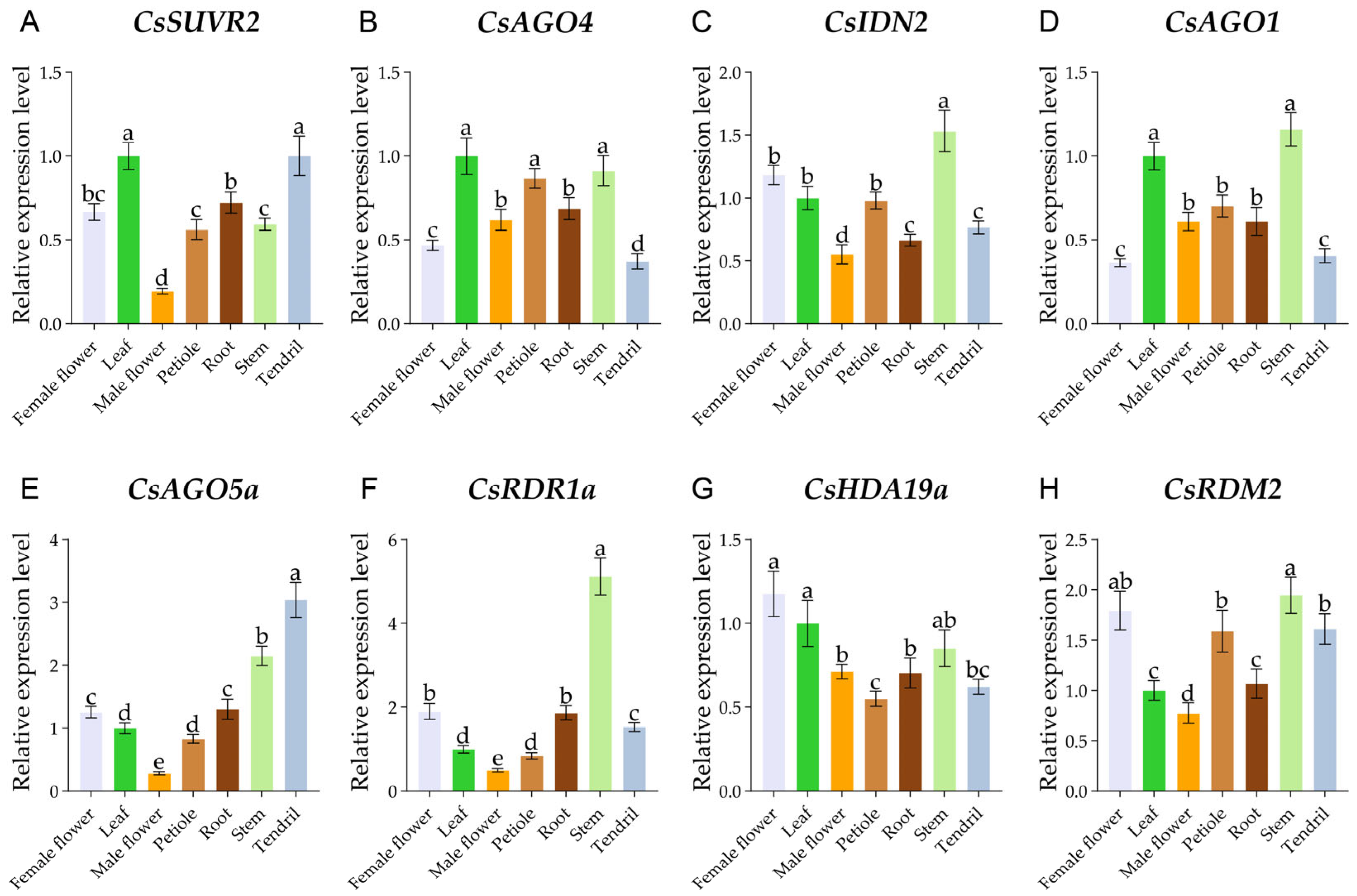
2.6. Expression Patterns of CsRdDMs in Different Tissues
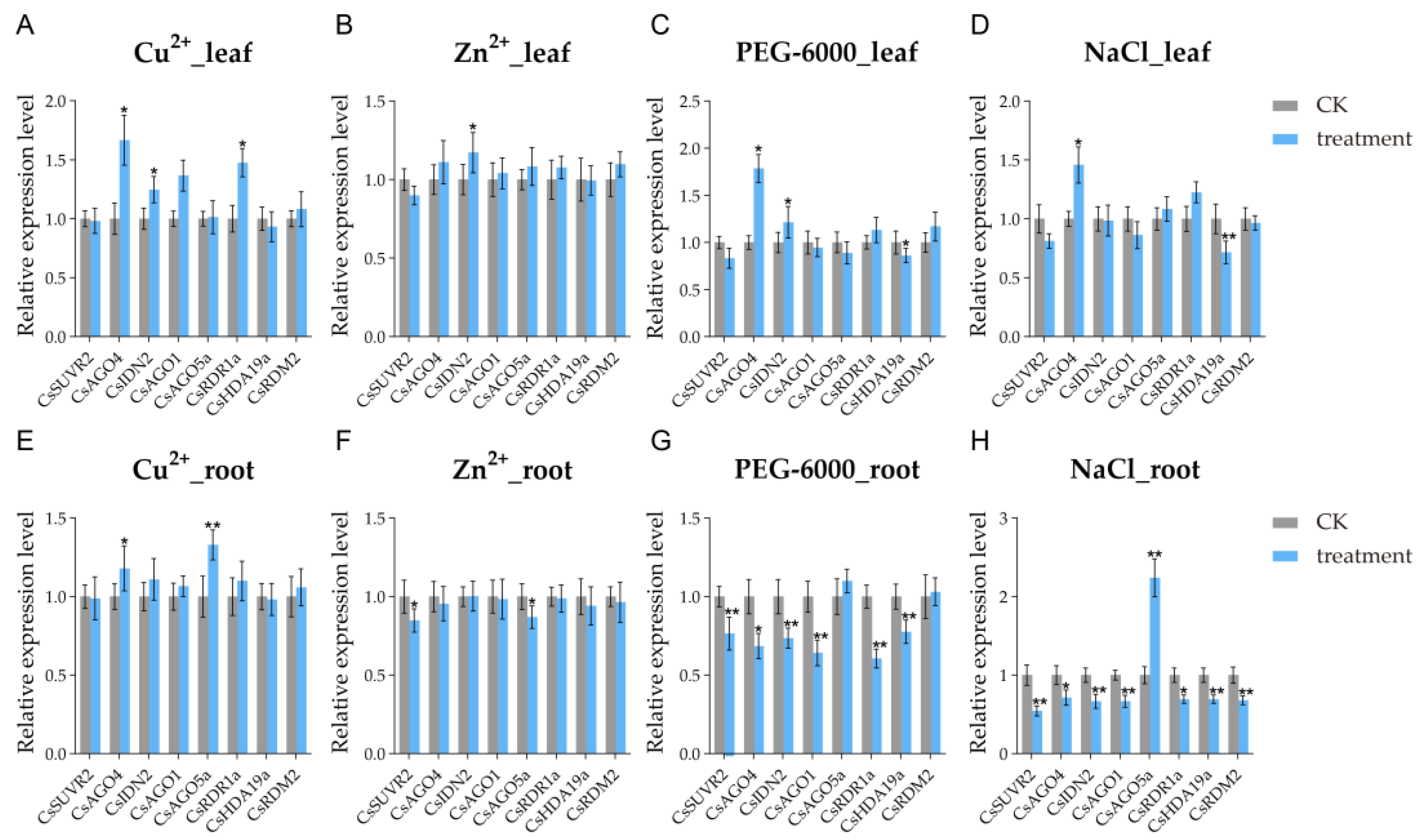
2.7. Expression Patterns of CsRdDMs Under Abiotic Stress
3. Discussion
4. Materials and Methods
4.1. Identification of CsRdDMs
4.2. Gene Structure, Physicochemical Properties, and Chromosomal Localization of CsRdDMs
4.3. Phylogenetic, Conserved Motif, and Domain Architecture Analysis
4.4. Cis-Regulatory Element Analysis in Promoters
4.5. Plant Material, Growth Conditions, and Abiotic Stress Treatments
4.6. RT-qPCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rai, A.; Chugh, V.; Pandey, S. Cucumber (Cucumis sativus L.): Genetic Improvement for Nutraceutical Traits; Springer Nature: Singapore, 2023; pp. 1527–1544. [Google Scholar]
- Ugwu, C.; Suru, S. Cosmetic, Culinary and Therapeutic Uses of Cucumber (Cucumis sativus L.); IntechOpen: London, UK, 2021. [Google Scholar]
- Gebretsadik, K.; Qiu, X.; Dong, S.; Miao, H.; Bo, K. Molecular research progress and improvement approach of fruit quality traits in cucumber. Theor. Appl. Genet. 2021, 134, 3535–3552. [Google Scholar] [CrossRef] [PubMed]
- Burzyński, M.; Kłobus, G. Changes of photosynthetic parameters in cucumber leaves under Cu, Cd, and Pb stress. Photosynthetica 2004, 42, 505–510. [Google Scholar] [CrossRef]
- Arshad, I. Effect of water stress on the growth and yield of greenhouse cucumber (Cucumis sativus L.). PSM Biol. Res. 2017, 2, 63–67. [Google Scholar]
- Chartzoulakis, K.S. Effects of NaCl salinity on germination, growth and yield of greenhouse cucumber. J. Hortic. Sci. 1992, 67, 115–119. [Google Scholar] [CrossRef]
- Hongal, D.A.; Munshi, A.D.; Talukdar, A.; Das, A.; Pradeepkumara, N.; Kumar, M.K.; Kumari, J.; Chinnusamy, V.; Ranjan, J.K.; Behera, T.K.; et al. Role of important physiological traits and development of heat tolerance index in a large set of diverse cucumber germplasm. Acta Physiol. Plant. 2024, 46, 112. [Google Scholar] [CrossRef]
- Mirouze, M.; Paszkowski, J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011, 14, 267–274. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.-K. Epigenetic gene regulation in plants and its potential applications in crop improvement. Nat. Rev. Mol. Cell Biol. 2025, 26, 51–67. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Le, H.; Simmons, C.H.; Zhong, X. Functions and mechanisms of histone modifications in plants. Annu. Rev. Plant Biol. 2025, 76, 551–578. [Google Scholar] [CrossRef]
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA METHylation. PLoS Genet. 2020, 16, e1009034. [Google Scholar] [CrossRef]
- Wendte, J.M.; Pikaard, C.S. The RNAs of RNA-directed DNA methylation. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2017, 1860, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2016, 2, 16163. [Google Scholar] [CrossRef] [PubMed]
- Popova, O.V.; Dinh, H.Q.; Aufsatz, W.; Jonak, C. The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol. Plant 2013, 6, 396–410. [Google Scholar] [CrossRef]
- Authier, A.; Cerdán, P.; Auge, G.A. Role of the RNA-directed DNA Methylation pathway in the regulation of maternal effects in Arabidopsis thaliana seed germination. Micropublication Biol. 2021, 2021, 10-17912. [Google Scholar]
- Liu, X.S.; Li, H.; Feng, S.J.; Yang, Z.M. A transposable element-derived siRNAs involve DNA hypermethylation at the promoter of OsGSTZ4 for cadmium tolerance in rice. Gene 2024, 892, 147900. [Google Scholar] [CrossRef]
- Huang, W.; Xian, Z.; Hu, G.; Li, Z. SlAGO4A, a core factor of RNA-directed DNA methylation (RdDM) pathway, plays an important role under salt and drought stress in tomato. Mol. Breed. 2016, 36, 28. [Google Scholar] [CrossRef]
- Bhattarai, K.; Poudel, M.R. DNA methylation and plants response to biotic and abiotic stress. Trends Sci. 2022, 19, 5102. [Google Scholar] [CrossRef]
- Kankel, M.W.; Ramsey, D.E.; Stokes, T.L.; Flowers, S.K.; Haag, J.R.; Jeddeloh, J.A.; Riddle, N.C.; Verbsky, M.L.; Richards, E.J. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 2003, 163, 1109–1122. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, N.; Chen, M.; Zhang, R.; Sun, Q.; Xu, H.; Zhang, Z.; Wang, Y.; Sui, X.; Wang, S.; et al. Methylation of MdMYB1 locus mediated by RdDM pathway regulates anthocyanin biosynthesis in apple. Plant Biotechnol J. 2020, 18, 1736–1748. [Google Scholar] [CrossRef]
- Ahmed, F.F.; Hossen, M.I.; Sarkar, M.A.R.; Konak, J.N.; Zohra, F.T.; Shoyeb, M.; Mondal, S. Genome-wide identification of DCL, AGO and RDR gene families and their associated functional regulatory elements analyses in banana (Musa acuminata). PLoS ONE 2021, 16, e0256873. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Carmel, L.; Csuros, M.; Koonin, E.V. Origin and evolution of spliceosomal introns. Biol. Direct 2012, 7, 11. [Google Scholar] [CrossRef]
- Chen, R.; Shen, X.; Zhang, S.; Zhao, H.; Chen, X.; Xu, X.; Huo, W.; Zhang, Z.; Lin, Y.; Lai, Z. Genome-wide identification and expression analysis of Argonaute gene family from longan embryogenic callus. J. Integr. Agric. 2021, 20, 2138–2155. [Google Scholar] [CrossRef]
- Yang, Y.; Tang, K.; Datsenka, T.U.; Liu, W.; Lv, S.; Lang, Z.; Wang, X.; Gao, J.; Wang, W.; Nie, W.; et al. Critical function of DNA methyltransferase 1 in tomato development and regulation of the DNA methylome and transcriptome. J. Integr. Plant Biol. 2019, 61, 1224–1242. [Google Scholar] [CrossRef] [PubMed]
- Xiang, R.; Ahmad, B.; Liang, C.; Shi, X.; Yang, L.; Du, G.; Wang, L. Systematic genome-wide and expression analysis of RNA-directed DNA methylation pathway genes in grapes predicts their involvement in multiple biological processes. Front. Plant Sci. 2022, 13, 1089392. [Google Scholar] [CrossRef]
- Duan, C.G.; Zhang, H.; Tang, K.; Zhu, X.; Qian, W.; Hou, Y.J.; Wang, B.; Lang, Z.; Zhao, Y.; Wang, X.; et al. Specific but interdependent functions for Arabidopsis AGO4 and AGO6 in RNA-directed DNA methylation. Embo J. 2015, 34, 581–592. [Google Scholar] [CrossRef]
- Conant, G.C.; Birchler, J.A.; Pires, J.C. Dosage, duplication, and diploidization: Clarifying the interplay of multiple models for duplicate gene evolution over time. Curr. Opin. Plant Biol. 2014, 19, 91–98. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of hy5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Jeynes-Cupper, K.; Catoni, M. Long distance signalling and epigenetic changes in crop grafting. Front. Plant Sci. 2023, 14, 1121704. [Google Scholar] [CrossRef]
- Wang, M.; Qi, Z.; Pei, W.; Cheng, Y.; Mao, K.; Ma, F. The apple Argonaute gene MdAGO1 modulates salt tolerance. Environ. Exp. Bot. 2023, 207, 105202. [Google Scholar] [CrossRef]
- Lang-Mladek, C.; Popova, O.; Kiok, K.; Berlinger, M.; Rakic, B.; Aufsatz, W.; Jonak, C.; Hauser, M.T.; Luschnig, C. Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol. Plant 2010, 3, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Bartolomé, J.; Gardiner, J.; Liu, W.; Papikian, A.; Ghoshal, B.; Kuo, H.Y.; Zhao, J.M.; Segal, D.J.; Jacobsen, S.E. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. USA 2018, 115, E2125–E21234. [Google Scholar] [CrossRef] [PubMed]
- Panda, K.; Mohanasundaram, B.; Gutierrez, J.; McLain, L.; Castillo, S.E.; Sheng, H.; Casto, A.; Gratacós, G.; Chakrabarti, A.; Fahlgren, N.; et al. The plant response to high CO2 levels is heritable and orchestrated by DNA methylation. New Phytol. 2023, 238, 2427–2439. [Google Scholar] [CrossRef]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER web server for protein sequence similarity search. Curr. Protoc. Bioinform. 2017, 60, 3.15.11–3.15.23. [Google Scholar] [CrossRef]
- Poole, R.L. The TAIR database. Methods Mol. Biol. 2007, 406, 179–212. [Google Scholar]
- Zheng, Y.; Wu, S.; Bai, Y.; Sun, H.; Jiao, C.; Guo, S.; Zhao, K.; Blanca, J.; Zhang, Z.; Huang, S.; et al. Cucurbit Genomics Database (CuGenDB): A central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 2019, 47, D1128-d36. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Mulder, N.; Apweiler, R. InterPro and InterProScan: Tools for protein sequence classification and comparison. Methods Mol. Biol. 2007, 396, 59–70. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988-d95. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- López-Moreno, M.L.; de la Rosa, G.; Cruz-Jiménez, G.; Castellano, L.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Effect of ZnO nanoparticles on corn seedlings at different temperatures; X-ray absorption spectroscopy and ICP/OES studies. Microchem. J. 2017, 134, 54–61. [Google Scholar] [CrossRef]
- Jiang, B.; Gao, G.; Ruan, M.; Bian, Y.; Geng, F.; Yan, W.; Xu, X.; Shen, M.; Wang, J.; Chang, R.; et al. Quantitative Assessment of Abiotic Stress on the Main Functional Phytochemicals and Antioxidant Capacity of Wheatgrass at Different Seedling Age. Front. Nutr. 2021, 8, 731555. [Google Scholar] [CrossRef]
- Wang, X.; Sun, H.; Lian, X.; Feng, J.; Zhao, J.; Wang, Y.; Liu, Y. Physiological and biochemical characteristics of cucumber seedlings under different levels drought stress (PEG 6000 concentrations). Hortic. Sci. 2024, 51, 202–211. [Google Scholar] [CrossRef]
- Sanchez-Munoz, R.; Depaepe, T.; Samalova, M.; Hejatko, J.; Zaplana, I.; Van Der Straeten, D. Machine-learning meta-analysis reveals ethylene as a central component of the molecular core in abiotic stress responses in Arabidopsis. Nat. Commun. 2025, 16, 4778. [Google Scholar] [CrossRef]
- Agarwal, T.; Wang, X.; Mildenhall, F.; Ibrahim, I.M.; Puthiyaveetil, S.; Varala, K. Chilling stress drives organ-specific transcriptional cascades and dampens diurnal oscillation in tomato. Hortic. Res. 2023, 10, uhad137. [Google Scholar] [CrossRef]
- Gan, J.; Qiu, Y.; Tao, Y.; Zhang, L.; Okita, T.W.; Yan, Y.; Tian, L. RNA-seq analysis reveals transcriptome reprogramming and alternative splicing during early response to salt stress in tomato root. Front. Plant Sci. 2024, 15, 1394223. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, L.; Li, Z.; Qiu, L.; Gu, J.; Shi, P.; Cao, X.; Zhang, X.; Xu, X.; Ma, Y. Genome-Wide Identification and Expression Profiling of the RNA-Directed DNA Methylation Pathway Genes in Cucumis sativus L. Plants 2025, 14, 2908. https://doi.org/10.3390/plants14182908
Ma L, Li Z, Qiu L, Gu J, Shi P, Cao X, Zhang X, Xu X, Ma Y. Genome-Wide Identification and Expression Profiling of the RNA-Directed DNA Methylation Pathway Genes in Cucumis sativus L. Plants. 2025; 14(18):2908. https://doi.org/10.3390/plants14182908
Chicago/Turabian StyleMa, Li, Ziyi Li, Lei Qiu, Jieni Gu, Piaopiao Shi, Xinyi Cao, Xinran Zhang, Xi Xu, and Yinbo Ma. 2025. "Genome-Wide Identification and Expression Profiling of the RNA-Directed DNA Methylation Pathway Genes in Cucumis sativus L." Plants 14, no. 18: 2908. https://doi.org/10.3390/plants14182908
APA StyleMa, L., Li, Z., Qiu, L., Gu, J., Shi, P., Cao, X., Zhang, X., Xu, X., & Ma, Y. (2025). Genome-Wide Identification and Expression Profiling of the RNA-Directed DNA Methylation Pathway Genes in Cucumis sativus L. Plants, 14(18), 2908. https://doi.org/10.3390/plants14182908





