Exploring Plastome Diversity and Molecular Evolution Within Genus Tortula (Family Pottiaceae, Bryophyta)
Abstract
1. Introduction
2. Results
2.1. Organelle Genome Characteristics
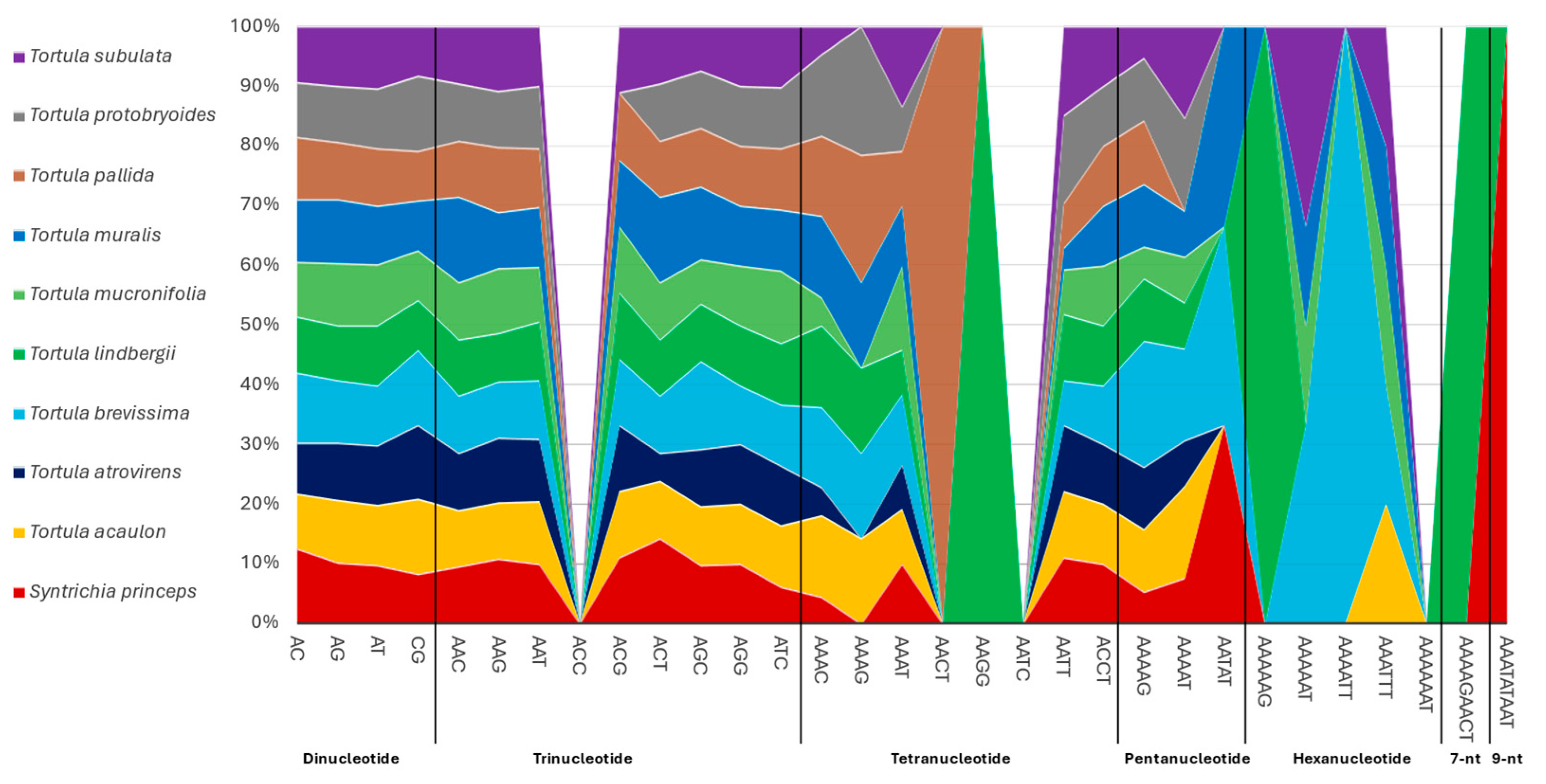
2.2. Tandem Repeats—Microsatellites
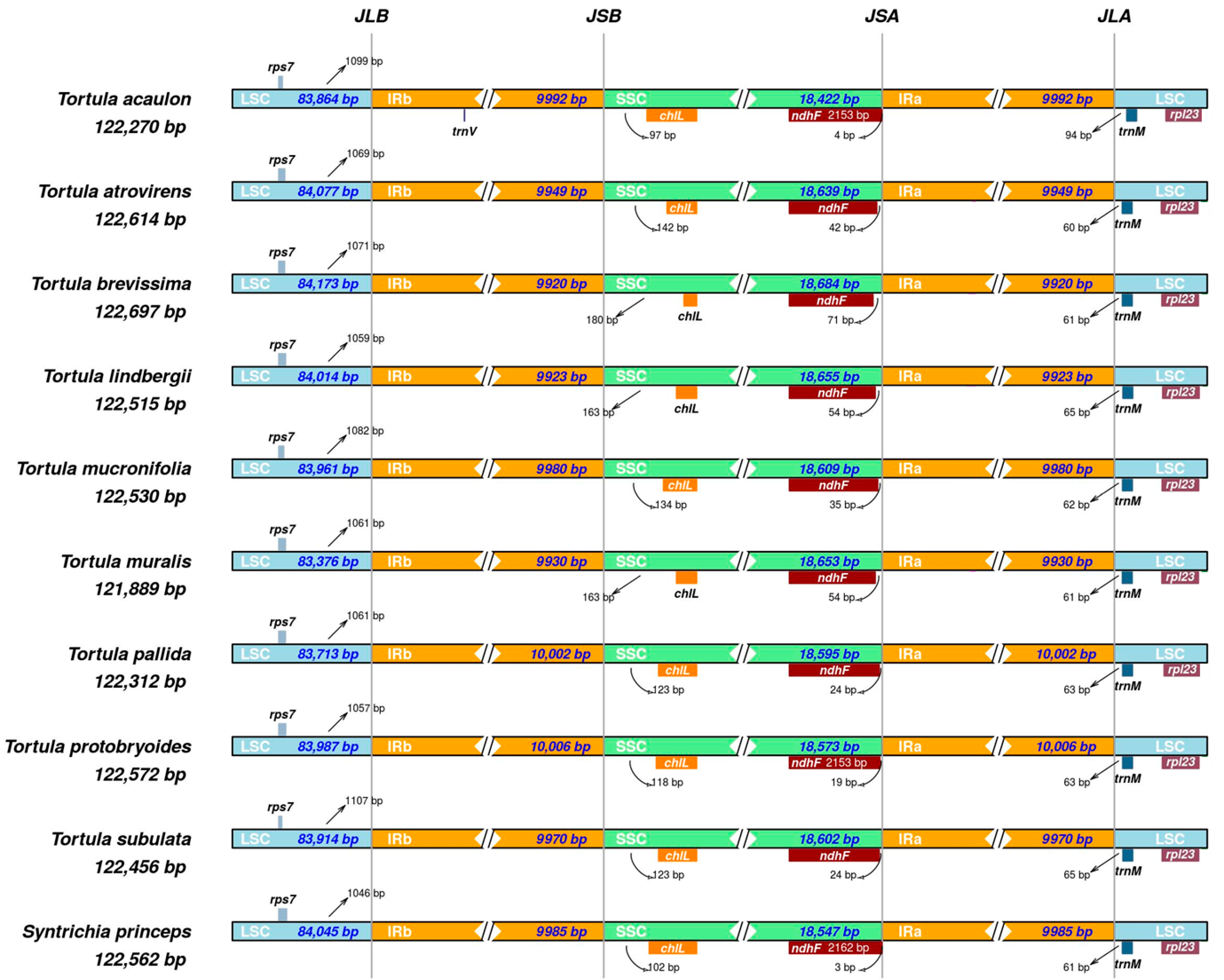
2.3. IR Boundaries Contraction and Expansion
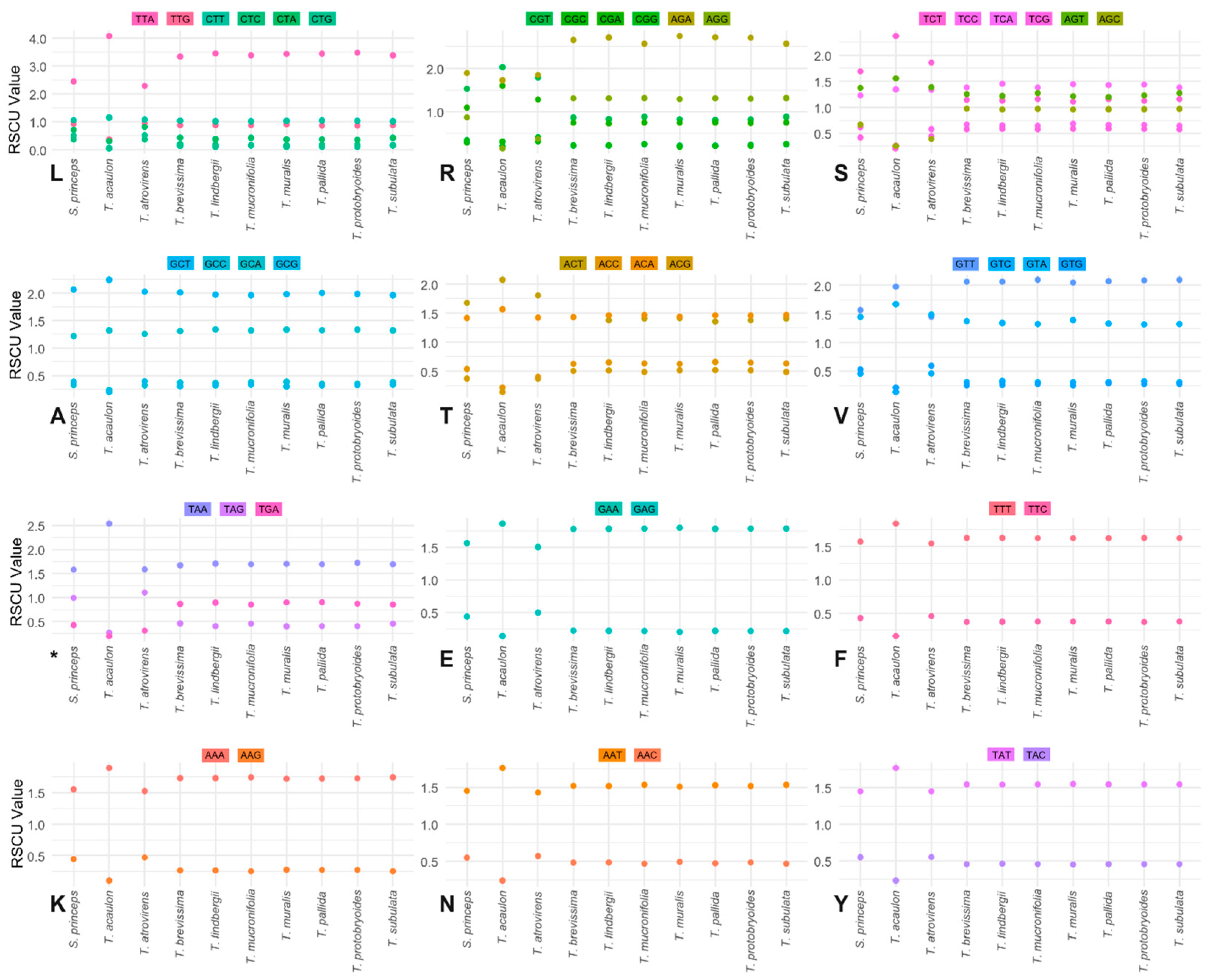
2.4. Relative Synonymous Codon Usage (RSCU) and Codon Bias
2.5. Identification of Hypervariable Regions and Barcode Candidates
2.5.1. Haplotype and Nucleotide Diversity in the Chloroplast Genome
2.5.2. Phylogenetic Analyses
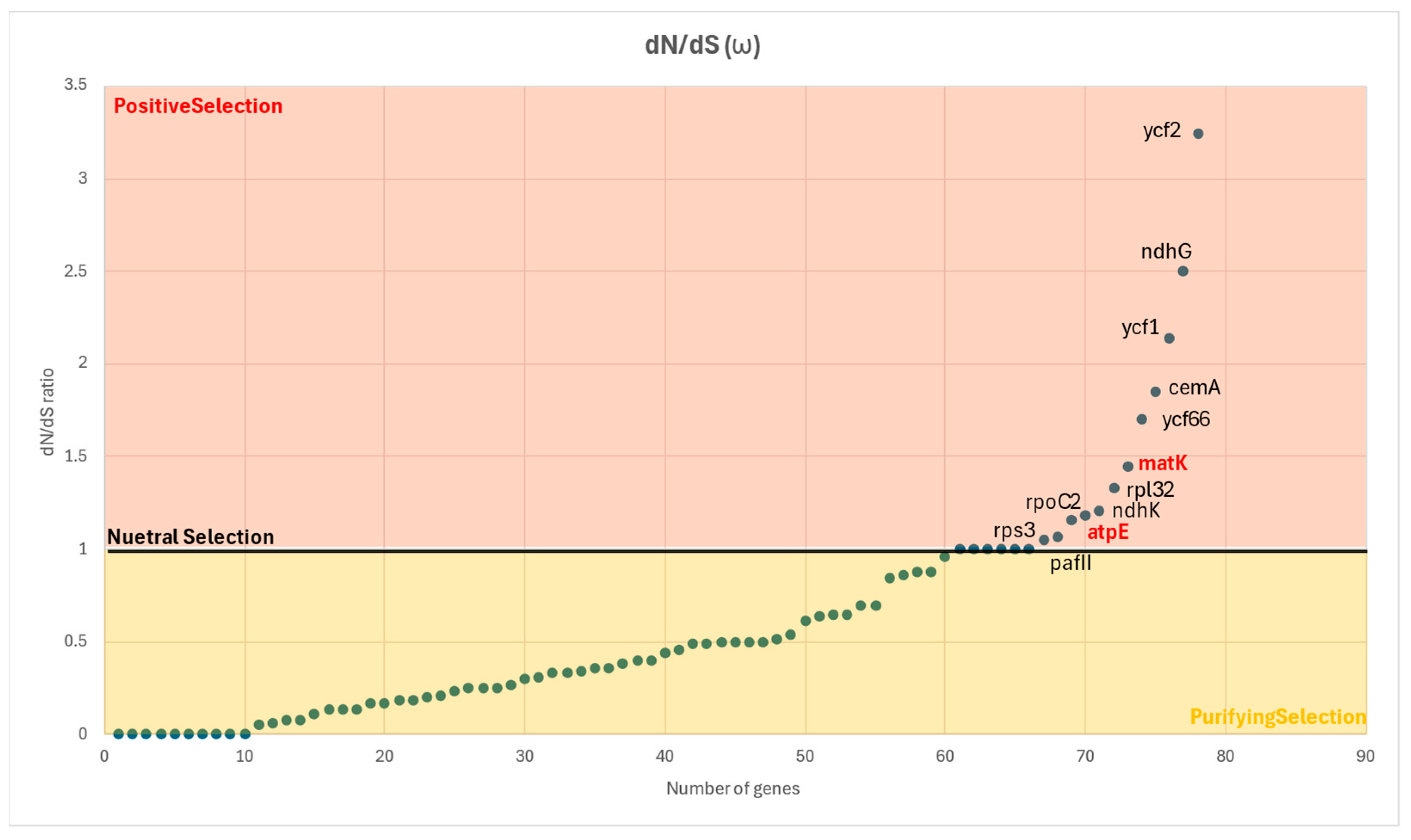
2.5.3. Nonsynonymous-to-Synonymous Substitutions Ratio (dN/dS Ratio) or ω
3. Discussion
4. Materials and Methods
4.1. Sampling and Axenic In Vitro Cultivation
4.2. DNA Extraction and Sequencing
4.3. Plastome Assembly and Annotation
4.4. Repetitive Sequences and Tandem Repeats Analysis
4.5. IR Boundaries Analysis
4.6. Codon Usage Preference
4.7. Identifying and Evaluating Hypervariable Regions
4.7.1. Haplotype and Nucleotide Diversity in the Chloroplast Genome
4.7.2. Phylogenetic Analyses
4.7.3. Nonsynonymous-to-Synonymous Substitutions Ratio (dN/dS Ratio) or ω
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Slate, M.L.; Sullivan, B.W.; Callaway, R.M. Desiccation and Rehydration of Mosses Greatly Increases Resource Fluxes That Alter Soil Carbon and Nitrogen Cycling. J. Ecol. 2019, 107, 1767–1778. [Google Scholar] [CrossRef]
- Schofield, W.B. Introduction to Bryology; Macmillan Publishing: New York, NY, USA, 1985; ISBN 978-0-02-949660-2. [Google Scholar]
- Goffinet, B.; Shaw, A.J. Bryophyte Biology, 2nd ed.; Cambridge University Press: Cambridge, UK, 2008; ISBN 978-0-521-69322-6. [Google Scholar]
- Crosby, M.R.; Magill, R.E.; Allen, B.; He, S. A Checklist of Mosses. Available online: https://www.mobot.org/MOBOT/tropicos/most/checklist.shtml (accessed on 27 June 2025).
- Zander, R.H. Genera of the Pottiaceae: Mosses of Harsh Environments. Bull. Buffalo Soc. Nat. Sci. 1993, 32, 1–378. [Google Scholar]
- Saito, K. A Monograph of Japanese Pottiaceae (Musci). J. Hattori Bot. Lab. 1975, 39, 373–537. [Google Scholar]
- Hedwig, J. Species Muscorum frondosorum: Descriptae et Tabulis Aeneis LXXVII Coloratis Illustratae; Schwaegrichen, F., Ed.; Joannis Ambrosii Barthii: Leipzig, Germany, 1801; Volume 1. [Google Scholar]
- Zander, R.H. Seven New Genera in Pottiaceae (Musci) and a Lectotype for Syntrichia. Phytologia 1989, 65, 424–436. [Google Scholar]
- Spagnuolo, V.; Caputo, P.; Cozzolino, S.; Castaldo, R.; De Luca, P. Patterns of Relationships in Trichostomoideae (Pottiaceae, Musci). Plant Syst. Evol. 1999, 216, 69–79. [Google Scholar] [CrossRef]
- Košnar, J.; Herbstová, M.; Kolář, F.; Koutecký, P.; Kučera, J. A Case Study of Intragenomic ITS Variation in Bryophytes: Assessment of Gene Flow and Role of Polyploidy in the Origin of European Taxa of the Tortula muralis (Musci: Pottiaceae) Complex. Taxon 2012, 61, 709–720. [Google Scholar] [CrossRef]
- Cox, C.J.; Li, B.; Foster, P.G.; Embley, T.M.; Civáň, P. Conflicting Phylogenies for Early Land Plants Are Caused by Composition Biases among Synonymous Substitutions. Syst. Biol. 2014, 63, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Werner, O.; Ros, R.M.; Cano, M.J.; Guerra, J. Tortula and Some Related Genera (Pottiaceae, Musci): Phylogenetic Relationships Based on Chloroplast rps4 Sequences. Plant Syst. Evol. 2002, 235, 197–207. [Google Scholar] [CrossRef]
- Werner, O.; Ros, R.M.; Cano, M.J.; Guerra, J. Molecular Phylogeny of Pottiaceae (Musci) Based on Chloroplast rps4 Sequence Data. Plant Syst. Evol. 2004, 243, 147–164. [Google Scholar] [CrossRef]
- Smith, A.J.E. The Moss Flora of Britain and Ireland, 2nd ed.; Cambridge University Press: Cambridge, UK, 2004; ISBN 978-0-521-54672-0. [Google Scholar]
- Flora of North America Editorial Committee. Flora of North America: North of Mexico; Oxford University Press: New York, NY, USA, 2007; Volume 27 Bryophyta, Part 1; ISBN 978-0-19-531823-4. [Google Scholar]
- Ochyra, R.; Żarnowiec, J.; Bednarek-Ochyra, H. Census Catalogue of Polish Mosses. In Biodiversity of Poland = Różnorodność Biologiczna Polski; Ochyra, R., Ed.; Polish Academy of Sciences, Institute of Botany: Kraków, Poland, 2003; Volume 3, pp. 1–372. ISBN 9788385444848. [Google Scholar]
- Frey, W.; Frahm, J.P.; Fischer, E.; Lobin, W. The Liverworts, Mosses and Ferns of Europe; Harley Books: Essex, UK, 2006; ISBN 0946589704. [Google Scholar]
- Ros, R.M.; Mazimpaka, V.; Abou-Salama, U.; Aleffi, M.; Blockeel, T.L.; Brugués, M.; Cros, R.M.; Dia, M.G.; Dirkse, G.M.; Draper, I.; et al. Mosses of the Mediterranean, an Annotated Checklist. Cryptogam. Bryol. 2013, 34, 99–283. [Google Scholar] [CrossRef]
- Garilleti, R.; Albertos, B. Atlas y Libro Rojo de los Briófitos Amenazados de España; Organismo Autónomo Parques Nacionales: Madrid, Spain, 2012; ISBN 978-84-8014-836-8. [Google Scholar]
- Aleffi, M.; Cogoni, A.; Poponessi, S. An Updated Checklist of the Bryophytes of Italy, Including the Republic of San Marino and Vatican City State. Plant Biosyst.-Int. J. Deal. Asp. Plant Biol. 2023, 157, 1259–1307. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast Genomes: Diversity, Evolution, and Applications in Genetic Engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, L.-N.; Roston, R.L.; Soll, J.; Gao, H. Editorial: Structure and Function of Chloroplasts—Volume II. Front. Plant Sci. 2020, 11, 620152. [Google Scholar] [CrossRef] [PubMed]
- Magdy, M.; El-Sherbeny, E.A.; Ramirez Sanchez, A. The Complete Chloroplast Genome of the Egyptian Henbane (Hyoscyamus muticus L., Solanaceae). Mitochondrial DNA Part B 2022, 7, 1109–1111. [Google Scholar] [CrossRef]
- Shen, Z.; Liu, Q.; Hao, J.; Bi, S.; Fu, Y.; Zhang, L. The Complete Chloroplast Genome Sequence of the Medicinal Moss Rhodobryum giganteum (Bryaceae, Bryophyta): Comparative Genomics and Phylogenetic Analyses. Genes 2024, 15, 900. [Google Scholar] [CrossRef] [PubMed]
- Safhi, F.A.; Jalal, A.S.; Alshegaihi, R.M.; Alshamrani, R.; Alamri, A.M.; Felemban, W.; Abuzaid, A.O.; Hussein, M.A.A.; Al Aboud, N.M.; Magdy, M.; et al. The Complete Chloroplast Genome of Psydrax latifolia: Evolutionary Dynamics, Comparative Genomics and Phylogeny. Front. Ecol. Evol. 2024, 12, 1416876. [Google Scholar] [CrossRef]
- Lubna; Asaf, S.; Jan, R.; Asif, S.; Bilal, S.; Khan, A.L.; Kim, K.-M.; Lee, I.-J.; AL-Harrasi, A. Plastome Diversity and Evolution in Mosses: Insights from Structural Characterization, Comparative Genomics, and Phylogenetic Analysis. Int. J. Biol. Macromol. 2024, 257, 128608. [Google Scholar] [CrossRef]
- Magdy, M.; Ou, L.; Yu, H.; Chen, R.; Zhou, Y.; Hassan, H.; Feng, B.; Taitano, N.; Van Der Knaap, E.; Zou, X.; et al. Pan-Plastome Approach Empowers the Assessment of Genetic Variation in Cultivated Capsicum Species. Hortic. Res. 2019, 6, 108. [Google Scholar] [CrossRef]
- Pandey, S.; Alam, A. Molecular Markers (RAPD and SSR) Based Characterisation of Genetic Diversity and Population Structure of Moss Hyophila involuta. Acta Bot. Hung. 2021, 63, 171–193. [Google Scholar] [CrossRef]
- Geng, Y.; Li, Y.; Yuan, X.; Luo, T.; Wang, Y. The Complete Chloroplast Genome Sequence of Fosbergia shweliensis, an Endemic Species to Yunnan of China. Mitochondrial DNA Part B 2020, 5, 1796–1797. [Google Scholar] [CrossRef]
- Yurina, N.P.; Sharapova, L.S.; Odintsova, M.S. Structure of Plastid Genomes of Photosynthetic Eukaryotes. Biochemistry 2017, 82, 678–691. [Google Scholar] [CrossRef]
- Bopp, M. Developmental Physiology of Bryophytes. In New Manual of Bryology; Schuster, R.M., Ed.; The Hattori Botanical Laboratory: Nichinan, Japan, 1983; Volume 1, pp. 276–324. ISBN 4-938163-3045. [Google Scholar]
- Mower, J.P.; Vickrey, T.L. Structural Diversity Among Plastid Genomes of Land Plants. In Advances in Botanical Research; Chaw, A.H., Jansen, R.K., Eds.; Elsevier: London, UK, 2018; Volume 85, pp. 263–292. ISBN 978-0-12-813457-3. [Google Scholar]
- Cevallos, M.A.; Guerrero, G.; Ríos, S.; Arroyo, A.; Villalobos, M.A.; Porta, H. The Chloroplast Genome of the Desiccation-Tolerant Moss Pseudocrossidium replicatum (Taylor) R.H. Zander. Genet. Mol. Biol. 2019, 42, 488–493. [Google Scholar] [CrossRef]
- Oliver, M.J.; Murdock, A.G.; Mishler, B.D.; Kuehl, J.V.; Boore, J.L.; Mandoli, D.F.; Everett, K.D.; Wolf, P.G.; Duffy, A.M.; Karol, K.G. Chloroplast Genome Sequence of the Moss Tortula ruralis: Gene Content, Polymorphism, and Structural Arrangement Relative to Other Green Plant Chloroplast Genomes. BMC Genom. 2010, 11, 143. [Google Scholar] [CrossRef]
- Inoue, Y.; Nakahara-Tsubota, M.; Tsubota, H. The Complete Chloroplast and Mitochondrial Genomes of Scopelophila cataractae (Mitt.) Broth. (Pottiaceae, Bryophyta). Mitochondrial DNA Part B 2022, 7, 125–127. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI). Didymodon constrictus Chloroplast Genome. Organelle Genome Resources. Available online: https://www.ncbi.nlm.nih.gov/datasets/organelle/?taxon=200710 (accessed on 1 April 2024).
- Kim, S.C.; Byun, M.Y.; Kim, J.H.; Lee, H. The Complete Chloroplast Genome of an Antarctic Moss Syntrichia filaris (Müll. Hal.) R.H. Zander. Mitochondrial DNA Part B 2019, 4, 2303–2304. [Google Scholar] [CrossRef]
- Chang, Y.; Graham, S.W. Inferring the Higher-order Phylogeny of Mosses (Bryophyta) and Relatives Using a Large, Multigene Plastid Data Set. Am. J. Bot. 2011, 98, 839–849. [Google Scholar] [CrossRef]
- Goffinet, B.; Wickett, N.J.; Werner, O.; Ros, R.M.; Shaw, A.J.; Cox, C.J. Distribution and Phylogenetic Significance of the 71-Kb Inversion in the Plastid Genome in Funariidae (Bryophyta). Ann. Bot. 2006, 99, 747–753. [Google Scholar] [CrossRef][Green Version]
- Qiu, Y.-L.; Li, L.; Hendry, T.A.; Li, R.; Taylor, D.W.; Issa, M.J.; Ronen, A.J.; Vekaria, M.L.; White, A.M. Reconstructing the Basal Angiosperm Phylogeny: Evaluating Information Content of Mitochondrial Genes. Taxon 2006, 55, 837–856. [Google Scholar] [CrossRef]
- Pandey, S.; Sharma, V.; Alam, A. Potential of Microsatellites Markers for the Genetic Analysis of Bryophytes. Not. Sci. Biol. 2016, 8, 37–46. [Google Scholar] [CrossRef]
- Magdy, M.; Werner, O.; Patiño, J.; Ros, R.M. Landscape Heterogeneity Drives Genetic Diversity in the Highly Dispersive Moss Funaria hygrometrica Hedw. Plants 2024, 13, 2785. [Google Scholar] [CrossRef]
- Dhyani, A.; Kasana, S.; Uniyal, P.L. From Barcodes to Genomes: A New Era of Molecular Exploration in Bryophyte Research. Front. Plant Sci. 2025, 15, 1500607. [Google Scholar] [CrossRef]
- Stech, M.; McDaniel, S.F.; Hernández-Maqueda, R.; Ros, R.M.; Werner, O.; Muñoz, J.; Quandt, D. Phylogeny of Haplolepideous Mosses—Challenges and Perspectives. J. Bryol. 2012, 34, 173–186. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, L.; Yang, M.; Qi, Q.; Yang, Q.; López-Pujol, J.; Wang, L.; Zhao, D. Complete Organelle Genome of the Desiccation-Tolerant (DT) Moss Tortula atrovirens and Comparative Analysis of the Pottiaceae Family. Genes 2024, 15, 782. [Google Scholar] [CrossRef]
- Ruhlman, T.A.; Zhang, J.; Blazier, J.C.; Sabir, J.S.M.; Jansen, R.K. Recombination-dependent Replication and Gene Conversion Homogenize Repeat Sequences and Diversify Plastid Genome Structure. Am. J. Bot. 2017, 104, 559–572. [Google Scholar] [CrossRef]
- Smith, D.R. Unparalleled GC Content in the Plastid DNA of Selaginella. Plant Mol. Biol. 2009, 71, 627–639. [Google Scholar] [CrossRef]
- Ślipiko, M.; Myszczyński, K.; Buczkowska, K.; Bączkiewicz, A.; Szczecińska, M.; Sawicki, J. Molecular Delimitation of European Leafy Liverworts of the Genus Calypogeia Based on Plastid Super-Barcodes. BMC Plant Biol. 2020, 20, 243. [Google Scholar] [CrossRef]
- Palidwor, G.A.; Perkins, T.J.; Xia, X. A General Model of Codon Bias Due to GC Mutational Bias. PLoS ONE 2010, 5, e13431. [Google Scholar] [CrossRef]
- Ohyama, K. Chloroplast and Mitochondrial Genomes from a Liverwort, Marchantia polymorpha—Gene Organization and Molecular Evolution. Biosci. Biotechnol. Biochem. 1996, 60, 16–24. [Google Scholar] [CrossRef]
- Sugiura, C.; Kobayashi, Y.; Aoki, S.; Sugita, C.; Sugita, M. Complete Chloroplast DNA Sequence of the Moss Physcomitrella Patens: Evidence for the Loss and Relocation of rpoA from the Chloroplast to the Nucleus. Nucleic Acids Res. 2003, 31, 5324–5331. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, H.; Cao, T.; Ge, X. Evaluation of 10 Plant Barcodes in Bryophyta (Mosses). J. Syst. Evol. 2010, 48, 36–46. [Google Scholar] [CrossRef]
- Werner, O.; Ros, R.M.; Grundmann, M. Molecular Phylogeny of Trichostomoideae (Pottiaceae, Bryophyta) Based on nrITS Sequence Data. Taxon 2005, 54, 361–368. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W. Rapid Isolation of High Molecular Weight Plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Werner, O.; Ros, R.M.; Guerra, J. Direct Amplification and NaOH Extraction: Two Rapid and Simple Methods for Preparing Bryophyte DNA for Polymerase Chain Reaction (PCR). J. Bryol. 2002, 24, 127–131. [Google Scholar] [CrossRef]
- FastQC V0.12. A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 April 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. Spades: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An Online Program to Visualize the Junction Sites of Chloroplast Genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Gómez-Rubio, V. Ggplot2—Elegant Graphics for Data Analysis (2nd Edition). J. Stat. Softw. 2017, 77, 1–3. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Akhunov, E.D.; Akhunova, A.R.; Anderson, O.D.; Anderson, J.A.; Blake, N.; Clegg, M.T.; Coleman-Derr, D.; Conley, E.J.; Crossman, C.C.; Deal, K.R.; et al. Nucleotide Diversity Maps Reveal Variation in Diversity among Wheat Genomes and Chromosomes. BMC Genom. 2010, 11, 702. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
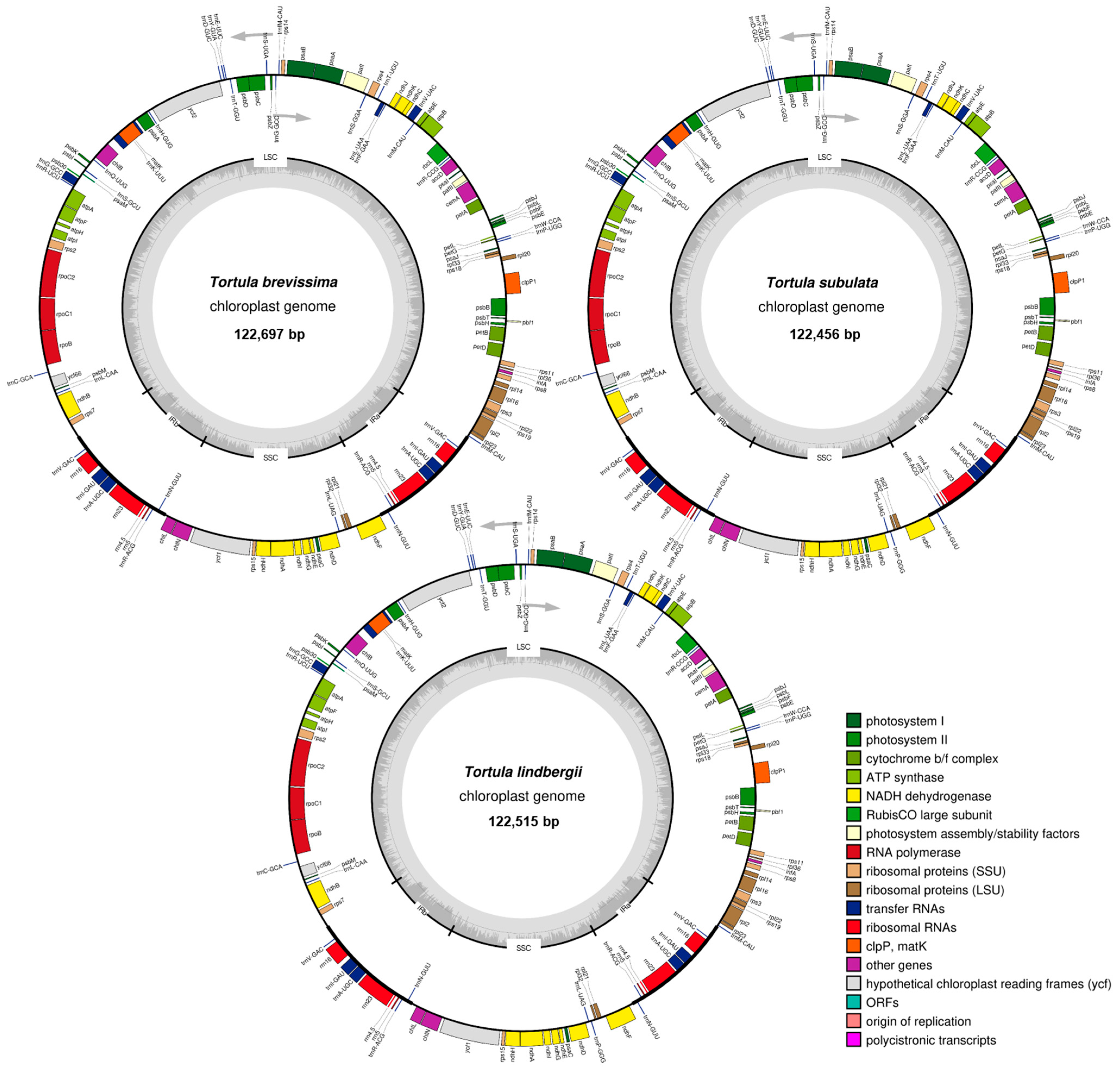

| Species | LSC (bp) | SSC (bp) | IR (bp) | Total (bp) | GC% |
|---|---|---|---|---|---|
| Syntrichia princeps | 84,045 | 18,547 | 9985 | 122,562 | 28.30% |
| Tortula acaulon | 83,864 | 18,422 | 9992 | 122,270 | 28.40% |
| Tortula atrovirens | 84,077 | 18,639 | 9949 | 122,614 | 28.50% |
| Tortula brevissima | 84,173 | 18,684 | 9920 | 122,697 | 28.30% |
| Tortula lindbergii | 84,014 | 18,655 | 9923 | 122,515 | 28.40% |
| Tortula mucronifolia | 83,961 | 18,609 | 9980 | 122,530 | 28.20% |
| Tortula muralis | 83,376 | 18,653 | 9930 | 121,889 | 28.40% |
| Tortula pallida | 83,713 | 18,595 | 10,002 | 122,312 | 28.50% |
| Tortula protobryoides | 83,987 | 18,573 | 10,006 | 122,572 | 28.40% |
| Tortula subulata | 83,914 | 18,602 | 9970 | 122,456 | 28.20% |
| Type | Length (bp) | S | Hap | Hd | π | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| CDS | 837.92 | 820.39 | 51.72 | 63.57 | 8.12 | 2.50 | 0.90 | 0.18 | 0.02 | 0.01 |
| IGS | 263.41 | 313.47 | 27.64 | 33.58 | 7.86 | 2.78 | 0.87 | 0.25 | 0.04 | 0.02 |
| rRNA | 1130.75 | 1197.19 | 8.00 | 11.29 | 4.50 | 3.42 | 0.54 | 0.42 | 0.00 | 0.00 |
| tRNA | 164.38 | 229.11 | 5.79 | 11.59 | 3.18 | 2.99 | 0.35 | 0.36 | 0.01 | 0.02 |
| Total | 459.41 | 633.80 | 31.50 | 45.90 | 7.17 | 3.24 | 0.79 | 0.32 | 0.03 | 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassannezhad, H.; Magdy, M.; Werner, O.; Ros, R.M. Exploring Plastome Diversity and Molecular Evolution Within Genus Tortula (Family Pottiaceae, Bryophyta). Plants 2025, 14, 2808. https://doi.org/10.3390/plants14172808
Hassannezhad H, Magdy M, Werner O, Ros RM. Exploring Plastome Diversity and Molecular Evolution Within Genus Tortula (Family Pottiaceae, Bryophyta). Plants. 2025; 14(17):2808. https://doi.org/10.3390/plants14172808
Chicago/Turabian StyleHassannezhad, Hamideh, Mahmoud Magdy, Olaf Werner, and Rosa M. Ros. 2025. "Exploring Plastome Diversity and Molecular Evolution Within Genus Tortula (Family Pottiaceae, Bryophyta)" Plants 14, no. 17: 2808. https://doi.org/10.3390/plants14172808
APA StyleHassannezhad, H., Magdy, M., Werner, O., & Ros, R. M. (2025). Exploring Plastome Diversity and Molecular Evolution Within Genus Tortula (Family Pottiaceae, Bryophyta). Plants, 14(17), 2808. https://doi.org/10.3390/plants14172808









