Transcriptomic Analysis of Differential Gene Expression in Hevea brasiliensis Under Short-Term Cold Stress
Abstract
1. Introduction
2. Results
2.1. RNA Sequencing Data Quality and Mapping
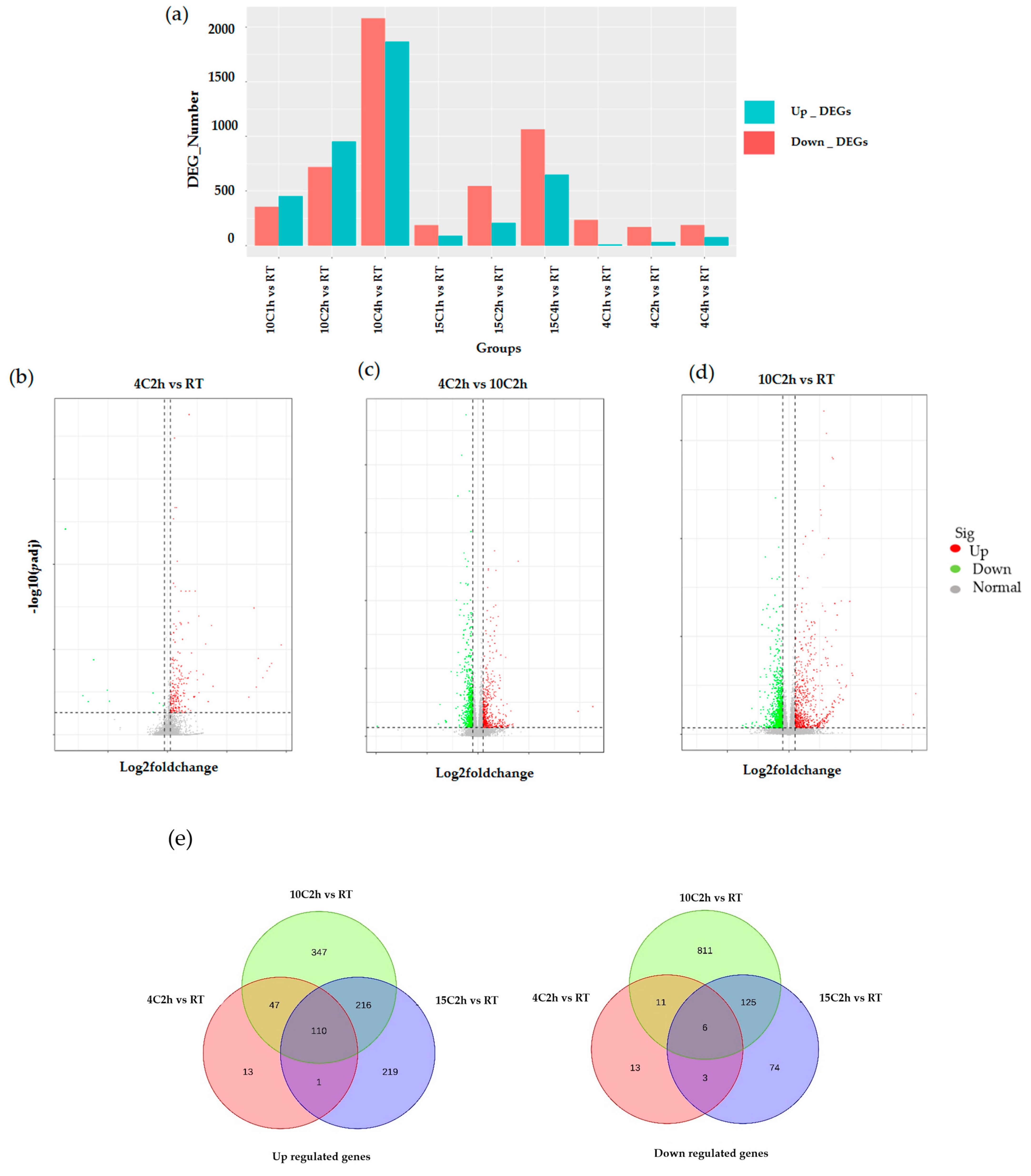
2.2. Identification of Differentially Expressed Genes (DEGs)
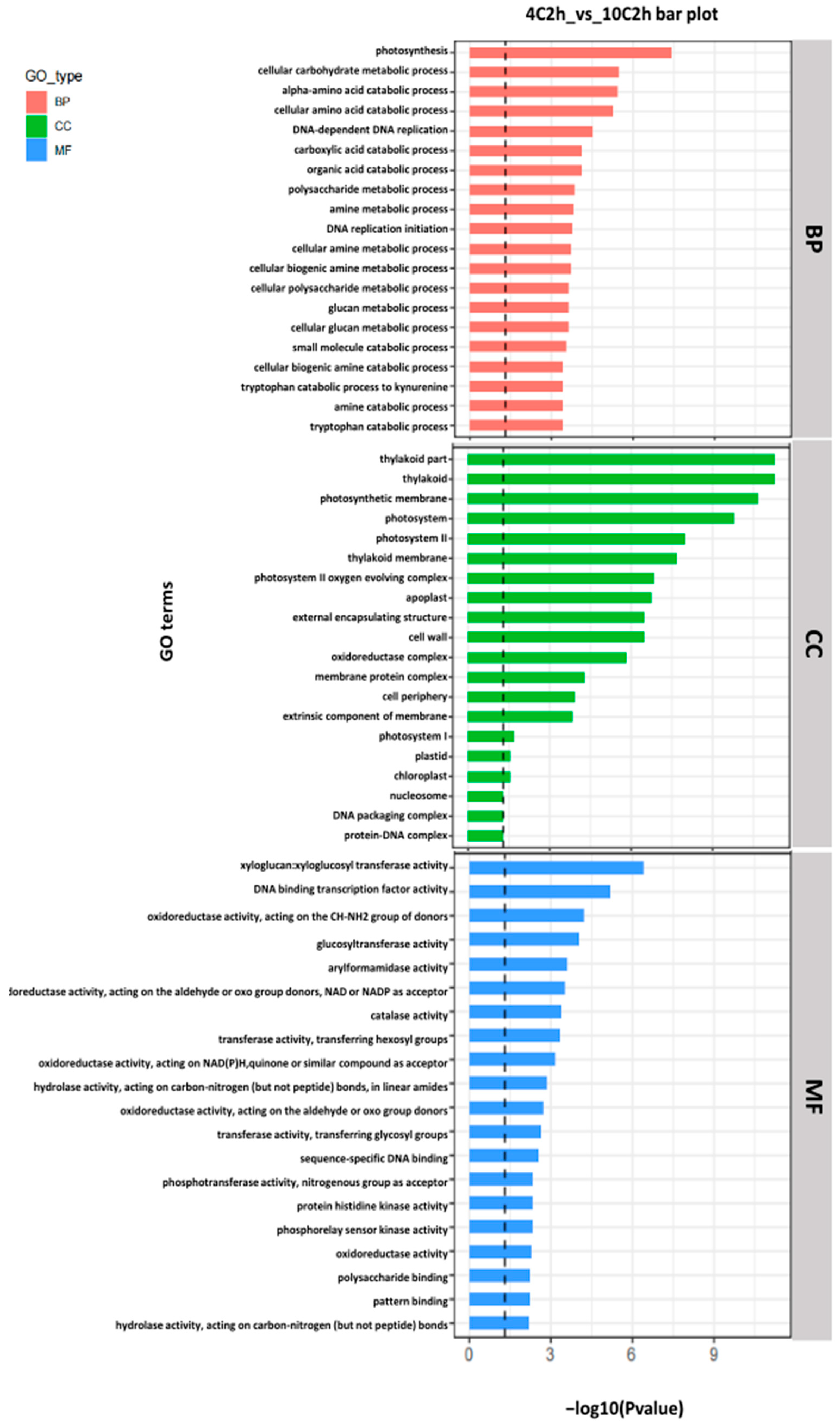
2.3. Gene Ontology (GO) Enrichment
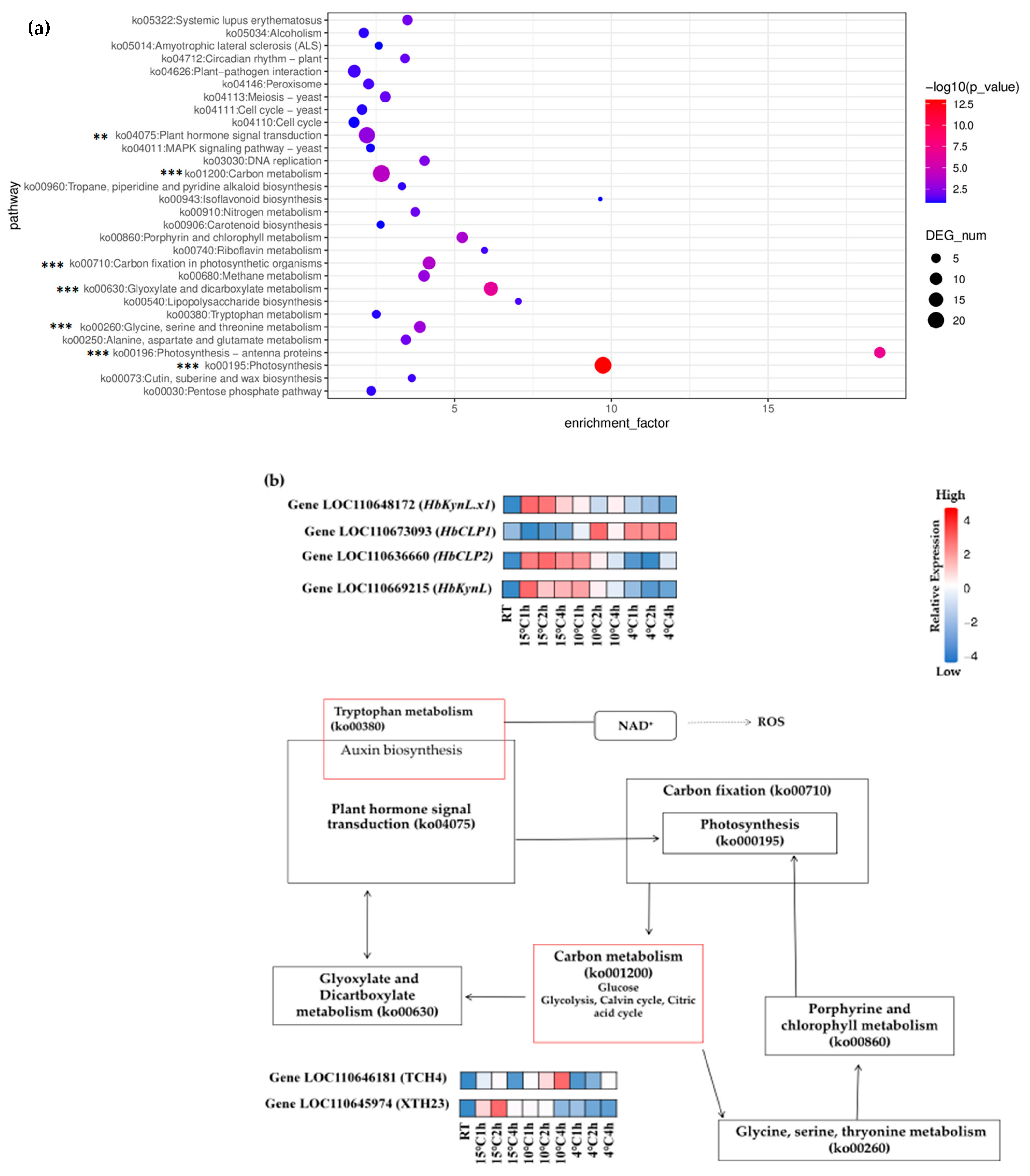
2.4. KEGG Pathways and Enrichment Analysis
2.5. Regulation of Carbon Metabolic Pathways Under Cold Stress in Hevea brasiliensis
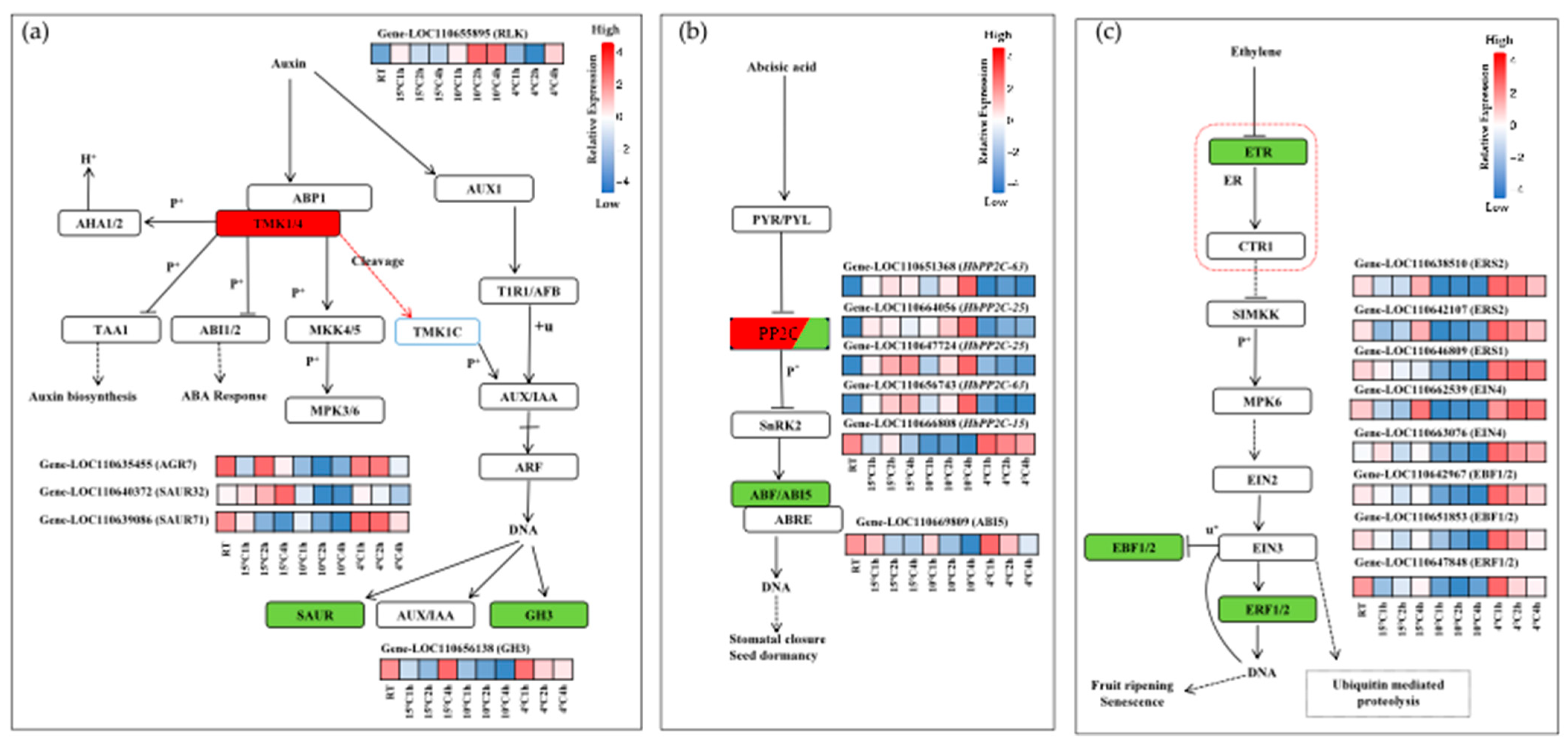
2.6. DEGs Involved in Phytohormone Signaling Transduction Under Cold Stress in Hevea brasiliensis
2.7. The Role of Different Protein Kinases in Cold Stress Tolerance in Hevea brasiliensis
2.8. Transcription Factors (TFs) Involved in the Response to Cold Stress in Hevea brasiliensis
2.9. Differentially Expressed Transcription Factors That Regulate Antioxidants in the Cold Stress
3. Discussion
4. Materials and Methods
4.1. Plant Material and Cold Treatment
4.2. RNA Extraction, Library Construction, and Sequencing
4.3. Sequence Data Processing and Differential Expression Analysis
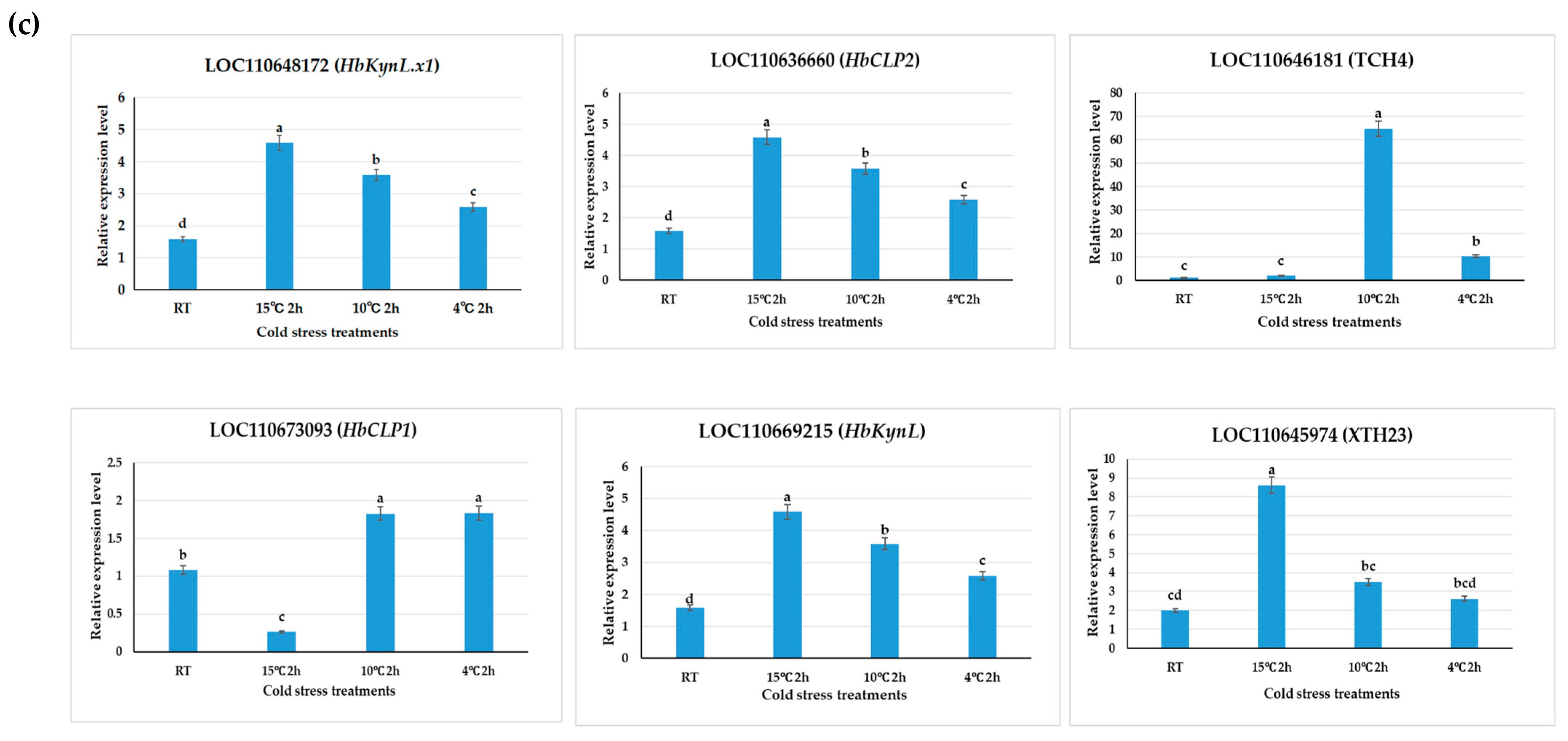
4.4. Quantitative Real-Time PCR (qRT-PCR) Validation
4.5. Gene Ontology (GO) Enrichment Analysis
4.6. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, B.; Wang, Y.; Xie, L.; Yu, W.; Lan, Q.; Wang, Y.; Chen, C.; Zhang, Y. Knocking out OsNAC050 Expression Causes Low-Temperature Tolerance in Rice by Regulating Photosynthesis and the Sucrose Metabolic Pathway. Agriculture 2023, 13, 1378. [Google Scholar] [CrossRef]
- Silva, C.C.; Mantello, C.C.; Campos, T.; Souza, L.M.; Gonçalves, P.S.; Souza, A.P. Leaf-, panel- and latex-expressed sequenced tags from the rubber tree (Hevea brasiliensis) under cold-stressed and suboptimal growing conditions: The development of gene-targeted functional markers for stress response. Mol. Breed. 2014, 34, 1035–1053. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.P.; Meenakumari, T. Natural Rubber (Hevea brasiliensis [Willd. ex A. Juss.] Müll. Arg.): History, Domestication, Genetic Diversity, Conservation and Cultivar Improvement. In Economically Important Trees: Origin, Evolution, Genetic Diversity and Ecology, 2nd ed.; Uthup, T.K., Karumamkandathil, R., Eds.; Springer: Singapore, 2024; Sustainable Development and Biodiversity; Volume 37, pp. 3–50. [Google Scholar]
- Guan, Y.; Hwarari, D.; Korboe, H.M.; Ahmad, B.; Cao, Y.; Movahedi, A.; Yang, L. Low temperature stress-induced perception and molecular signaling pathways in plants. Environ. Exp. Bot. 2023, 207, 105190. [Google Scholar] [CrossRef]
- Yuan, H.; Sheng, Y.; Chen, W.; Lu, Y.; Tang, X.; Huang, X. Overexpression of Hevea brasiliensis HbICE1 Enhances Cold Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1462. [Google Scholar] [CrossRef]
- Ruiz-Aguilar, B.; Torres-Serrallonga, N.B.; Ortega-Amaro, M.A.; Duque-Ortiz, A.; Ovando-Vázquez, C.; Jiménez-Bremont, J.F. Transcriptome Analysis Reveals Genes Responsive to Three Low-Temperature Treatments in Arabidopsis thaliana. Plants 2024, 13, 3127. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, A.; Oelmüller, R. Emerging roles of receptor-like protein kinases in plant response to abiotic stresses. Int. J. Mol. Sci. 2023, 24, 14762. [Google Scholar] [CrossRef]
- Guo, J.; Liu, S.; Li, X.; Liu, F. Crop exposure to cold stress responses in physiological, biochemical and molecular levels. In Sustainable Crop Productivity and Quality Under Climate Change: Responses of Crop Plants to Climate Change, 2nd ed.; Liu, F., Li, X., Hogy, P., Jiang, D., Brestic, M., Liu, B., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 1–19. [Google Scholar]
- Wang, Y.; Wang, J.; Sarwar, R.; Zhang, W.; Geng, R.; Zhu, K.M.; Tan, X.L. Research progress on the physiological response and molecular mechanism of cold response in plants. Front. Plant Sci. 2024, 15, 1333654. [Google Scholar] [CrossRef]
- Ding, Z.; Fu, L.; Tan, D.; Sun, X.; Zhang, J. An integrative transcriptomic and genomic analysis reveals novel insights into the hub genes and regulatory networks associated with rubber synthesis in H. brasiliensis. Ind. Crops Prod. 2020, 153, 112562. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, X.; Liu, B.; Wang, J.; Shan, J.; Wang, J.; Zhang, Y.; Li, G.; Jia, Y.; Wang, R. Transcriptome and metabolome reveal the primary and secondary metabolism changes in Larix gmelinii seedlings under abiotic stress. BMC Plant Biol. 2024, 24, 583. [Google Scholar] [CrossRef]
- Li, M.; Yue, T.; Han, J.; Jiao, J.; Xiao, H.; Shang, F. Exogenous glucose irrigation alleviates cold stress by regulating soluble sugars, ABA and photosynthesis in melon seedlings. Plant Physiol. Biochem. 2024, 217, 109214. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; He, L.; Li, F. Understanding cold stress response mechanisms in plants: An overview. Front. Plant Sci. 2024, 15, 1443317. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ullah, F.; Zou, J.; Zeng, X. Molecular and Physiological Responses of Plants that Enhance Cold Tolerance. Int. J. Mol. Sci. 2025, 26, 1157. [Google Scholar] [CrossRef]
- Wang, X.; Liu, W.-C.; Zeng, X.-W.; Yan, S.; Qiu, Y.-M.; Wang, J.-B.; Huang, X.; Yuan, H.-M. HbSnRK2.6 Functions in ABA-Regulated Cold Stress Response by Promoting HbICE2 Transcriptional Activity in Hevea brasiliensis. Int. J. Mol. Sci. 2021, 22, 12707. [Google Scholar] [CrossRef]
- Raza, A.; Salehi, H.; Rahman, M.A.; Zahid, Z.; Madadkar Haghjou, M.; Charagh, S.; Osman, H.S.; Albaqami, M.; Zhuang, Y.; Siddique, K.H.; et al. Plant hormones and neurotransmitter interactions mediate antioxidant defenses under induced oxidative stress in plants. Front. Plant Sci. 2022, 13, 961872. [Google Scholar] [CrossRef]
- Tang, X.; Wang, Q.; Yuan, H.; Huang, X. Chilling-induced DNA Demethylation is associated with the cold tolerance of Hevea brasiliensis. BMC Plant Biol. 2018, 18, 70. [Google Scholar] [CrossRef]
- Qiu, Y.-M.; Guo, J.; Jiang, W.-Z.; Ding, J.-H.; Song, R.-F.; Zhang, J.-L.; Huang, X.; Yuan, H.-M. HbBIN2 Functions in Plant Cold Stress Resistance through Modulation of HbICE1 Transcriptional Activity and ROS Homeostasis in Hevea brasiliensis. Int. J. Mol. Sci. 2023, 24, 15778. [Google Scholar] [CrossRef]
- Gong, X.X.; Yan, B.Y.; Hu, J.; Yang, C.P.; Li, Y.J.; Liu, J.P.; Liao, W.B. Transcriptome profiling of rubber tree (Hevea brasiliensis) discovers candidate regulators of the cold stress response. Genes Genom. 2018, 40, 1181–1197. [Google Scholar] [CrossRef]
- Yu, M.; Luobu, Z.; Zhuoga, D.; Wei, X.; Tang, Y. Advances in plant response to low-temperature stress. Plant Growth Regul. 2025, 105, 167–185. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, S.; Gao, Y.; Kan, C.; Wang, L.; Yang, Q.; Xia, X.; Ishida, T.; Sawa, S.; Guo, H.; et al. CLE42 delays leaf senescence by antagonizing ethylene pathway in Arabidopsis. New Phytol. 2022, 235, 550–562. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Chu, X.; Cheng, Y.; Hu, B.; Li, Q.; Peng, W.; Yang, Q.; Zhang, X.; Li, Y. UV-B pre-irradiation—Promoted flavonoid accumulation enhances cold tolerance in tea plants by mediating CsHY5 pathways. Sci. Hortic. 2025, 345, 114145. [Google Scholar] [CrossRef]
- Deng, X.; Wang, J.; Li, Y.; Wu, S.; Yang, S.; Chao, J.; Chen, Y.; Zhang, S.; Shi, M.; Tian, W. Comparative transcriptome analysis reveals phytohormone signalings, heat shock module and ROS scavenger mediate the cold-tolerance of rubber tree. Sci. Rep. 2018, 8, 4931. [Google Scholar] [CrossRef]
- Yang, B.; Shi, X.; Guo, T.; Liang, M. De novo transcriptome analysis in Hevea brasiliensis to unveil genes involved in low temperature stress response. Phyton 2023, 92, 559–575. [Google Scholar] [CrossRef]
- Mantello, C.C.; Boatwright, L.; da Silva, C.C.; Scaloppi, E.J.; de Souza Goncalves, P.; Barbazuk, W.B.; de Souza, A.P. Deep expression analysis reveals distinct cold-response strategies in rubber tree (Hevea brasiliensis). BMC Genom. 2019, 20, 455. [Google Scholar] [CrossRef]
- Chen, J.; Zeng, H.; Yan, F.; Jiang, Z.; Chen, J.; Wang, W.; Zhu, Q. Identification of the WRKY gene family in Bergenia purpurascens and functional analysis of BpWRKY13 under cold stress. Plant Physiol. Biochem. 2025, 223, 109832. [Google Scholar] [CrossRef]
- Li, Y.; Zong, Y.; Li, W.; Guo, G.; Zhou, L.; Xu, H.; Gao, R.; Liu, C. Transcriptomics integrated with metabolomics reveals the effect of cold stress on rice microspores. BMC Plant Biol. 2023, 23, 521. [Google Scholar] [CrossRef]
- Fürtauer, L.; Weiszmann, J.; Weckwerth, W.; Nägele, T. Dynamics of Plant Metabolism during Cold Acclimation. Int. J. Mol. Sci. 2019, 20, 5411. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, J.; Xu, J.; Zhang, X.; Xie, Z.; Li, Z. Effect of cold stress on photosynthetic physiological characteristics and molecular mechanism analysis in cold-resistant cotton (ZM36) seedlings. Front. Plant Sci. 2024, 15, 1396666. [Google Scholar] [CrossRef]
- Gall, H.L.; Philippe, F.; Domon, M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell Wall Metabolism in Response to Abiotic Stress. Plants 2015, 4, 112. [Google Scholar] [CrossRef] [PubMed]
- Novaković, L.; Guo, T.; Bacic, A.; Sampathkumar, A.; Johnson, K.L. Hitting the Wall—Sensing and Signaling Pathways Involved in Plant Cell Wall Remodeling in Response to Abiotic Stress. Plants 2018, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Willick, I.R.; Kasuga, J.; Livingston, D.P. Responses of the plant cell wall to sub-zero temperatures: A brief update. Plant Cell Physiol. 2021, 62, 1858–1866. [Google Scholar] [CrossRef]
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The Plant Cell Wall: A Complex and Dynamic Structure as Revealed by the Responses of Genes under Stress Conditions. Front. Plant Sci. 2016, 7, 984. [Google Scholar] [CrossRef]
- Shi, H.; Ye, T.; Zhong, B.; Liu, X.; Jin, R.; Chan, Z. AtHAP5A modulates freezing stress resistance in arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol. 2014, 203, 554–567. [Google Scholar] [CrossRef]
- Ishida, K.; Yokoyama, R. Reconsidering the function of the xyloglucan endotransglucosylase/hydrolase family. J. Plant Res. 2022, 135, 145–156. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, W.; Chang, Y.; Ma, Y.; Deng, Y.; Zhang, N.; Bai, Y.; Jiang, Z.; Hu, T. Cloning, Characterization, and Expression Pattern Analysis of the BBM Gene in Tree Peony (Paeonia ostii). Forests 2024, 15, 36. [Google Scholar] [CrossRef]
- Kamiyama, Y.; Katagiri, S.; Umezawa, T. Growth Promotion or Osmotic Stress Response: How SNF1-Related Protein Kinase 2 (SnRK2) Kinases Are Activated and Manage Intracellular Signaling in Plants. Plants 2021, 10, 1443. [Google Scholar] [CrossRef]
- Zhang, J.; Shao, W.; Xu, Y.; Tian, F.; Chen, J.; Wang, D.; Lin, X.; He, C.; Yang, X.; Staiger, D.; et al. Unveiling the regulatory role of GRP7 in ABA signal-mediated mRNA translation efficiency regulation. Nat. Commun. 2025, 16, 3947. [Google Scholar] [CrossRef]
- Bi, B.; Shao, L.; Xu, T.; Du, H.; Li, D. Transcriptomic Analysis Reveals Calcium and Ethylene Signaling Pathway Genes in Response to Cold Stress in Cinnamomum camphora. Horticulturae 2024, 10, 995. [Google Scholar] [CrossRef]
- Zeng, X.W.; Jiang, W.Z.; Zhang, J.L.; Ding, J.H.; Qiu, Y.M.; Wen, W.; Yang, H.; Zhang, Q.Y.; Yuan, H.M. Ethylene negatively regulates cold tolerance through HbEIN3—HbICE2 regulatory module in Hevea brasiliensis. Plant Physiol. Biochem. 2025, 219, 109397. [Google Scholar] [CrossRef]
- Dou, X.; Xie, S.; Wang, J.; Shen, X.; Liu, S.; Tian, N. Genome-wide identification of F-box-LRR gene functional analysis of CsFBXL13 transcription factor in tea plants. Funct. Integr. Genom. 2025, 25, 57. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, J.; Zhou, X.; Liu, S.; Zhuang, Y. Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. PeerJ 2020, 8, e8777. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, L.; Wong, D.C.J.; Wang, Y.; Zhu, Z.; Xu, G.; Wang, Q.; Li, S.; Liang, Z.; Xin, H. The ethylene response factor VaERF092 from Amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J. 2019, 99, 988–1002. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhao, A.; Han, J.; Lu, Y.; Chen, X.; Cheng, Z.; Hou, L.; Lv, L. Single-cell transcriptome reveals the cellular response to PEG-induced stress in wheat leaves. J. Agric. Food Chem. 2025, 73, 10764–10777. [Google Scholar] [CrossRef]
- Han, L.; Ma, K.; Zhao, Y.; Mei, C.; Mamat, A.; Wang, J.; Qin, L.; He, T. The cold stress responsive gene DREB1A involved in low temperature tolerance in Xianjiang wild walnut. PeerJ 2022, 10, e14021. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; You, X.; Gao, L.; Wen, W.; Song, Y.; Shen, Z.; Xing, Q.; An, Y.; Zhou, P. Functional analysis of AtDPBF3, encoding a key member of the ABI5 subfamily involved in ABA signaling, in Arabidopsis thaliana under salt stress. Plant Physiol. Biochem. 2025, 220, 109494. [Google Scholar] [CrossRef]
- Kang, H.; Thomas, H.R.; Xia, X.; Shi, H.; Zhang, L.; Hong, J.; Shi, K.; Zhou, J.; Yu, J.; Zhou, Y. An integrative overview of cold response and regulatory pathways in horticultural crops. J. Integr. Plant Biol. 2025, 67, 1028–1059. [Google Scholar] [CrossRef] [PubMed]
- Fedoreyeva, L.I.; Kononenko, N.V. Peptides and Reactive Oxygen Species Regulate Root Development. Int. J. Mol. Sci. 2025, 26, 2995. [Google Scholar] [CrossRef]
- Ben Saad, R.; Ben Romdhane, W.; Baazaoui, N.; Bouteraa, M.T.; Chouaibi, Y.; Mnif, W.; Ben Hsouna, A.; Kačániová, M. Functional Characterization of Lobularia maritima LmTrxh2 Gene Involved in Cold Tolerance in Tobacco through Alleviation of ROS Damage to the Plasma Membrane. Int. J. Mol. Sci. 2023, 24, 3030. [Google Scholar] [CrossRef]
- Zhou, W.; Du, J.; Jiao, R.; Wang, X.; Fang, T.; Huang, G. Genome-wide identification of CAMTA gene family in teak (Tectona grandis) and functional characterization of TgCAMTA1 and TgCAMTA3 in cold tolerance. BMC Plant Biol. 2025, 25, 35. [Google Scholar] [CrossRef]
- Satyakam; Zinta, G.; Singh, R.K.; Kumar, R. Cold adaptation strategies in plants: An emerging role of epigenetics and antifreeze proteins to engineer cold resilient plants. Front. Genet. 2022, 13, 909007. [Google Scholar] [CrossRef]
- Zhang, B.-Q.; Huang, Y.-X.; Zhou, Z.-F.; Zhou, S.; Duan, W.-X.; Yang, C.-F.; Gao, Y.-J.; Zhang, G.-M.; Song, X.-P.; Zhang, X.-Q.; et al. Cold-Induced Physiological and Biochemical Alternations and Proteomic Insight into the Response of Saccharum spontaneum to Low Temperature. Int. J. Mol. Sci. 2022, 23, 14244. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Yang, M.; Fang, Y.; Luo, Y.; Gao, S.; Xiao, X.; An, Z.; Zhou, B.; Zhang, B.; Tan, X.; et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Mao, C.; Li, L.; Yang, T.; Gui, M.; Li, X.; Zhang, F.; Zhao, Q.; Wu, Y. Transcriptomics integrated with widely targeted metabolomics reveals the cold resistance mechanism in Hevea brasiliensis. Front. Plant Sci. 2023, 13, 1092411. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
| Gene Name | Gene Id | Forward/Reverse Primer | Primer Sequence (5′ to 3′) |
|---|---|---|---|
| TCH4 | LOC110646181 | qLOC110646181-F | CGCATCCCATCTCCTCACTAC |
| qLOC110646181-R | TGGCTGCCATGGAAGAGAAG | ||
| DREB 1A/CBF1 | LOC110641584 | qLOC110641584-F | GCGTCTAGTCATGAGGGCTG |
| qLOC110641584-R | AATCCGAACCAAGACGAGAAT | ||
| HbKynL | LOC110669215 | qLOC110669215-F | GGCTACTGATTGATGTGCCA |
| qLOC110669215-R | TCTGAAATTTACCTGTCGGTG | ||
| CLP1 | LOC110673093 | qLOC110673093-F | CTTTCAGGACGCTGACAGAA |
| qLOC110673093-R | TGGCACTTGCTATACTCTCAA | ||
| CLP2 | LOC110636660 | qLOC110636660-F | GATTCTTCCCTCTCCACAGC |
| qLOC110636660-R | CGGTGATGGAATGTAGCAGA | ||
| CLP2 | LOC110648172 | qLOC110648172-F | CCTTTCTTGCCTCTCCACCAC |
| qLOC110648172-R | CGGCGGACTGGGATTAGATT | ||
| XTH23 | LOC110645974 | qLOC110645974-F | CTTCCTCTGTTGGCTATTCAGC |
| qLOC110645974-R | GGTTGTCACAGCTCTCAGGTATT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weeramange, M.V.; Gu, C.; Xin, S.; Gu, X.; Yi, B.; Huang, T. Transcriptomic Analysis of Differential Gene Expression in Hevea brasiliensis Under Short-Term Cold Stress. Plants 2025, 14, 2900. https://doi.org/10.3390/plants14182900
Weeramange MV, Gu C, Xin S, Gu X, Yi B, Huang T. Transcriptomic Analysis of Differential Gene Expression in Hevea brasiliensis Under Short-Term Cold Stress. Plants. 2025; 14(18):2900. https://doi.org/10.3390/plants14182900
Chicago/Turabian StyleWeeramange, Madushi Vishmitha, Chenrui Gu, Shichao Xin, Xiaochuan Gu, Bin Yi, and Tiandai Huang. 2025. "Transcriptomic Analysis of Differential Gene Expression in Hevea brasiliensis Under Short-Term Cold Stress" Plants 14, no. 18: 2900. https://doi.org/10.3390/plants14182900
APA StyleWeeramange, M. V., Gu, C., Xin, S., Gu, X., Yi, B., & Huang, T. (2025). Transcriptomic Analysis of Differential Gene Expression in Hevea brasiliensis Under Short-Term Cold Stress. Plants, 14(18), 2900. https://doi.org/10.3390/plants14182900






